Abstract
New insights into the early viral evolution and cellular immune response during acute hepatitis C virus (HCV) infection are being gained following a global outbreak in human immunodeficiency virus-1 (HIV)-positive men who have sex with men. Cross-sectional and longitudinal sequence analysis at both the population and individual level have facilitated tracking of the HCV epidemic across the world and enabled the development of tests of viral diversity in individual patients in order to predict spontaneous clearance of HCV and response to treatment. Immunological studies in HIV-positive cohorts have highlighted the role of the CD4+ T-cell response in the control of early HCV infection and will increase the opportunity for the identification of protective epitopes that could be used in future vaccine development.
Introduction
Studies of the natural history of acute hepatitis C virus (HCV) infection have been restricted in number due to the silent nature of early infection. Insights have been gained from studies in humans and the chimpanzee model, but these have been limited in size due to the lack of availability of subjects and restrictions on the use of primates in laboratory experiments. The global rapid rise of acute HCV in human immunodeficiency virus-1 (HIV-1)-infected men who have sex with men (MSM) is providing the opportunity to study the natural history of acute HCV infection in greater detail and on a larger scale than has been possible previously. Understanding the determinants of spontaneous clearance and response to treatment is important and may provide opportunities for vaccine development as well as new treatment strategies. While the impact of HIV on the evolution of HCV should not be underestimated, new insights from studies in the HIV-positive population are likely to apply also to HIV-negative patients with HCV. We review advances in the understanding of the evolution of HCV in HIV-infected patients at a population and individual level and discuss how further knowledge may be gained from the ongoing study of such patient cohorts.
Acute HCV in HIV-positive men; a global epidemic
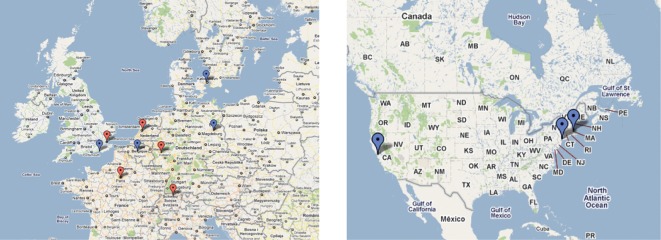
Reports of sexually transmitted acute HCV infection occurring in HIV-positive MSM in The Netherlands, France, Germany, Switzerland and the UK first appeared in the scientific literature between 2004 and 2005 (Ruys et al., 2004; Ghosn et al., 2004; Vogel et al., 2005; Rauch et al., 2005; Gilleece et al., 2005). Subsequently, outbreaks have been reported in other Northern European countries (Peters et al., 2006; Bottieau et al., 2010) as well as the East and West coasts of the USA (Luetkemeyer et al., 2006; Fierer et al., 2008), and Australia (Nguyen et al., 2007). In total, more than 1000 cases of acute HCV in HIV-positive MSM have been reported worldwide (Vogel et al., 2011). Such patients are commonly asymptomatic and are identified by routine screening at HIV follow-up clinics (Fig. 1).
Fig. 1.
Geographical distribution of acute HCV in HIV-positive MSM in Europe and the USA. A map of Northern Europe and USA showing cities where acute HCV cohorts have been reported. Centres where infection was reported between 2004 and 2005 are shown in red, while those reported in 2006 or later are highlighted in blue.
This remarkable rise in the incidence of acute HCV in HIV-positive MSM is continuing and is likely to herald a serious health burden over the coming years. Already, infection with HCV is emerging as a major cause of morbidity and mortality in individuals infected with HIV and is set to surpass AIDS-defining illnesses as a cause of death in patient populations who have access to highly active antiretroviral therapy (HAART; Bica et al., 2001; Arnold et al., 2006). In Amsterdam, before 2000, the prevalence of HCV in HIV-positive MSM was 1–4 %, while in 2007–2008, the prevalence was 15–21 % (Urbanus et al., 2009). In London, 10 % of MSM HIV seroconverters recruited to an HIV treatment study (SPARTAC) subsequently became infected with acute HCV over a 5 year period (Fox et al., 2008).
Phylogenetic analysis of HCV strains obtained from such patients has shown that a number of monophyletic lineages exist, distinguishing infections in HIV-positive MSM from other risk groups. These lineages cross international boundaries revealing networks of infection occurring in urban centres across Europe and Australia (van de Laar, 2009a). Surprisingly, while intravenous drug use is a risk factor in a minority of cases within each cohort, statistically, the strongest risk factors for transmission of HCV are unprotected anal sex and the use of permucosal (but not intravenous) recreational drugs (Danta et al., 2007; Fierer et al., 2008; Matthews et al., 2009). Preceding medical instrumentation or sexual practices that result in rectal bleeding are particularly strongly associated with transmission (Schmidt et al., 2011).
Molecular clock data points to the origins of the outbreak occurring in the mid-1990s (Danta et al., 2007; de Bruijne et al., 2009). This may reflect a change in sexual risk behaviour and recreational drug use around this time. Most cohorts include a number of individuals who are intravenous drug users (IDUs); although these individuals are in the minority, it is likely that transmission events between IDU and MSM populations have occurred, resulting in a new ecological niche of sexually transmitted HCV in HIV-positive MSM. A feature of the European outbreak has been a high prevalence of genotype 4d HCV (Serpaggi et al., 2006; de Bruijne et al., 2009). Genotype 4 HCV (HCV-4) is the most common circulating strain in North Africa and the Middle East with a decreasing geographical prevalence in countries further north. In Southern European countries, HCV-4 infection is less prevalent but occurs in migrants from North Africa and in IDUs, while in Northern Europe this genotype was previously rare but is increasingly associated with the HIV-positive MSM outbreak. In Amsterdam, two phylogenetically distinct populations of genotype 4d-infected individuals exist; Dutch IDUs and HIV-positive MSM. Molecular clock analysis has shown that the emergence of genotype 4d HCV in IDUs between 1960 and 1990 coincides with increasing recreational drug use in The Netherlands associated with permissive drug legislation (de Bruijne et al., 2009). Later expansion of a subcluster of HCV-4d strains in HIV-positive MSM from 1996 onwards coincides with the availability of HAART and may reflect an increase in unprotected sexual contact due to a reduction in fear of developing AIDS-related illnesses (Stolte et al., 2004). Thus, some of the earliest transmission events from IDUs into MSM may have occurred in Amsterdam. Similar events are likely to have taken place also in other urban centres such as London, Berlin, Paris, San Francisco, New York and Sydney. Strains obtained from patients in Sydney, for example, are largely distinct from those identified in the European outbreak (van de Laar et al., 2009a).
At present, the outbreak of acute HCV appears to be mainly confined to HIV-positive MSM. However, further transmission into other patient groups, for example HIV-negative MSM or heterosexual individuals could provide a potential public health problem in the future. Furthermore, if transmission continues at a high rate in this population, transmission of drug-resistant variants could reduce the future success rate of therapeutic interventions.
Acute HCV; evolution within the host
Viral load (VL) dynamics in humans and chimpanzees
A prominent feature of early infection with HCV in both humans and chimpanzees is a peak and then dip in VL (Fig. 2). During spontaneous clearance, this downward descent continues rapidly until viraemia is no longer detectable. In contrast, progression to chronicity is associated with recrudescent viraemia (Dahari et al., 2005; Thomson et al., 2011; Loomba et al., 2011). In human studies, a dip is not always seen. This may not always occur or could reflect the length of sampling windows used to measure sequential VLs in human studies.
Fig. 2.
