Abstract
The largest documented outbreak of Chikungunya virus (CHIKV) disease occurred in the Indian Ocean islands and India during 2004–2007. The magnitude of this outbreak led to speculation that a new variant of the virus had emerged that was either more virulent or more easily transmitted by mosquito vectors. To study this assertion, it is important to know the origin of the virus and how the particular strain circulating during the outbreak is related to other known strains. This study genetically characterized isolates of CHIKV obtained from Mombasa and Lamu Island, Kenya, during 2004, as well as strains from the 2005 outbreak recorded in Comoros. The results of these analyses demonstrated that the virus responsible for the epidemic that spread through the Indian Ocean originated in coastal Kenya during 2004 and that the closest known ancestors are members of the Central/East African clade. Genetic elements that may be responsible for the scope of the outbreak were also identified.
INTRODUCTION
Chikungunya virus (CHIKV) was responsible for explosive outbreaks during 2004–2007 in the islands of the Indian Ocean and India (Borgherini et al., 2007; Chretien et al., 2007; Mishra & Ratho, 2006). Estimates of the number of affected individuals in the islands of the Indian Ocean have reached several hundred thousand and serological survey data suggest that over 60 % of the regional populations have been exposed to the virus (Sergon et al., 2007, 2008). The first reports of the epidemic in the Indian Ocean occurred during the spring of 2005 when over 5000 residents of the Union of the Comoros acquired CHIKV disease (Sergon et al., 2007). Later, the virus was detected on the island of Réunion, where over 200 000 people were infected (Josseran et al., 2006). From Réunion, the virus spread to the Seychelles, Mauritius, Madagascar and finally India, with equally phenomenal numbers of infections (Ravi, 2006; Saxena et al., 2006). For example in India, it is estimated that over 1.3 million people across 13 states were infected (Arankalle et al., 2007). In addition, exposed travellers returning from the affected areas to Europe, the USA, Canada, Hong Kong and numerous other countries carried the virus into these new niches where the imported cases were subsequently identified (CDC, 2006; Druce et al., 2007; Hochedez et al., 2007; Lanciotti et al., 2007; Lee et al., 2006; Simon et al., 2007; Taubitz et al., 2007; Watson, 2007).
An important question is the origin of the virus that initiated the outbreaks in the Indian Ocean. Given the enhanced morbidity following infection with the virus, one suggestion has been that mutations in the virus led to increased virulence and/or transmissibility during the course of the epidemic (Mishra & Ratho, 2006; Powers & Logue, 2007). For example, although only 1400 CHIKV cases were identified in the Kenyan islands in 2004, a serosurvey estimated a 75 % attack rate on Lamu Island, suggesting that over 13 000 cases occurred on the small island of 18 000 residents (Sergon et al., 2008). A similar serosurvey performed in Comoros revealed a 60 % infection rate (Sergon et al., 2007). As mutations in the virus may potentially have led to altered virulence, we sequenced the complete genome of isolates obtained from the Lamu, Mombasa and Comoros outbreaks and compared these genetic sequences with a later strain from the outbreak to determine whether changes had indeed arisen during the course of the epidemic spread.
METHODS
Source of CHIKV isolates.
The five CHIKV isolates sequenced in this analysis were obtained from the serum of infected individuals or recovered from a pool of mosquitoes collected during the peak of the outbreaks. Mosquitoes were collected from bedrooms and living rooms in the homes of clinically ill patients. The isolates are described in Table 1.
Table 1.
Isolates used in this study
| Strain | Origin of isolation | Month and year of isolation | Source of material |
|---|---|---|---|
| KPA15 | Mombasa, Kenya | October 2004 | Human |
| LAMU33 | Lamu Island, Kenya | July 2004 | Human |
| COMJ | Grande Comore, Comoros | March 2005 | Human |
| COM25 | Moroni, Comoros | March 2005 | Human |
| COM125 | Moroni, Comoros | March 2005 | A. aegypti mosquitoes |
Method of virus isolation.
