Abstract
Endonuclease cleavage was one of the first identified mechanisms of mRNA decay but until recently it was thought to play a minor role to the better-known processes of deadenylation, decapping and exonuclease-catalyzed decay. Most of the early examples of endonuclease decay came from studies of a particular mRNA whose turnover changed in response to hormone, cytokine, developmental or nutritional stimuli. Only a few of these examples of endonuclease-mediated mRNA decay progressed to the point where the enzyme responsible for the initiating event was identified and studied in any detail. The discovery of microRNAs and RISC-catalyzed endonuclease cleavage followed by the identification of PIN (pilT N-terminal) domains that impart endonuclease activity to a number of the proteins involved in mRNA decay has led to a resurgence of interest in endonuclease-mediated mRNA decay. PIN domains show no substrate selectivity, and their involvement in a number of decay pathways highlights a recurring theme that the context in which an endonuclease functions is a primary factor in determining whether any given mRNA will be targeted for decay by this or the default exonuclease-mediated decay processes.
Keywords: endonuclease, cleavage, deadenylation, NMD, mRNA decay
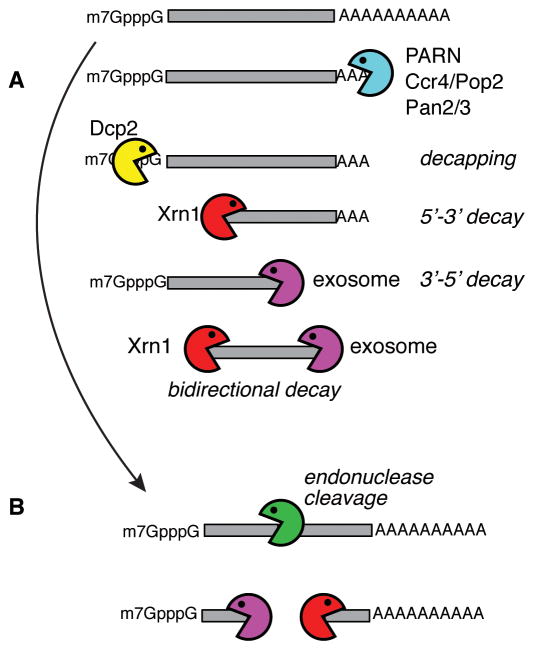
The default process of metazoan mRNA decay begins with poly(A) shortening, followed by removal of the cap by Dcp2 and exonuclease decay from the 5′ end catalyzed by Xrn1 and from the 3′ end catalyzed by one or more of the ribonuclease activities that associate with the eukaryotic exosome [1] (Fig. 1A). However as early as the 1980s there were reports of unstable mRNAs that did not undergo deadenylation prior to decay. The turnover of these mRNAs was initiated by endonuclease cleavage either within the body of the mRNA or in the 3′-UTR, with the resulting fragments cleared by exonuclease digestion (Fig. 1B). Some of the historical evidence for endonuclease decay is likely due to targeted cleavage by microRNAs in association with RISC, and support for this notion comes from recent results using deep sequencing to look for evidence of endonuclease cleavage [2, 3]. While these studies identified a large number of mRNAs that are targeted by microRNAs and a smaller number targeted by Drosha, there were nonetheless a large number of mRNAs that undergo cleavage by other endonucleases that were not identified in those studies. By starting with poly(A) RNA both papers selectively enriched for mRNAs whose decay is at least in part independent of deadenylation, and in doing so show that much of the transcriptome is subject to endonuclease-mediated decay. Finally, the lack of sequence specificity for most mRNA endonucleases makes it likely that their action is heavily influenced by context of their associated proteins and sequence or structural determinants within substrate mRNAs.
Figure 1.
While there have been many reviews covering the overall field of mRNA decay this is the first to focus exclusively on endonuclease-mediated mRNA decay. Because this is still an emerging field I have tried to be as comprehensive as possible, both to inform and to provide perspectives to help guide future work in this area. There are three main sections to this review. The first is a historical perspective of endonuclease-mediated mRNA decay and the second describes the known mRNA endonucleases in some detail. I close with a summary of the major unanswered questions and some perspectives of where this field may be going in the future.
Historical evidence for endonuclease-mediated mRNA decay
The first reports describing endonuclease cleavage appeared in the literature around the same time as the early observations linking deadenylation to mRNA decay [4], making endonuclease decay among the earliest known decay processes. Ironically, only one of the enzymes involved in these early studies has been identified (see Ape1). For most of the following the site of endonuclease cleavage lies in a highly structured region. While these structures may set up the context for cleavage it seems likely that products were detected because these structures inhibit exonuclease degradation of the primary cleavage products.
Apo-very low density lipoprotein II mRNA
The earliest evidence for vertebrate endonuclease decay came from work studying apo-very low density lipoprotein II (apo-II) mRNA in avian liver. In male birds estrogen induces the transcription of apo- II and the yolk protein precursor vitellogenin, and in the continued presence of estrogen the half-lives of both mRNAs are on the order of 15–24 hr [5, 6]. For these studies estrogen was implanted in a pellet, and when it was removed the authors observed a rapid loss of each of these mRNAs, and calculated that their half-lives decreased 7-fold. The rapid decay of apo- II mRNA was independent of the length of the poly(A) tail, and two groups identified endonuclease cleavage within the 3′-UTR as the activating event [7, 8]. The relatively small size of apo-II mRNA enabled determination of its entire secondary structure [9, 10], which in turn made it possible to map endonuclease cleavage with single nucleotide resolution to several sites within the loop of a stable stem-loop structure in the 3′-UTR [7]. Proteins binding to different regions of the 3′-UTR also influenced the decay process [11], but unfortunately this work was not continued and the identities of these proteins and the estrogen-regulated endonuclease remain unknown.