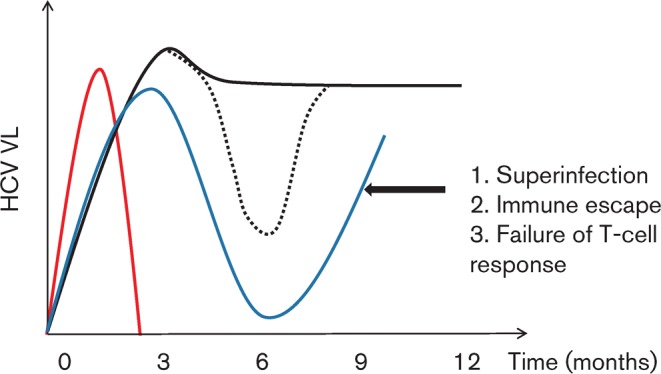
Patterns of viraemia during acute HCV infection. Three patterns of viraemia are seen in acute HCV; spontaneous clearance (red; 15 %) or progression towards chronicity associated with either a plateau viraemia (PV; black; 45 %) or fluctuating viraemia (FV; blue 40 %). Patients with PV may also have FV at an unsampled time point (dashed black line).
VL dynamics can be monitored in order to predict outcome in patients with acute HCV. Chimpanzees experimentally infected with the H77 HCV strain (genotype 1a) had a lower peak VL and a shorter time to peak viraemia in evolving spontaneous clearance than progressing infection. In humans, viral clearance usually occurs within 3 months of diagnosis in both HIV-infected and uninfected cohorts (Santantonio et al., 2006; Page et al., 2009). In a UK-based cohort of 112 HIV-positive patients, the maximum drop in HCV VL within 100 days of the first positive test was strongly predictive of spontaneous clearance [odds ratio (OR) per log10 drop = 1.78, P<0.0001; Thomson et al., 2011]. Patients who subsequently cleared infection had an earlier and lower peak HCV VL and a steeper gradient of descent than evolving progressor patients, even if the VL became transiently negative. The fluctuating viraemia pattern observed during early HCV infection could represent loss of immune control of circulating virus due to viral escape, superinfection with new strains and/or failure of the T-cell immune response. Studies employing longitudinal viral sequence analysis and T-cell function during early HCV infection in HIV-positive MSM are providing new information about the mechanisms that define continued decrease or increase in VL at early pivotal time points.
Viral diversification within the host
HCV replicates rapidly (the estimated daily production of virus is 1012 HCV viral copies in an infected host) and the viral RNA polymerase does not have proof-reading capacity so the possibility of variation within the HCV genome is extremely high (Neumann et al., 1998). The resulting viral quasispecies consists of a rapidly expanding group of related but genetically distinct viral variants. During early infection, viable viral variants are subject to selection pressures from the innate and acquired immune system as well as other growth restraints such as variation in the concentration of circulating fats (Bader et al., 2008; Felmlee et al., 2010).
Viral genetic sequence analysis can be used to estimate the nature of the prevailing evolutionary force at any point in time. Genetic distance (the percentage difference in nucleotides between quasispecies strains) and measures of diversity such as the Hamming distance (the mean number of amino acid differences between strains) may be used to quantify the composition of the quasispecies population. Selection pressure can be quantified by examining the ratio of non-synonymous substitutions per non-synonymous site (dN) to synonymous substitutions per synonymous site (dS); the dN/dS ratio. HCV may be subject to positive (dN/dS >1), purifying (dN/dS <1) or neutral (dN/dS = 1) selection pressures and these may change at different times during infection. The dN/dS ratio model relies on the assumption that silent synonymous mutations are not subject to selection pressure. While these do not result in amino acid changes, regions of the genome that display synonymous variation may have secondary non-coding functions. HCV core and NS5B RNA, for example, contain multiple stem–loops that are found in all HCV genotypes throughout both regions, as well as several strikingly conserved unpaired regions, one of which coincides with a region of the genome to which ribosomal access is required for translation initiation (Tuplin et al., 2004). Variation within the HCV genome is currently considered to be predominantly clonal. Viable recombinant strains has been identified (Kalinina et al., 2002) and the presence of recombinant and parent strains in individual patients have also been reported (Sentandreu et al., 2008). Thus in a minority of cases, recombination events may also contribute to viral diversity.
Genetic variation of HCV is most evident in the hypervariable region 1 (HVR-1), a 28 aa structure within the HCV E2 envelope protein (Weiner et al., 1991) Overall, the rate of nucleotide substitution in HCV is 1×10−3 nt per site per year and within the HVR-1 of the envelope gene, this is as high as 2.5–6.9×10−3 nt per site per year (Gray et al., 2011) The E2 gene is often targeted for the analysis of viral structure for this reason. Selection within this region is largely antibody-mediated (Farci et al., 2000; Gaud et al., 2003). It also contains CD4+ and CD8+ T-cell epitopes (Shirai et al., 1999; Lauer et al., 2002; Sarobe et al., 2006). Changes within this region may also reflect CD4+ or CD8+ cell-mediated variation in regions outwith envelope such as the non-structural genes through a genetic hitchhiking effect or antibody-mediated variation following failure of the cellular immune response (Klenerman et al., 2000).
The early evolution of HCV
During early infection with HCV, most newly formed viral variants contain potentially detrimental mutations and are subject to strong selection constraints. As a result, purifying selection (dN/dS <1) is the major force moulding viral population structure at this time in HIV-negative (Farci et al., 2000; Kuntzen et al., 2007) and HIV-positive individuals (Thomson et al., 2011). Swinging shifts in viral quasispecies structure are evident as the virus adapts to the environment within a newly infected host (Smith et al., 2010; Thomson et al., 2011). Progression to chronicity is associated with a switch from purifying to positive selection as well-adapted variants outgrow constrained ones. During spontaneous clearance, in contrast, this switch to positive selection does not occur.
The evolution of the viral quasispecies is emerging as key in defining the outcome of infection and response to treatment. Small studies of acute HCV infection have shown that the evolution of HVR-1 is associated with progressive disease, while a narrowed quasispecies repertoire occurs in patients who subsequently spontaneously clear infection due to the gradual elimination of viral variants (Farci et al., 2000; Sheridan et al., 2004; Thomson et al., 2011). In contrast, patients that progress to chronicity exhibit higher viral diversity secondary to viral variation driven by a partially effective immune response. Patients with immunodeficiency; such as a low CD4 count (<200 cells mm−3) or agammaglobulinaemia exhibit lower genetic diversity, however, due to a lack of positive immune selection pressure (Gaud et al., 2003; López-Labrador et al., 2007).
Is spontaneous clearance defined by the transmission bottleneck? Superinfection versus varying dominance during early infection
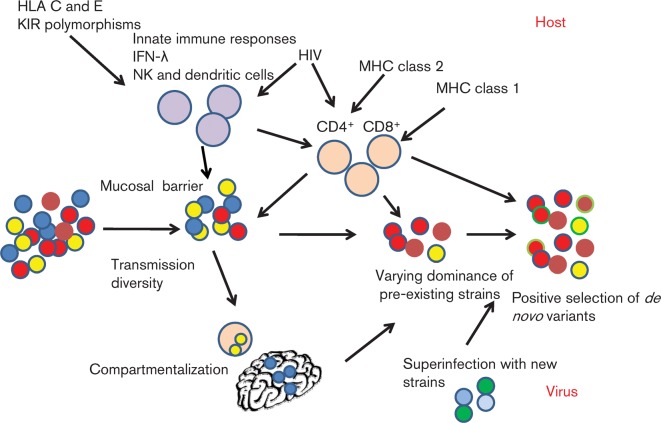
During transmission, infection may be initiated by a single or multiple infectious particles. Measures of viral diversity around this time could potentially be employed to predict the individual risk of progressing to chronicity. However, simple calculations of genetic diversity and positive or negative selection are confounded by the fact that viral diversification during early HCV infection is a non-linear process characterized by superinfection, varying dominance, compartmentalization as well as de novo immune escape mutations (Fig. 3).