For virus isolation from human sera, Vero cells were grown to about 95 % confluency in minimum essential media (MEM) containing 10 % fetal bovine serum, 2 % l-glutamine and 1 % antibiotics in T-25 tissue culture flasks. Human serum (100 μl) was mixed with 300 μl MEM and inoculated onto the Vero cells. The inoculated cells were incubated at 37 °C with 5 % CO2 for up to 12 days and monitored daily for the development of a cytopathic effect (CPE). When there was CPE in over 90 % of the cells, the flasks were frozen at −80 °C and freeze–thawed several times to facilitate cell lysis and virus release, followed by centrifugation at 3000 r.p.m. in a microfuge for 10 min to remove cellular debris. The virus-containing supernatant was aliquotted into cryovials and stored at −80 °C. For virus isolation from mosquitoes, the mosquitos were pooled and ground in 1 ml Dulbecco's MEM. The homogenate was clarified by centrifugation and the suspension inoculated onto confluent Vero cells. Supernatant was recovered from flasks exhibiting CPE, centrifuged and aliquotted prior to storage at −70 °C.
RNA extraction, cDNA synthesis and amplification.
An aliquot of supernatant from infected Vero cells was mixed at a ratio of 1 : 4 with AVL buffer from a QIAamp Viral RNA Extraction kit (Qiagen) and RNA extraction performed according to the manufacturer's instructions. RT-PCR assays were performed using a Titan One Tube RT-PCR kit (Roche Molecular Biochemicals) according to the manufacturer's protocol. Each reaction contained 10 μl RNA and 20 pmol each CHIKV-specific forward and reverse primer in a 50 μl total volume. The primers used for genome amplification were as follows: CHIKV301(+): 5′-CAGGAAGTACCACTGCGTCTGCC-3′, CHIKV662(−): 5′-GTACGAGGGGTATGCACCTGC-3′, CHIKV2625(+): 5′-CTGCCTGTGACTGCCATTGTGTC-3′, CHIKV2820(−): 5′-GCTGCTGTCATGACCTCGTGTCCAC-3′, CHIKV4625(+): 5′-TTGGCAGGCAGAAAAGGATACAG-3′, CHIKV4782(−): 5′-CCAGGGCATATAGGCAGACTTG-3′, CHIKV7028(+): 5′-TGCGCGGCCTTCATCGGCGACTAC-3′, CHIKV7275(−): 5′-GCTAGCGGTTTGCCCAGTT-3′, CHIKV9222(+): 5′-GCCGCGGTCACCAATCACA-3′, CHIKV9647(−): 5′-GTCATAGTAGGGTACAGCTC-3′, T25v(−): 5′-T25GAAATATTAAAAACAAAATAACATCTCC-3′.
The cDNA amplicons were gel purified and sequencing reactions were performed on each.
Sequencing and genomic analysis.
The samples were resuspended in formamide and loaded onto a 96-well plate; the sequence was obtained using an ABI 3100 Prism Genetic Analyzer. Data were collected and analysed using ABI data collection software v2.0 and sequence analysis software v5.1.1. Further data analyses including nucleotide sequence editing and prediction of amino acid sequences were performed using Lasergene and dnastar software.
The cDNA sequences obtained in this study were aligned with each other and all available sequences in GenBank (including those from Réunion and India in 2006–2007) using the gap and pileup programs in the Genetics Computer Group package (Pustell & Kafatos, 1982) or clustal_x software (Thompson et al., 1997). Comparisons included our full-length genomes plus the complete genomes of strains Reu and S27 from GenBank, as well as alignments of the E1 coding region of our sequences plus those available in GenBank (over 50 strains selected). The E1 comparisons were performed to increase the number of strains under examination and to compare more completely the relationships among the variants. Because the sequences were highly conserved, phylogenetic analyses were performed to optimize identification of the most likely relationships for closely related taxa, and multiple analyses within the paup v4.0b software package were performed to analyse the data. Maximum parsimony was employed using a heuristic algorithm with unordered characters. A West African genotype sequence was used to root the phylograms under the parsimony criterion. Two neighbour-joining distance matrix algorithms (Kimura two-parameter and F84 corrections) were completed and a maximum-likelihood model using the HYK85 substitution variant without enforcing a molecular clock was also utilized. Bootstrap resampling (1000 replicates) provided estimates of confidence on the groups generated in each analysis.