Fig. 3.
Host and virus factors defining progression to chronicity or spontaneous clearance of acute HCV. The early evolution of HCV is shaped by an arms race between the host and virus. Viral diversification occurs as a result of varying dominance of pre-existing strains and immune selection of de novo variants formed during inaccurate viral replication and is aided by replication within compartments such as the brain and PBMCs.
HIV-infected patients with sexually acquired HCV are more likely to spontaneously clear infection than those infected via intravenous drug use (Shores et al., 2008). In this multicentre study of 769 HIV-infected individuals, patients with a history of sexual transmission spontaneously cleared the HCV infection more often than injection drug users (21.9 versus 11.6 %; P = 0.004). This supports the hypothesis that individuals who acquire HCV sexually are subject to a smaller, less diverse inoculum, possibly due to local effects at the mucosal level, although the effect of recreational drugs on the immune system could also partly explain this difference.
Clonal sequence analysis has provided clear evidence for the transmission of multiple HCV strains in both IDUs and sexually infected HIV-positive MSM (Pham et al., 2010; Smith et al., 2010; Thomson et al., 2011). Much of the early variation seen in infected individuals may be explained by varying dominance of strains acquired at the point of transmission. In the chimpanzee model, variation within the HVR-1 region is largely due to the selection of pre-existing variants rather than de novo mutations (Fernandez et al., 2004). In humans, this is also the case; in a study of 10 patients predominantly infected by IDU, major shifts in viral diversity occurred with the varying dominant strains (often of different genotypes) switching to and fro over short periods of time (Smith et al., 2010). Both sequential replacement by previously undetected variants and a dominance of variants seen earlier during infection, but not detected in intervening samples, were responsible for this phenomenon. Similar findings were evident in a longitudinal study of 50 HIV-1-infected MSM in which the same primer sets were used (Thomson et al., 2011) and in a cohort of 59 IDU seroconverters (13 of whom were HIV infected) (van de Laar et al. 2009b). In the former study, fluctuating changes in viraemia were significantly associated with evidence of superinfection or switching of dominant strains in 63 % of cases. Thus, a diverse infecting population structure is not uncommon and may at least in part define the final outcome of infection.
The narrower quasispecies repertoire evident in those who subsequently spontaneously clear the virus is in keeping with the hypothesis that transmission diversity defines the final outcome. However, using clonal sequence analysis, viral diversity at the earliest sampled time point in a new host does not reliably predict outcome. In a study of 12 HIV-negative individuals with acute HCV, higher diversity was seen only after seroconversion in evolving progressors (Farci et al., 2000) and in a cohort of 50 HIV-positive patients, higher diversity was evident only after two sequential samples taken within 150 days of infection were analysed (Thomson et al., 2011). These results may be biased by the limitations of clonal analysis which may underestimate true viral diversity. Using this technique, variants may be missed that are present in small numbers at transmission (<5 % if 20 clones are sampled at a single time point). Other approaches that are more sensitive than clonal analysis such as pyrosequencing and strain-specific PCR may in the future increase the detection of minority variant strains in baseline samples that later become dominant. In the previously described study of HIV-positive MSM with acute HCV, 36 % of patients who were labelled as superinfected using clonal analysis had a positive PCR result for the apparently superinfecting strain at the onset of infection when tested with strain-specific primers (Thomson et al., 2011). Studies employing pyrosequence analysis may also be useful in the future for distinguishing reinfection with new strains from the emergence of a pre-existing minority variant. However, based on current evidence, it is likely that superinfection and varying dominance of strains both play important roles during early HCV infection.
Compartmentalization
Detection of minority variants is further compounded by the fact that viral strains may also be compartmentalized and replicate in peripheral blood mononuclear cells (PBMCs; Maggi et al., 1997; Roque Alfonso et al., 1999) and the central nervous system (Forton et al., 2004). Compartmentalization of PBMC-specific strains occurs commonly in HIV-infected individuals with distinct genetic sequences from those detected in plasma (Blackard et al., 2007). Some of the virus detected in such patients may in fact be attached to the surface of NK cells, monocytes and B-cells and be dispersed via this route (Natarajan et al., 2010).
Predicting clinical outcome using viral diversity measures
Higher viral diversity is likely to increase the risk of progression to chronicity as well as lack of response to treatment due to an increased prevalence of escape mutations and resistant strains.
The point at which such diversity arises after acute infection has not been fully defined but is significantly different in evolving progression and spontaneous clearance by 150 days of infection (Thomson et al., 2011). Those with low Hamming distance and dN/dS ratios are significantly more likely to spontaneously clear the virus than those with high measures of genetic diversity.
Acute HCV is curable in the majority of HIV-negative patients but is less easy to treat in HIV-positive individuals (Jaeckel et al., 2001; Gilleece et al., 2005; Serpaggi et al., 2006; Dominguez et al., 2006). Early diagnosis and treatment of HIV-infected patients with peginterferon (IFN)-α and ribavirin results in improved sustained virological response (SVR) rates [59 % in acutely versus 40 % in chronically infected patients (Gilleece et al., 2005; Torriani et al., 2004)], but does not reach the 98 % treatment success rate reported in HIV-negative individuals (Jaeckel et al., 2001). The reason for this difference is not fully understood but could be partially explained by a higher rate of viral evolution and the formation of multiple variants due to HIV-related impaired immune control.
In keeping with the viral diversity theory, studies in HCV monoinfection have shown that two regions of the genome correlate with the likelihood of SVR in patients with chronic HCV (genotypes 1 and 3); the E2 HVR-1 and the IFN sensitivity determining region (ISDR) within NS5A (Enomoto et al., 1995; Enomoto et al., 1996; Ueda et al., 2004; Shire et al., 2006; Moreau et al., 2008). Increased diversity within E2 is associated with lower SVR. The mean number of mutations within the ISDR that differ from resistant prototype genotype 1a, 1b and 3a strains increases the likelihood of SVR (a mean two to four mutations difference confers protection; Pascu et al., 2004; MacQuillan et al., 2004; Nakagawa et al., 2010). A mutation within HCV core – the R70Q substitution is also strongly associated with resistance and lack of early virological response (EVR; Enomoto & Maekawa, 2010). The role of these resistance markers in acute infection and in HIV-infected individuals is as yet unknown.
Viral diversity is governed by the immune response
The key role of both innate and acquired immune responses in the control of early HCV infection is supported by a large body of evidence (recently reviewed by this journal; Jo et al., 2011). Different areas of the viral genome are subject to different selection pressures; the E2 HVR-1 is largely subject to antibody responses, while core and the non-structural proteins are moulded by the CD4+ and CD8+ acquired immune response.
Innate immune responses
While the key role of innate immunity in controlling HCV infection is well established, the impact of innate responses on viral diversity remains relatively unexplored. Single nucleotide polymorphisms near or within the IL-28B locus (the gene that encodes IFN-λ3) are associated with spontaneous clearance and response to treatment in HIV-positive and -negative individuals (Thomas et al., 2009; Tanaka et al., 2009). It is not clear yet whether IFN-λ encoded by this region is responsible for such effects. Among HIV-infected patients with chronic HCV, those bearing the IL-28B genotype CC were more commonly infected with genotype 3 than subjects with non-CC genotypes; thus, the protective effect of the CC genotype is mainly exerted in patients infected with HCV genotypes 1 or 4 (Neukam et al., 2011). While favourable IL-28B polymorphisms are associated with SVR in chronically infected HIV-positive patients, they are not associated with SVR in acute HCV (Nattermann et al., 2011).
Natural killer (NK) cell responses are also strongly associated with outcome during acute HCV (recently reviewed by Cheent & Khakoo, 2011); for example, spontaneous clearance is associated with the presence of the inhibitory receptor KIR2DL3 and the HLA-C1 ligand. This gene combination may be protective because the KIR2DL3 binds HLA-C with a lower avidity than other inhibitory KIRs, and thus NK cells expressing this specific inhibitory receptor have a lower threshold for activation.
Studies investigating the impact of such host genetic polymorphisms on viral diversification are awaited.
Acquired immune responses
The impact of both B- and T-cell responses on viral diversity is well-established. Immune pressure may result in swings in dominant quasispecies and the selection of HLA-defined epitopes. The role of individual epitopes in defining the final outcome is less clear.
B-cell responses
As discussed above, a gradual increase in genetic diversity and dN/dS ratio in the E2 HVR-1 gene is likely to be mediated predominantly by antibody responses (Farci et al., 2000; Gaud et al., 2003). High-resolution analysis of positively selected codons during early HCV infection show that these gradually accumulate in progressors (Sheridan et al., 2004) but not in clearers. Low CD4 count in HIV-positive patients is also associated with lower HVR-1 diversity, while higher CD4 counts following treatment of HIV with HAART is associated with an increase in HVR-1 diversity (Bernini et al., 2011). This may not be protective; the HVR-1 could be a decoy site; increasing diversity within the HVR-1 in this study was also significantly associated with an increase in HCV VL.
T-cell responses during co-infection with HCV and HIV
Both CD4+ and CD8+ responses are critical in defining the final outcome of HCV infection. Evidence for this comes from the chimpanzee model, epidemiological HLA-association studies and functional and phenotypic studies of T-cells in acutely infected individuals. Protective immunity in HIV-negative individuals is known to be associated with multi-specific CD4+ and CD8+ T-cell responses (Cooper et al., 1999; Lechner et al., 2000; Harcourt et al., 2001; Cox et al., 2005). In patients with chronic HCV, both the magnitude and breadth of CD4+ and CD8+ responses are lower in HIV-infected patients (Capa et al., 2007). The role of CD4+ T-cells is likely to be of particular relevance in patients who are HIV infected.
In chimpanzees, depletion of CD4+ cells results in immune escape and persistent infection (Grakoui et al., 2003). In humans, MHC class 2 associations also provide evidence for the role of CD4+ cells in spontaneous clearance; HLA DRB1*0101, DR1B1*1101 (OR = 2.14) and DQB1*0301 (OR = 2.22) are associated with spontaneous resolution of acute HCV (Thursz et al., 1999) and an improved immune response (Harcourt et al., 2001), while HLA-DRB1*0701 is associated with viral persistence (Fanning et al., 2001).
As might be expected, CD4+ responses to acute HCV are weaker in HIV-positive individuals. In particular, significantly reduced CD4+ IFN-γ responses occur in HIV-positive patients, especially those directed against NS3-5 (Danta et al., 2008). Targeting of NS proteins is associated with spontaneous clearance in both HIV-negative and HIV-positive individuals, while responses to core are more common in chronically evolving infection (Gerlach et al., 2005; Aberle et al., 2006; Ruys et al., 2008; van den Berg et al., 2009; Schnuriger et al., 2009).
A key role of CD4+ cells is to provide support for CD8+ T-cells and CD4+ cell dysfunction impacts on the CD8+ response in both chimpanzees and humans. CD4+ T-cell help is critical in maintaining long-term protective CD8+ T-cell immunity in chimpanzees (Zajac et al., 1998; Grakoui et al., 2003) and in humans (Urbani et al., 2006). In humans, a cross-sectional analysis of HCV-specific CD8+ cell responses during chronic HCV and HIV infection revealed that HIV-related CD4 cell depletion was associated with significantly lower HCV-specific CD8+ responses (P<0.0001; Kim et al., 2005).
This is important, as CD8+ T-cells also play a key role in HCV outcome. Depletion of CD8+ cells in chimpanzees is associated with viral persistence (Shoukry et al., 2004). In humans, MHC class 1 associations (particularly HLA B27) with spontaneous clearance provide evidence of the role of the CD8+ response in defining outcome (McKiernan et al., 2004; Neumann-Haefelin et al., 2007). Footprints of the HLA-defined T-cell response (HLA-defined viral escape mutations from CD8+ T-cell responses) may be found within the viral quasispecies in the HCV structural and NS proteins (Cox et al., 2005). The number of mutations occurring in the viral population at any time due to CD8+ selection pressure is as much as 32 %; this is significant, but lower than that observed in early HIV infection (30–60 %; Pfafferott et al., 2011). This figure is likely to be lower in HIV-infected individuals as HCV-specific CD8+ responses are weaker (Capa et al., 2007). Interestingly, reversion from HLA-restricted epitopes does not appear to be common in HCV following transmission. This may be due to the co-existence of deleterious and compensatory mutations in differing parts of the viral genome. Such epistatic mutations are evident in HCV cell culture and in natural infection; thus evolution of HCV is constrained not just by the immune response but also by other fitness considerations and evolution of the genome occurs in a co-ordinated fashion (Campo et al., 2008).
Summary and future perspectives
The emerging epidemic of acute HCV in HIV-positive MSM has allowed the study of acute HCV infection on a large scale. Studies of viral evolution have shown that immune control of infection is hampered by rapid diversification due to switching dominance of viral quasispecies, superinfection, generation of de novo escape variants and compartmentalization of the virus. The key role of the CD4+ response in controlling infection is highlighted in HIV-positive cohorts. Many questions remain unanswered however. Firstly, the predictive role of baseline viral diversity in defining spontaneous clearance may be increased using techniques in which viral genetic diversity may be assessed in greater depth. Such analyses may also assist in predicting the response to treatment. The time at which the quasispecies diversity reaches a threshold level may define the suitability and timing of treatment and with the advent of newer treatments for HCV such as the protease inhibitors bocepravir and telapravir could potentially be used to target treatment for appropriate individual patients. Finally, the availability of larger cohorts of patients may facilitate the definition of particular protective epitopes which may be used in future vaccine development.
Acknowledgements
Funding sources: P. K., Wellcome Trust (WT091663MA), MRC, NIHR Biomedical Research Centre, Oxford, the James Martin School for 21st Century, Oxford, and the NIH NIAID 1U19AI082630-01; E. T., Wellcome Trust (WT083690); J. S., the James Martin School for 21st Century, Oxford.