RESULTS
Complete genome sequences of five strains of CHIKV from the earliest identified epidemic activity in coastal Kenya in 2004 through the peak of the outbreak on Comoros in 2005 were obtained and compared with each other and with an isolate from Réunion from March 2006 [strain REU (LR2006_OPY1)]. Comparisons of our five isolates revealed a total of 16 aa that were variable in any of the strains examined (Table 2) and which were distributed across all regions of the genome. The only true cluster of differences was in the nsP3 gene where three variable sites were present in an 83 aa stretch (Fig. 1). Not unexpectedly, pairwise comparisons indicated that the most closely related isolates were those from a single site (Comoros) and those closest together in time. However, the five 2004–2005 isolates we sequenced were extremely closely related, with pairwise differences ranging from only 5 to 17 nt. This level of conservation and the inability to readily distinguish the early isolates (strains KPA15 and LAMU33) from later isolates from Comoros and Réunion suggested that the outbreaks in Comoros, which subsequently moved to Réunion and India, probably originated from the 2004 outbreaks in coastal Kenya.
Table 2.
Amino acid differences associated with the CHIKV sequences analysed
| Gene | Polyprotein aa position | KPA15 (Mombasa, Kenya) | LAMU33 (Lamu Island, Kenya) | COM25 (Comoros – human) | COMJ (Comoros – human) | COM125 (Comoros – mosquito) | REU (Réunion) |
|---|---|---|---|---|---|---|---|
| nsP1 | 72 | Met | Met | Met | Arg | Met | Met |
| 82 | Cys | Cys | Cys | Ser | Cys | Cys | |
| 171 | Arg | Arg | Arg | Gln | Arg | Arg | |
| 326 | Val | Val | Val | Val | Val | Val or Met | |
| 376 | Thr | Met | Thr | Thr | Thr | Thr | |
| 458 | Thr | Ser | Ser | Ser | Ser | Ser | |
| nsP2 | 688 | Ala | Val | Val | Val | Val | Val |
| nsP3 | 1364 | Asp | Asp | Asp | Asp | Asp | Asp or Gly |
| 1609 | Leu | Leu | Leu | Leu | Leu and Phe | Leu | |
| 1658 | Arg | Arg | Arg | Arg | Arg | Arg or Ile | |
| 1691 | Ser | Ser | Ser | Ser | Pro | Ser | |
| nsP3/nsP4 junction | 1857 | Opal | Opal | Opal | Arg | Opal | Opal |
| nsP4 | 2330 | Tyr | Tyr | Asp | Asp | Tyr | Asp |
| E2 | 536 | Thr | Thr | Ile | Ile | Thr | Thr |
| 630 | Asn | Asn | Asn | Asn | His | Asn | |
| E1 | 1035 | Ala | Ala | Ala | Ala | Ala | Val |
Fig. 1.
Depiction of the locations of amino acid differences identified among Indian Ocean island sequences (bottom) and between early Indian Ocean isolates and older Central/East African genotype strains (top) examined in this study. Numbers indicate non-structural (ns) or structural polyprotein amino acid positions.