References
- Aberle J. H., Formann E., Steindl-Munda P., Weseslindtner L., Gurguta C., Perstinger G., Grilnberger E., Laferl H., Dienes H. P., Popowkraupp T. (2006). Prospective study of viral clearance and CD4+ T-cell response in acute hepatitis C primary infection and reinfection. J Clin Virol 36, 24–31 10.1016/j.jcv.2005.12.010 [DOI] [PubMed] [Google Scholar]
- Arnold D. M., Julian J. A., Walker I. R., Association of Hemophilia Clinic Directors of Canada (2006). Mortality rates and causes of death among all HIV-positive individuals with hemophilia in Canada over 21 years of follow-up. Blood 108, 460–464 10.1182/blood-2005-11-4407 [DOI] [PubMed] [Google Scholar]
- Bader T., Fazili J., Madhoun M., Aston C., Hughes D., Rizvi S., Seres K., Hasan M. (2008). Fluvastatin inhibits hepatitis C replication in humans. Am J Gastroenterol 103, 1383–1389 10.1111/j.1572-0241.2008.01876.x [DOI] [PubMed] [Google Scholar]
- Bernini F., Ebranati E., De Maddalena C., Shkjezi R., Milazzo L., Lo Presti A., Ciccozzi M., Galli M., Zehender G. (2011). Within-host dynamics of the hepatitis C virus quasispecies population in HIV-1/HCV coinfected patients. PLoS ONE 6, e16551 10.1371/journal.pone.0016551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bica I., McGovern B., Dhar R., Stone D., McGowan K., Scheib R., Snydman D. R. (2001). Increasing mortality due to end-stage liver disease in patients with human immunodeficiency virus infection. Clin Infect Dis 32, 492–497 10.1086/318501 [DOI] [PubMed] [Google Scholar]
- Blackard J. T., Hiasa Y., Smeaton L., Jamieson D. J., Rodriguez I., Mayer K. H., Chung R. T. (2007). Compartmentalization of hepatitis C virus (HCV) during HCV/HIV coinfection. J Infect Dis 195, 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottieau E., Apers L., Van Esbroeck M., Vandenbruaene M., Florence E. (2010). Hepatitis C virus infection in HIV-infected men who have sex with men: sustained rising incidence in Antwerp, Belgium, 2001–2009. Euro Surveill 15, 19673. [PubMed] [Google Scholar]
- Campo D. S., Dimitrova Z., Mitchell R. J., Lara J., Khudyakov Y. (2008). Coordinated evolution of the hepatitis C virus. Proc Natl Acad Sci U S A 105, 9685–9690 10.1073/pnas.0801774105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capa L., Soriano V., García-Samaniego J., Nuñez M., Romero M., Cascajero A., Muñoz F., González-Lahoz J., Benito J. M. (2007). Influence of HCV genotype and co-infection with human immunodeficiency virus on CD4+ and CD8+ T-cell responses to hepatitis C virus. J Med Virol 79, 503–510 10.1002/jmv.20856 [DOI] [PubMed] [Google Scholar]
- Cheent K., Khakoo S. I. (2011). Natural killer cells and hepatitis C: action and reaction. Gut 60, 268–278 10.1136/gut.2010.212555 [DOI] [PubMed] [Google Scholar]
- Cooper S., Erickson A. L., Adams E. J., Kansopon J., Weiner A. J., Chien D. Y., Houghton M., Parham P., Walker C. M. (1999). Analysis of a successful immune response against hepatitis C virus. Immunity 10, 439–449 10.1016/S1074-7613(00)80044-8 [DOI] [PubMed] [Google Scholar]
- Cox A. L., Mosbruger T., Lauer G. M., Pardoll D., Thomas D. L., Ray S. C. (2005). Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology 42, 104–112 10.1002/hep.20749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahari H., Major M., Zhang X., Mihalik K., Rice C. M., Perelson A. S., Feinstone S. M., Neumann A. U. (2005). Mathematical modeling of primary hepatitis C infection: noncytolytic clearance and early blockage of virion production. Gastroenterology 128, 1056–1066 10.1053/j.gastro.2005.01.049 [DOI] [PubMed] [Google Scholar]
- Danta M., Brown D., Bhagani S., Pybus O. G., Sabin C. A., Nelson M., Fisher M., Johnson A. M., Dusheiko G. M., HIV and Acute HCV (HAAC) group (2007). Recent epidemic of acute hepatitis C virus in HIV-positive men who have sex with men linked to high-risk sexual behaviours. AIDS 21, 983–991 10.1097/QAD.0b013e3281053a0c [DOI] [PubMed] [Google Scholar]
- Danta M., Semmo N., Fabris P., Brown D., Pybus O. G., Sabin C. A., Bhagani S., Emery V. C., Dusheiko G. M., Klenerman P. (2008). Impact of HIV on host-virus interactions during early hepatitis C virus infection. J Infect Dis 197, 1558–1566 10.1086/587843 [DOI] [PubMed] [Google Scholar]
- de Bruijne J., Schinkel J., Prins M., Koekkoek S. M., Aronson S. J., van Ballegooijen M. W., Reesink H. W., Molenkamp R., van de Laar T. J. (2009). Emergence of hepatitis C virus genotype 4: phylogenetic analysis reveals three distinct epidemiological profiles. J Clin Microbiol 47, 3832–3838 10.1128/JCM.01146-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez S., Ghosn J., Valantin M. A., Schruniger A., Simon A., Bonnard P., Caumes E., Pialoux G., Benhamou Y., et al. & other authors (2006). Efficacy of early treatment of acute hepatitis C infection with pegylated interferon and ribavirin in HIV-infected patients. AIDS 20, 1157–1161 10.1097/01.aids.0000226956.02719.fd [DOI] [PubMed] [Google Scholar]
- Enomoto N., Maekawa S. (2010). HCV genetic elements determining the early response to peginterferon and ribavirin therapy. Intervirology 53, 66–69 10.1159/000252787 [DOI] [PubMed] [Google Scholar]
- Enomoto N., Sakuma I., Asahina Y., Kurosaki M., Murakami T., Yamamoto C., Izumi N., Marumo F., Sato C. (1995). Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest 96, 224–230 10.1172/JCI118025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto N., Sakuma I., Asahina Y., Kurosaki M., Murakami T., Yamamoto C., Ogura Y., Izumi N., Marumo F., Sato C. (1996). Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med 334, 77–82 10.1056/NEJM199601113340203 [DOI] [PubMed] [Google Scholar]
- Fanning L. J., Levis J., Kenny-Walsh E., Whelton M., O’Sullivan K., Shanahan F. (2001). HLA class II genes determine the natural variance of hepatitis C viral load. Hepatology 33, 224–230 10.1053/jhep.2001.20642 [DOI] [PubMed] [Google Scholar]
- Farci P., Shimoda A., Coiana A., Diaz G., Peddis G., Melpolder J. C., Strazzera A., Chien D. Y., Munoz S. J., et al. & other authors (2000). The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288, 339–344 10.1126/science.288.5464.339 [DOI] [PubMed] [Google Scholar]
- Felmlee D. J., Sheridan D. A., Bridge S. H., Nielsen S. U., Milne R. W., Packard C. J., Caslake M. J., McLauchlan J., Toms G. L., et al. & other authors (2010). Intravascular transfer contributes to postprandial increase in numbers of very-low-density hepatitis C virus particles. Gastroenterology 139, 1774–1783.e6 10.1053/j.gastro.2010.07.047 [DOI] [PubMed] [Google Scholar]