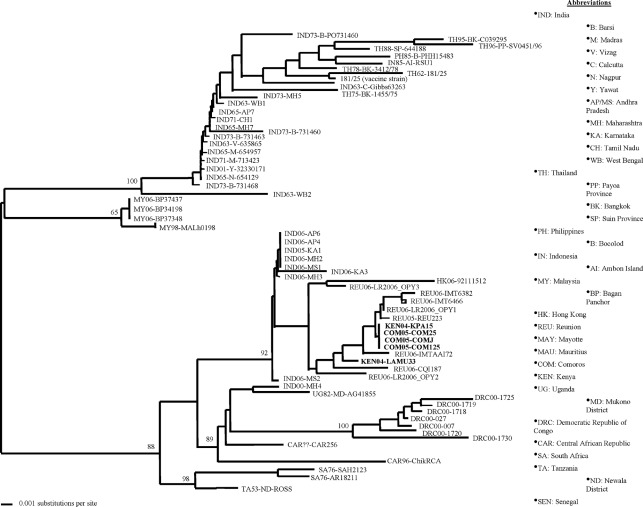
To ascertain further the origin of the CHIKV variant that caused these massive outbreaks, phylogenetic comparisons were performed using nucleotide sequences derived from the six isolates described above with other sequences from earlier CHIKV strains available in GenBank (Figs 2 and 3). In addition to full genome nucleotide phylogenetic analyses (Fig. 3), additional phylogenies were constructed using partial nucleotide sequences derived from the E2 and/or E1 3′ non-coding regions of over 50 other strains. All analyses clearly put all of the 2004–2006 isolates we evaluated in the Central/East African clade with strains from Tanzania, South Africa, Central African Republic and the Democratic Republic of the Congo (Fig. 2). The isolates from 2004 to 2006 were virtually identical and formed a monophyletic cluster within the Central/East African genotype with robust bootstrap support. Attempts to delineate further the temporal characteristics of the 2004–2006 Indian Ocean isolates revealed relatively low bootstrap support (<55), indicating the uncertainty of the genetic relationships among these strains based on short sequence regions. In fact, analyses such as maximum parsimony were unable to resolve the sequences of isolates from the 2004–2007 outbreaks and consistently generated a polytomy for these sequences. With only 16 aa differences present throughout the entire genome among all of these strains, a clear resolution of the evolutionary positions of these strains may only be available with additional genomes possessing a greater temporal (and therefore genetic) distance.
Fig. 2.
Distance matrix cladogram based on CHIKV E1 nucleotide sequences. The analysis included samples from the 2004 to 2007 outbreaks in Kenya, Comoros, Réunion and India, along with historical isolates from the Central/East African (lower clade) and Asian (upper clade) genotypes. Isolates are designated by country of origin, year of isolation, province or city of infection, and strain name or number (e.g. strain 1455/76 isolated from Bangkok, Thailand, in 1975 is designated TH75-BK-1455/75). Unknown data are designated by ‘??’. Isolates sequenced in this study are indicated in bold. Bootstrap values based on 1000 replicates are indicated at the nodes.
Fig. 3.
Phylogram of the full-length CHIKV genomic sequence demonstrating the extreme genetic conservation seen during the course of the outbreak. Complete nucleotide sequences of isolates from coastal Kenya, the Indian Ocean and India from 2004 to 2007 were compared using a neighbour-joining algorithm. The prototype strain (S27) was used to represent the historical Central/East African genotype. Bootstrap values are indicated at the nodes.
A comparison of the earliest sequences from the Indian Ocean areas (Mombasa and Lamu Island) with the older sequences available from the Central/East African genotype revealed a total of only 16 aa that differed (Table 3). The differences in the older Central/East African strains were more evenly distributed across the genome than the mutations present in only the Indian Ocean isolates (Fig. 1). When these changes associated with the emergence of the Mombasa/Lamu consensus sequence were compared with the later isolates obtained from the Indian Ocean (i.e. the sequence of strain REU from Réunion), there were no differences present; in other words, all changes identified from the early Central/East African genotype and found in the Mombasa strain were also present in the Réunion sequence. Furthermore, when the Mombasa and Lamu sequences were compared with the consensus sequence obtained from the Comoros samples, no amino acid changes were identified. There were, however, possible additional changes associated with the Réunion strain that were not present in either Mombasa, Lamu or the historical Central/East African sequences. Thus, additional mutations detected as the outbreak expanded could have served to exacerbate the epidemic. Some of the additional amino acid changes, such as those at aa 326, 1364 and 1658 of the non-structural polyprotein, appeared as ambiguities in the GenBank sequence. These could represent mixed nucleotide populations where the true nucleotide changes would result in amino acid substitutions, possibly resulting in phenotypic changes. Other changes found only in the REU strain of our analysis, such as the switch from alanine to valine at aa 1035 of the structural polyprotein, have been shown to confer a replicative advantage in Aedes albopictus mosquitoes (Tsetsarkin et al., 2007; Vazeille et al., 2007).