- Fernandez J., Taylor D., Morhardt D. R., Mihalik K., Puig M., Rice C. M., Feinstone S. M., Major M. E. (2004). Long-term persistence of infection in chimpanzees inoculated with an infectious hepatitis C virus clone is associated with a decrease in the viral amino acid substitution rate and low levels of heterogeneity. J Virol 78, 9782–9789 10.1128/JVI.78.18.9782-9789.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer D. S., Uriel A. J., Carriero D. C., Klepper A., Dieterich D. T., Mullen M. P., Thung S. N., Fiel M. I., Branch A. D. (2008). Liver fibrosis during an outbreak of acute hepatitis C virus infection in HIV-infected men: a prospective cohort study. J Infect Dis 198, 683–686 10.1086/590430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forton D. M., Karayiannis P., Mahmud N., Taylor-Robinson S. D., Thomas H. C. (2004). Identification of unique hepatitis C virus quasispecies in the central nervous system and comparative analysis of internal translational efficiency of brain, liver, and serum variants. J Virol 78, 5170–5183 10.1128/JVI.78.10.5170-5183.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J., Nastouli E., Thomson E., Muir D., McClure M., Weber J., Fidler S. (2008). Increasing incidence of acute hepatitis C in individuals diagnosed with primary HIV in the United Kingdom. AIDS 22, 666–668 10.1097/QAD.0b013e3282f5f4cf [DOI] [PubMed] [Google Scholar]
- Gaud U., Langer B., Petropoulou T., Thomas H. C., Karayiannis P. (2003). Changes in hypervariable region 1 of the envelope 2 glycoprotein of hepatitis C virus in children and adults with humoral immune defects. J Med Virol 69, 350–356 10.1002/jmv.10296 [DOI] [PubMed] [Google Scholar]
- Gerlach J. T., Ulsenheimer A., Grüner N. H., Jung M. C., Schraut W., Schirren C. A., Heeg M., Scholz S., Witter K., et al. & other authors (2005). Minimal T-cell-stimulatory sequences and spectrum of HLA restriction of immunodominant CD4+ T-cell epitopes within hepatitis C virus NS3 and NS4 proteins. J Virol 79, 12425–12433 10.1128/JVI.79.19.12425-12433.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn J., Pierre-François S., Thibault V., Duvivier C., Tubiana R., Simon A., Valantin M. A., Dominguez S., Caumes E., Katlama C. (2004). Acute hepatitis C in HIV-infected men who have sex with men. HIV Med 5, 303–306 10.1111/j.1468-1293.2004.00225.x [DOI] [PubMed] [Google Scholar]
- Gilleece Y. C., Browne R. E., Asboe D., Atkins M., Mandalia S., Bower M., Gazzard B. G., Nelson M. R. (2005). Transmission of hepatitis C virus among HIV-positive homosexual men and response to a 24-week course of pegylated interferon and ribavirin. J Acquir Immune Defic Syndr 40, 41–46 10.1097/01.qai.0000174930.64145.a9 [DOI] [PubMed] [Google Scholar]
- Grakoui A., Shoukry N. H., Woollard D. J., Han J. H., Hanson H. L., Ghrayeb J., Murthy K. K., Rice C. M., Walker C. M. (2003). HCV persistence and immune evasion in the absence of memory T cell help. Science 302, 659–662 10.1126/science.1088774 [DOI] [PubMed] [Google Scholar]
- Gray R. R., Parker J., Lemey P., Salemi M., Katzourakis A., Pybus O. G. (2011). The mode and tempo of hepatitis C virus evolution within and among hosts. BMC Evol Biol 11, 131 10.1186/1471-2148-11-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt G., Hellier S., Bunce M., Satsangi J., Collier J., Chapman R., Phillips R., Klenerman P. (2001). Effect of HLA class II genotype on T helper lymphocyte responses and viral control in hepatitis C virus infection. J Viral Hepat 8, 174–179 10.1046/j.1365-2893.2001.00289.x [DOI] [PubMed] [Google Scholar]
- Jaeckel E., Cornberg M., Wedemeyer H., Santantonio T., Mayer J., Zankel M., Pastore G., Dietrich M., Trautwein C., et al. & other authors (2001). Treatment of acute hepatitis C with interferon alfa-2b. N Engl J Med 345, 1452–1457 10.1056/NEJMoa011232 [DOI] [PubMed] [Google Scholar]
- Jo J., Lohmann V., Bartenschlager R., Thimme R. (2011). Experimental models to study the immunobiology of hepatitis C virus. J Gen Virol 92, 477–493 10.1099/vir.0.027987-0 [DOI] [PubMed] [Google Scholar]
- Kalinina O., Norder H., Mukomolov S., Magnius L. O. (2002). A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol 76, 4034–4043 10.1128/JVI.76.8.4034-4043.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A. Y., Lauer G. M., Ouchi K., Addo M. M., Lucas M., Schulze Zur Wiesch J., Timm J., Boczanowski M., Duncan J. E., et al. & other authors (2005). The magnitude and breadth of hepatitis C virus-specific CD8+ T cells depend on absolute CD4+ T-cell count in individuals coinfected with HIV-1. Blood 105, 1170–1178 10.1182/blood-2004-06-2336 [DOI] [PubMed] [Google Scholar]
- Klenerman P., Lechner F., Kantzanou M., Ciurea A., Hengartner H., Zinkernagel R. (2000). Viral escape and the failure of cellular immune responses. Science 289, 2003 10.1126/science.289.5487.2003a [DOI] [PubMed] [Google Scholar]
- Kuntzen T., Timm J., Berical A., Lewis-Ximenez L. L., Jones A., Nolan B., Schulze zur Wiesch J., Li B., Schneidewind A., et al. & other authors (2007). Viral sequence evolution in acute hepatitis C virus infection. J Virol 81, 11658–11668 10.1128/JVI.00995-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer G. M., Ouchi K., Chung R. T., Nguyen T. N., Day C. L., Purkis D. R., Reiser M., Kim A. Y., Lucas M., et al. & other authors (2002). Comprehensive analysis of CD8+-T-cell responses against hepatitis C virus reveals multiple unpredicted specificities. J Virol 76, 6104–6113 10.1128/JVI.76.12.6104-6113.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner F., Gruener N. H., Urbani S., Uggeri J., Santantonio T., Kammer A. R., Cerny A., Phillips R., Ferrari C., et al. & other authors (2000). CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur J Immunol 30, 2479–2487 [DOI] [PubMed] [Google Scholar]
- Loomba R., Rivera M. M., McBurney R., Park Y., Haynes-Williams V., Rehermann B., Alter H. J., Herrine S. K., Liang T. J., et al. & other authors (2011). The natural history of acute hepatitis C: clinical presentation, laboratory findings and treatment outcomes. Aliment Pharmacol Ther 33, 559–565 10.1111/j.1365-2036.2010.04549.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Labrador F. X., Dove L., Hui C. K., Phung Y., Kim M., Berenguer M., Wright T. L. (2007). Trends for genetic variation of hepatitis C Virus quasispecies in Human Immunodeficiency virus-1 coinfected patients. Virus Res 130, 285–291 10.1016/j.virusres.2007.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luetkemeyer A., Hare C. B., Stansell J., Tien P. C., Charlesbois E., Lum P., Havlir D., Peters M. (2006). Clinical presentation and course of acute hepatitis C infection in HIV-infected patients. J Acquir Immune Defic Syndr 41, 31–36 10.1097/01.qai.0000191281.77954.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQuillan G. C., Niu X., Speers D., English S., Garas G., Harnett G. B., Reed W. D., Allan J. E., Jeffrey G. P. (2004). Does sequencing the PKRBD of hepatitis C virus NS5A predict therapeutic response to combination therapy in an Australian population? J Gastroenterol Hepatol 19, 551–557 10.1111/j.1440-1746.2003.03319.x [DOI] [PubMed] [Google Scholar]
- Maggi F., Fornai C., Vatteroni M. L., Giorgi M., Morrica A., Pistello M., Cammarota G., Marchi S., Ciccorossi P., et al. & other authors (1997). Differences in hepatitis C virus quasispecies composition between liver, peripheral blood mononuclear cells and plasma. J Gen Virol 78, 1521–1525 [DOI] [PubMed] [Google Scholar]
- Matthews G. V., Hellard M., Haber P., Yeung B., Marks P., Baker D., McCaughan G., Sasadeusz J., White P., et al. & other authors (2009). Characteristics and treatment outcomes among HIV-infected individuals in the Australian Trial in Acute Hepatitis C. Clin Infect Dis 48, 650–658 10.1086/596770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKiernan S. M., Hagan R., Curry M., McDonald G. S., Kelly A., Nolan N., Walsh A., Hegarty J., Lawlor E., Kelleher D. (2004). Distinct MHC class I and II alleles are associated with hepatitis C viral clearance, originating from a single source. Hepatology 40, 108–114 10.1002/hep.20261 [DOI] [PubMed] [Google Scholar]
- Moreau I., Levis J., Crosbie O., Kenny-Walsh E., Fanning L. J. (2008). Correlation between pre-treatment quasispecies complexity and treatment outcome in chronic HCV genotype 3a. Virol J 5, 78 10.1186/1743-422X-5-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Sakamoto N., Ueyama M., Mogushi K., Nagaie S., Itsui Y., Azuma S., Kakinuma S., Tanaka H., et al. & other authors (2010). Mutations in the interferon sensitivity determining region and virological response to combination therapy with pegylated-interferon alpha 2b plus ribavirin in patients with chronic hepatitis C-1b infection. J Gastroenterol 45, 656–665 10.1007/s00535-009-0195-7 [DOI] [PubMed] [Google Scholar]
- Natarajan V., Kottilil S., Hazen A., Adelsberger J., Murphy A. A., Polis M. A., Kovacs J. A. (2010). HCV in peripheral blood mononuclear cells are predominantly carried on the surface of cells in HIV/HCV co-infected individuals. J Med Virol 82, 2032–2037 10.1002/jmv.21906 [DOI] [PubMed] [Google Scholar]
- Nattermann J., Vogel M., Nischalke H. D., Danta M., Mauss S., Stellbrink H. J., Baumgarten A., Mayr C., Bruno R., et al. & other authors (2011). Genetic variation in IL28B and treatment-induced clearance of hepatitis C virus in HIV-positive patients with acute and chronic hepatitis C. J Infect Dis 203, 595–601 10.1093/infdis/jiq098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukam K., Nattermann J., Rallón N., Rivero A., Caruz A., Macías J., Vogel M., Benito J., Camacho A., et al. & other authors (2011). Different distributions of hepatitis C virus genotypes among HIV-infected patients with acute and chronic hepatitis C according to interleukin-28B genotype. HIV Med no 10.1111/j.1468-1293.2011.00912.x [DOI] [PubMed] [Google Scholar]
- Neumann A. U., Lam N. P., Dahari H., Gretch D. R., Wiley T. E., Layden T. J., Perelson A. S. (1998). Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282, 103–107 10.1126/science.282.5386.103 [DOI] [PubMed] [Google Scholar]
- Neumann-Haefelin C., Spangenberg H. C., Blum H. E., Thimme R. (2007). Host and viral factors contributing to CD8+ T cell failure in hepatitis C virus infection. World J Gastroenterol 13, 4839–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen O. K., Dore G. J., Kaldor J. M., Hellard M. E., ATAHC Protocol Steering Committee (2007). Recruitment and follow-up of injecting drug users in the setting of early hepatitis C treatment: insights from the ATAHC study. Int J Drug Policy 18, 447–451 10.1016/j.drugpo.2007.01.007 [DOI] [PubMed] [Google Scholar]
- Page K., Hahn J. A., Evans J., Shiboski S., Lum P., Delwart E., Tobler L., Andrews W., Avanesyan L., et al. & other authors (2009). Acute hepatitis C virus infection in young adult injection drug users: a prospective study of incident infection, resolution, and reinfection. J Infect Dis 200, 1216–1226 10.1086/605947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascu M., Martus P., Höhne M., Wiedenmann B., Hopf U., Schreier E., Berg T. (2004). Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: a meta-analysis focused on geographical differences. Gut 53, 1345–1351 10.1136/gut.2003.031336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L., Weis N. M., Lindhardt B. O. (2006). [Sexual transmission of hepatitis C among HIV-positive men]. Ugeskr Laeger 168, 3630–3631 (in Danish). [PubMed] [Google Scholar]
- Pfafferott K., Gaudieri S., Ulsenheimer A., James I., Heeg M., Nolan D., John M., Rauch A., Mallal S., et al. & other authors (2011). Constrained pattern of viral evolution in acute and early HCV infection limits viral plasticity. PLoS ONE 6, e16797 10.1371/journal.pone.0016797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham S. T., Bull R. A., Bennett J. M., Rawlinson W. D., Dore G. J., Lloyd A. R., White P. A. (2010). Frequent multiple hepatitis C virus infections among injection drug users in a prison setting. Hepatology 52, 1564–1572 10.1002/hep.23885 [DOI] [PubMed] [Google Scholar]
- Rauch A., Martin M., Weber R., Hirschel B., Tarr P. E., Bucher H. C., Vernazza P., Bernasconi E., Zinkernagel A. S., et al. & other authors (2005). Unsafe sex and increased incidence of hepatitis C virus infection among HIV-infected men who have sex with men: the Swiss HIV Cohort Study. Clin Infect Dis 41, 395–402 10.1086/431486 [DOI] [PubMed] [Google Scholar]
- Roque Afonso A. M., Jiang J., Penin F., Tareau C., Samuel D., Petit M. A., Bismuth H., Dussaix E., Féray C. (1999). Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets. J Virol 73, 9213–9221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruys T. A., den Hollander J. G., Beld M. G., van der Ende M. E., van der Meer J. T. (2004). [Sexual transmission of hepatitis C in homosexual men]. Ned Tijdschr Geneeskd 148, 2309–2312 (in Dutch). [PubMed] [Google Scholar]
- Ruys T. A., Nanlohy N. M., van den Berg C. H., Hassink E., Beld M., van de Laar T., Bruisten S., Wit F., Krol A., et al. & other authors (2008). HCV-specific T-cell responses in injecting drug users: evidence for previous exposure to HCV and a role for CD4+ T cells focussing on nonstructural proteins in viral clearance. J Viral Hepat 15, 409–420 10.1111/j.1365-2893.2007.00963.x [DOI] [PubMed] [Google Scholar]
- Santantonio T., Medda E., Ferrari C., Fabris P., Cariti G., Massari M., Babudieri S., Toti M., Francavilla R., et al. & other authors (2006). Risk factors and outcome among a large patient cohort with community-acquired acute hepatitis C in Italy. Clin Infect Dis 43, 1154–1159 10.1086/507640 [DOI] [PubMed] [Google Scholar]
- Sarobe P., Lasarte J. J., García N., Civeira M. P., Borrás-Cuesta F., Prieto J. (2006). Characterization of T-cell responses against immunodominant epitopes from hepatitis C virus E2 and NS4a proteins. J Viral Hepat 13, 47–55 10.1111/j.1365-2893.2005.00653.x [DOI] [PubMed] [Google Scholar]
- Schmidt A. J., Rockstroh J. K., Vogel M., An der Heiden M., Baillot A., Krznaric I., Radun D. (2011). Trouble with bleeding: risk factors for acute hepatitis C among HIV-positive gay men from Germany–a case-control study. PLoS ONE 6, e17781 10.1371/journal.pone.0017781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnuriger A., Dominguez S., Guiguet M., Harfouch S., Samri A., Ouazene Z., Slama L., Simon A., Valantin M. A., et al. & other authors (2009). Acute hepatitis C in HIV-infected patients: rare spontaneous clearance correlates with weak memory CD4 T-cell responses to hepatitis C virus. AIDS 23, 2079–2089 10.1097/QAD.0b013e328330ed24 [DOI] [PubMed] [Google Scholar]