Table 3.
Differences between Mombasa/Lamu Island isolates and older Central/East African genotype strains
| Gene | Nt position | Mombasa/Lamu | Central/East African genotype consensus |
|---|---|---|---|
| nsP1 | 488 | Arg | Gln |
| nsP2 | 589 | Asn | Ser |
| 909 | Tyr | His | |
| 1328 | Val | Ala | |
| nsP3 | 1550 | His | Tyr |
| 1670 | Ile | Thr | |
| 1794 | Pro | Leu | |
| 1804 | Ser | Pro | |
| nsP4 | 1938 | Ala | Thr |
| 2117 | Ala | Thr | |
| 2330 | Tyr | Asp | |
| E2 | 637 | Met | Thr |
| 700 | Thr | Ser | |
| 711 | Ala | Val | |
| 6K | 756 | Ile | Val |
| E1 | 1093 | Glu | Asp |
DISCUSSION
During the course of the large epidemic of CHIKV illness in the Indian Ocean islands and India during 2005–2007, virologists and epidemiologists worked to develop a detailed picture of how a CHIKV epidemic spreads and amplifies. Unfortunately, no clear understanding of what initiates an epidemic of this scope has been recorded to date. To accomplish this objective, extensive field studies to monitor human illness, non-human seroprevalence rates and mosquito infection rates would be ideal. However, as field investigations of this nature are unlikely to be supported, particularly during inter-epidemic periods, molecular genetics approaches using existing viral isolates may provide some clues as to the origins of outbreak strains and help to assess more effectively viral movement and infection patterns. Prior to this epidemic, there were only six distinct full-length CHIKV genomes available. Since the start of the epidemic, additional studies have generated 18 complete genomic sequences; however, because several of these were obtained from the same time and geographical locations, all of the genomes available represent only nine distinct outbreak events. None of these was a clear precursor to the initiation of the 2004–2007 epidemic. Therefore, we examined five isolates of CHIKV from very early in the outbreak in an attempt to ascertain relationships among them; we also looked at the most closely related historical sequences to determine the origins of the outbreak strains.
Our genetic analyses demonstrated that the CHIKV isolates obtained during this epidemic (which began on the east coast of Africa) were monophyletic, forming a single clade within the Central/East African genotype (Powers et al., 2000). Not unexpectedly, the viruses maintained a high degree of identity and had over 99 % identity at the nucleic acid level. Amino acid divergence was virtually non-existent, with only four changes identified from our earliest isolate collected on Lamu Island to the last one examined from the Indian Ocean area (strain COM125). These findings are similar to those published recently comparing isolates from Réunion and India, and others that used partial sequences from a number of isolates in the Indian Ocean region (Arankalle et al., 2007; Yergolkar et al., 2006). Given the extreme genetic conservation exhibited among all of the strains examined, it is clear that the timing of any particular sample collected, combined with the origin of the source host and their movement patterns, could influence the genetic patterns seen with these viruses.
As a specific example of the need to evaluate individual changes between isolates, the alanine to valine switch at aa 1035 of E1 was considered. It is possible that a variant of CHIKV from Comoros or Lamu Island may have had a valine residue at aa 1035 of E1, as seen in the Réunion sequence, but limited sampling simply did not identify it, as it was not selected for in these ecological climates. Alternatively, this mutation may truly have arisen in Réunion where the primary vector species has been suggested to be A. albopictus as an adaptation to this alternate mosquito host (Delatte et al., 2008). Aedes aegypti was the primary vector in Lamu Island, Mombasa and Comoros, so it has been hypothesized that the E1 valine was only beneficial after moving to an ecological niche where different mosquito vectors with alternative cholesterol usage requirements were predominant. Laboratory testing with valine- versus alanine-containing strains in both mosquito types has indicated that this is a likely hypothesis (Tsetsarkin et al., 2007; Vazeille et al., 2007). Thus, this single change identified among only a handful of amino acid differences did indeed appear to result in enhanced transmission in an alternative mosquito vector present in Réunion.