- Sentandreu V., Jiménez-Hernández N., Torres-Puente M., Bracho M. A., Valero A., Gosalbes M. J., Ortega E., Moya A., González-Candelas F. (2008). Evidence of recombination in intrapatient populations of hepatitis C virus. PLoS ONE 3, e3239 10.1371/journal.pone.0003239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpaggi J., Chaix M. L., Batisse D., Dupont C., Vallet-Pichard A., Fontaine H., Viard J. P., Piketty C., Rouveix E., et al. & other authors (2006). Sexually transmitted acute infection with a clustered genotype 4 hepatitis C virus in HIV-1-infected men and inefficacy of early antiviral therapy. AIDS 20, 233–240 10.1097/01.aids.0000200541.40633.56 [DOI] [PubMed] [Google Scholar]
- Sheridan I., Pybus O. G., Holmes E. C., Klenerman P. (2004). High-resolution phylogenetic analysis of hepatitis C virus adaptation and its relationship to disease progression. J Virol 78, 3447–3454 10.1128/JVI.78.7.3447-3454.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai M., Arichi T., Chen M., Masaki T., Nishioka M., Ikeda K., Takahashi H., Enomoto N., Saito T., et al. & other authors (1999). T cell recognition of hypervariable region-1 from hepatitis C virus envelope protein with multiple class II MHC molecules in mice and humans: preferential help for induction of antibodies to the hypervariable region. J Immunol 162, 568–576 [PubMed] [Google Scholar]
- Shire N. J., Horn P. S., Rouster S. D., Stanford S., Eyster M. E., Sherman K. E., Multicenter Hemophilia Cohort HCV Study Group (2006). HCV kinetics, quasispecies, and clearance in treated HCV-infected and HCV/HIV-1-coinfected patients with hemophilia. Hepatology 44, 1146–1157 10.1002/hep.21374 [DOI] [PubMed] [Google Scholar]
- Shores N. J., Maida I., Soriano V., Núnez M. (2008). Sexual transmission is associated with spontaneous HCV clearance in HIV-infected patients. J Hepatol 49, 323–328 10.1016/j.jhep.2008.04.010 [DOI] [PubMed] [Google Scholar]
- Shoukry N. H., Cawthon A. G., Walker C. M. (2004). Cell-mediated immunity and the outcome of hepatitis C virus infection. Annu Rev Microbiol 58, 391–424 [DOI] [PubMed] [Google Scholar]
- Smith J. A., Aberle J. H., Fleming V. M., Ferenci P., Thomson E. C., Karayiannis P., McLean A. R., Holzmann H., Klenerman P. (2010). Dynamic coinfection with multiple viral subtypes in acute hepatitis C. J Infect Dis 202, 1770–1779 10.1086/657317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolte I. G., Dukers N. H., Geskus R. B., Coutinho R. A., de Wit J. B. (2004). Homosexual men change to risky sex when perceiving less threat of HIV/AIDS since availability of highly active antiretroviral therapy: a longitudinal study. AIDS 18, 303–309 10.1097/00002030-200401230-00021 [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Nishida N., Sugiyama M., Kurosaki M., Matsuura K., Sakamoto N., Nakagawa M., Korenaga M., Hino K., et al. & other authors (2009). Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 41, 1105–1109 10.1038/ng.449 [DOI] [PubMed] [Google Scholar]
- Thomas D. L., Thio C. L., Martin M. P., Qi Y., Ge D., O’Huigin C., Kidd J., Kidd K., Khakoo S. I., et al. & other authors (2009). Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461, 798–801 10.1038/nature08463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E. C., Fleming V. M., Main J., Klenerman P., Weber J., Eliahoo J., Smith J., McClure M. O., Karayiannis P. (2011). Predicting spontaneous clearance of acute hepatitis C virus in a large cohort of HIV-1-infected men. Gut 60, 837–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursz M., Yallop R., Goldin R., Trepo C., Thomas H. C., The HENCORE Group (1999). Influence of MHC class II genotype on outcome of infection with hepatitis C virus. Lancet 354, 2119–2124 10.1016/S0140-6736(99)91443-5 [DOI] [PubMed] [Google Scholar]
- Torriani F. J., Rodriguez-Torres M., Rockstroh J. K., Lissen E., Gonzalez-García J., Lazzarin A., Carosi G., Sasadeusz J., Katlama C., et al. & other authors (2004). Peginterferon Alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N Engl J Med 351, 438–450 10.1056/NEJMoa040842 [DOI] [PubMed] [Google Scholar]
- Tuplin A., Evans D. J., Simmonds P. (2004). Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J Gen Virol 85, 3037–3047 10.1099/vir.0.80141-0 [DOI] [PubMed] [Google Scholar]
- Ueda E., Enomoto N., Sakamoto N., Hamano K., Sato C., Izumi N., Watanabe M. (2004). Changes of HCV quasispecies during combination therapy with interferon and ribavirin. Hepatol Res 29, 89–96 10.1016/j.hepres.2004.02.014 [DOI] [PubMed] [Google Scholar]
- Urbani S., Amadei B., Tola D., Massari M., Schivazappa S., Missale G., Ferrari C. (2006). PD-1 expression in acute hepatitis C virus (HCV) infection is associated with HCV-specific CD8 exhaustion. J Virol 80, 11398–11403 10.1128/JVI.01177-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus A. T., van de Laar T. J., Stolte I. G., Schinkel J., Heijman T., Coutinho R. A., Prins M. (2009). Hepatitis C virus infections among HIV-infected men who have sex with men: an expanding epidemic. AIDS 23, F1–F7 10.1097/QAD.0b013e32832e5631 [DOI] [PubMed] [Google Scholar]
- van de Laar T., Pybus O., Bruisten S., Brown D., Nelson M., Bhagani S., Vogel M., Baumgarten A., Chaix M. L., et al. & other authors (2009a). Evidence of a large, international network of HCV transmission in HIV-positive men who have sex with men. Gastroenterology 136, 1609–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Laar T. J., Molenkamp R., van den Berg C., Schinkel J., Beld M. G., Prins M., Coutinho R. A., Bruisten S. M. (2009b). Frequent HCV reinfection and superinfection in a cohort of injecting drug users in Amsterdam. J Hepatol 51, 667–674 10.1016/j.jhep.2009.05.027 [DOI] [PubMed] [Google Scholar]
- van den Berg C. H., Ruys T. A., Nanlohy N. M., Geerlings S. E., van der Meer J. T., Mulder J. W., Lange J. A., van Baarle D. (2009). Comprehensive longitudinal analysis of hepatitis C virus (HCV)-specific T cell responses during acute HCV infection in the presence of existing HIV-1 infection. J Viral Hepat 16, 239–248 10.1111/j.1365-2893.2009.01076.x [DOI] [PubMed] [Google Scholar]
- Vogel M., Bieniek B., Jessen H., Schewe C. K., Hoffmann C., Baumgarten A., Kroidl A., Bogner J. R., Spengler U., Rockstroh J. K. (2005). Treatment of acute hepatitis C infection in HIV-infected patients: a retrospective analysis of eleven cases. J Viral Hepat 12, 207–211 10.1111/j.1365-2893.2005.00580.x [DOI] [PubMed] [Google Scholar]
- Vogel M., Boesecke C., Rockstroh J. K. (2011). Acute hepatitis C infection in HIV-positive patients. Curr Opin Infect Dis 24, 1–6 10.1097/QCO.0b013e3283422e09 [DOI] [PubMed] [Google Scholar]
- Weiner A. J., Brauer M. J., Rosenblatt J., Richman K. H., Tung J., Crawford K., Bonino F., Saracco G., Choo Q. L., Houghton M. (1991). Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology 180, 842–848 10.1016/0042-6822(91)90104-J [DOI] [PubMed] [Google Scholar]
- Zajac A. J., Blattman J. N., Murali-Krishna K., Sourdive D. J., Suresh M., Altman J. D., Ahmed R. (1998). Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 188, 2205–2213 10.1084/jem.188.12.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]