Significantly, the molecular epidemiology patterns identified here clearly show that the changes associated with the explosive magnitude of the 2004–2007 outbreaks preceded the introduction of the variant to Lamu Island, Kenya, in 2004. Prior to the epidemics in Kenya in 2004, the most recent documented outbreaks of CHIKV occurred in 1996–1997 in Senegal, in 1998–1999 in Malaysia, in 1999–2000 in the Central African Republic (CAR) and the Democratic Republic of the Congo (DRC), and in 2000–2003 in Indonesia. Most of these outbreaks, including those in CAR and DRC, consisted of strains falling within the Central/East African genotype. However, none of these sequences was a clear ancestor of the 2004–2007 outbreak strains, and thus the strains initiating the outbreak on Lamu Island could have come from any one of a number of sites in Africa or Asia where strains in this clade have been circulating. There are currently no sequence data available from the recent Indonesian outbreak, so it is conceivable that the origin of the Indian Ocean outbreak could have been from this region. The CHIKV strains responsible for cases in France traced from travellers from Réunion were also variant, emerging from the Central/East African evolutionary lineage (Parola et al., 2006). Obtaining complete genetic data from each outbreak is imperative to follow the movement of particular strains of virus. However, perhaps even more important is obtaining inter-epidemic strains, as it is in these strains that critical mutations occur, leading to future outbreaks or novel ecological transmission patterns.
Acknowledgments
The authors wish to thank Clayton Onyango, Sam Konongoi, Victor Ofula, Victor Omballa and Solomon Gikundi for technical assistance. This work was supported in part by NIH grant U54 AI-65357.
References
- Arankalle, V. A., Shrivastava, S., Cherian, S., Gunjikar, R. S., Walimbe, A. M., Jadhav, S. M., Sudeep, A. B. & Mishra, A. C. (2007). Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J Gen Virol 88, 1967–1976. [DOI] [PubMed] [Google Scholar]
- Borgherini, G., Poubeau, P., Staikowsky, F., Lory, M., Le Moullec, N., Becquart, J. P., Wengling, C., Michault, A. & Paganin, F. (2007). Outbreak of chikungunya on Réunion Island: early clinical and laboratory features in 157 adult patients. Clin Infect Dis 44, 1401–1407. [DOI] [PubMed] [Google Scholar]
- CDC (2006). Chikungunya fever diagnosed among international travelers – United States, 2005–2006. MMWR Morb Mortal Wkly Rep 55, 1040–1042. [PubMed] [Google Scholar]
- Chretien, J. P., Anyamba, A., Bedno, S. A., Breiman, R. F., Sang, R., Sergon, K., Powers, A. M., Onyango, C. O., Small, J. & other authors (2007). Drought-associated chikungunya emergence along coastal East Africa. Am J Trop Med Hyg 76, 405–407. [PubMed] [Google Scholar]
- Delatte, H., Dehecq, J. S., Thiria, J., Domerg, C., Paupy, C. & Fontenille, D. (2008). Geographic distribution and developmental sites of Aedes albopictus (Diptera: Culicidae) during a Chikungunya epidemic event. Vector Borne Zoonotic Dis 8, 25–34. [DOI] [PubMed] [Google Scholar]
- Druce, J. D., Johnson, D. F., Tran, T., Richards, M. J. & Birch, C. J. (2007). Chikungunya virus infection in traveler to Australia. Emerg Infect Dis 13, 509–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedez, P., Hausfater, P., Jaureguiberry, S., Gay, F., Datry, A., Danis, M., Bricaire, F. & Bossi, P. (2007). Cases of chikungunya fever imported from the islands of the South West Indian Ocean to Paris, France. Euro Surveill 12, Article7 [Google Scholar]
- Josseran, L., Paquet, C., Zehgnoun, A., Caillere, N., Le Tertre, A., Solet, J. L. & Ledrans, M. (2006). Chikungunya disease outbreak, Réunion Island. Emerg Infect Dis 12, 1994–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti, R. S., Kosoy, O. L., Laven, J. J., Panella, A. J., Velez, J. O., Lambert, A. J. & Campbell, G. L. (2007). Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis 13, 764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, N., Wong, C. K., Lam, W. Y., Wong, A., Lim, W., Lam, C. W., Cockram, C. S., Sung, J. J., Chan, P. K. & Tang, J. W. (2006). Chikungunya fever, Hong Kong. Emerg Infect Dis 12, 1790–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, B. & Ratho, R. K. (2006). Chikungunya re-emergence: possible mechanisms. Lancet 368, 918. [DOI] [PubMed] [Google Scholar]
- Parola, P., de Lamballerie, X., Jourdan, J., Rovery, C., Vaillant, V., Minodier, P., Brouqui, P., Flahault, A., Raoult, D. & Charrel, R. N. (2006). Novel chikungunya virus variant in travelers returning from Indian Ocean islands. Emerg Infect Dis 12, 1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, A. M. & Logue, C. H. (2007). Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol 88, 2363–2377. [DOI] [PubMed] [Google Scholar]
- Powers, A. M., Brault, A. C., Tesh, R. R. & Weaver, S. C. (2000). Re-emergence of Chikungunya and O'nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol 81, 471–479. [DOI] [PubMed] [Google Scholar]
- Pustell, J. & Kafatos, F. C. (1982). A convenient and adaptable package of DNA sequence analysis programs for microcomputers. Nucleic Acids Res 10, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi, V. (2006). Re-emergence of chikungunya virus in India. Indian J Med Microbiol 24, 83–84. [DOI] [PubMed] [Google Scholar]
- Saxena, S. K., Singh, M., Mishra, N. & Lakshmi, V. (2006). Resurgence of chikungunya virus in India: an emerging threat. Euro Surveill 11, E060810–E060812. [DOI] [PubMed] [Google Scholar]
- Sergon, K., Yahaya, A. A., Brown, J., Bedja, S. A., Mlindasse, M., Agata, N., Allaranger, Y., Ball, M. D., Powers, A. M. & other authors (2007). Seroprevalence of Chikungunya virus infection on Grande Comore Island, Union of the Comoros, 2005. Am J Trop Med Hyg 76, 1189–1193. [PubMed] [Google Scholar]
- Sergon, K., Njuguna, C., Kalani, R., Ofula, V., Onyango, C., Konongoi, L. S., Bedno, S., Burke, H., Dumilla, A. M. & other authors (2008). Seroprevalence of Chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. Am J Trop Med Hyg 78, 333–337. [PubMed] [Google Scholar]
- Simon, F., Parola, P., Grandadam, M., Fourcade, S., Oliver, M., Brouqui, P., Hance, P., Kraemer, P., Ali Mohamed, A. & other authors (2007). Chikungunya infection: an emerging rheumatism among travelers returned from Indian Ocean islands. Report of 47 cases. Medicine (Baltimore) 86, 123–137. [DOI] [PubMed] [Google Scholar]
- Taubitz, W., Cramer, J. P., Kapaun, A., Pfeffer, M., Drosten, C., Dobler, G., Burchard, G. D. & Loscher, T. (2007). Chikungunya fever in travelers: clinical presentation and course. Clin Infect Dis 45, e1–e4. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997). The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsetsarkin, K. A., Vanlandingham, D. L., McGee, C. E. & Higgs, S. (2007). A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3, e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazeille, M., Moutailler, S., Coudrier, D., Rousseaux, C., Khun, H., Huerre, M., Thiria, J., Dehecq, J. S., Fontenille, D. & other authors (2007). Two chikungunya isolates from the outbreak of La Réunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS ONE 2, e1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, R. (2007). Europe witnesses first local transmission of chikungunya fever in Italy. BMJ 335, 532–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergolkar, P. N., Tandale, B. V., Arankalle, V. A., Sathe, P. S., Sudeep, A. B., Gandhe, S. S., Gokhle, M. D., Jacob, G. P., Hundekar, S. L. & Mishra, A. C. (2006). Chikungunya outbreaks caused by African genotype, India. Emerg Infect Dis 12, 1580–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]