Hoxb7 (Xlhbox2b) mRNA
The stability of both endogenous and injected mRNAs in Xenopus oocytes is one of the reasons they were used extensively for early studies in gene expression. An exception to this is Xenopus Hoxb7 (originally termed Xlhbox2) mRNA. Like most transcripts in Xenopus laevis Hoxb7 is encoded by two genes, one producing a stable mRNA (Xlhbox2A mRNA), and a second bearing an additional 123 nt in the 3′-UTR. This mRNA is lost as oocytes progress from the previtellogenic stage to later stages in development. Xlhbox2B mRNA is cleaved here within 30 min after injection, generating an uncapped 3′ product that was less stable than the capped 5′ product [12]. The authors mapped the site of endonuclease cleavage to a repeating series of ACCTACCTA elements within a 90 nt region, and this region was sufficient to confer sensitivity to endonuclease cleavage when placed in the context of another mRNA. Endonuclease cleavage was independent of translation and could be blocked by co-injection of an RNA antisense to the cleavage site, thus implicating a requirement for a single-stranded target. While it is tempting to consider this as a target for microRNA silencing a bioinformatics search failed to identify sites for known microRNAs. This is supported by subsequent biochemical studies in which endonuclease cleavage was replicated in vitro using Xenopus oocyte extracts [13]. Those experiments also identified an endogenous inhibitor that binds over the endonuclease cleavage site, providing the first evidence for stabilization by such a mechanism. The sequence requirements for binding by the protective factor differed from those required for endonuclease cleavage, suggesting a role for secondary and/or tertiary structural features. The amount of protective factor decreased during oocyte development, raising the possibility that this factor functions as part of a regulatory mechanism controlling the level of maternal Hoxb7 mRNA. Moreover this appears to be conserved as a similar endonuclease activity was also identified in Drosophila embryo extracts. The authors of that study named this enzyme xixernase, and showed it eluted from a gel filtration column at 123 kDa. Xixernase was never purified to the point where it could be identified and to this date both the endonuclease and the protective factor remain unknown.
9E3 (interleukin 8) mRNA
In chickens the gene for IL8 was originally called 9E3. Treating chick embryo fibroblasts with high levels of serum or infection with Rous Sarcoma Virus increased the transcription of the 9E3/IL8 gene and stabilized 9E3/IL8 mRNA, increasing its half-life from 1–2 hr in control to 8 hr in serum-stimulated or RSV infected cells. Northern blots of RNA from untreated cells revealed two smaller species, the sizes of which added up to that of the parent mRNA [14]. Primer extension and S1 protection showed that the 3′ end of the upstream fragment and the 5′ end of the downstream fragment corresponded to cleavage of a single phosphodiester bond, thus confirming that these were endonuclease cleavage products. The 3′ fragment retained its poly(A) tail, and in keeping with their production during the decay process the relative amount of each of the decay products correlated inversely with the amount and stability of 9E3 mRNA. Treating cells with cycloheximide increased the amount of each of the decay products, a result the authors of that study attributed to their stabilization. In light of our current understanding of mRNA decay it seems more likely that freezing ribosomes in 9E3/IL8 mRNA increased the opportunity for cleavage by a polysome-bound endonuclease. As with most of the mRNAs in this section no further work was done on 9E3/IL8 mRNA decay and the endonuclease has yet to be identified. Of note, the ability to identify stable fragments upstream and downstream of endonuclease cleavage without inactivating Xrn1 or the exosome (neither of which were known at the time) suggests there is something unusual about 9E3/IL8 mRNA.
Insulin-like growth factor II mRNA
Insulin-like growth factor II (IGF-II) is a key regulator of embryonic development encoded by multiple transcripts arising from alternative promoter use and alternative splicing. The various forms of IGF-II mRNA appear in different tissues at different times in development, but Meinsma et al. [15] and Nielsen and Christiansen [16] also identified a stable, 1.8 kb truncated form of IGF-II mRNA that consists solely of the 3′ terminus of the transcript. Both groups presented in vivo evidence that the 1.8 kb product was generated by endonuclease cleavage, and the latter group also showed this could be generated in vitro. IGF-II mRNA is unique in that endonuclease decay depends on specific interacting secondary (and likely tertiary) structural elements that are separated by almost 2 kb and lie some distance from the cleavage site [17]. It is the interaction of these elements and a G-quadruplex formed as part of this larger structure that establishes the structural context for endonuclease cleavage after G2183 in an unpaired GA dinucleotide in the 3′-UTR [18–20].
Growth conditions influence the rate of IGF-II mRNA decay, with serum deprivation stabilizing IGF-II mRNA and addition of serum, IGF-I or IGF-II to serum-starved cells activating decay via endonucleolytic cleavage [21]. Endonuclease cleavage occurs in the cytoplasm, and the level of IGF-II mRNA correlates inversely with the quantity of a 50 kDa cytoplasmic protein(s) (now known as IMPs) that binds in vivo and in vitro to the extended secondary structure formed by the widely-separated elements. [19, 22]. Recent work using embryo fibroblasts from mice knocked out for G3BP suggest this protein might be involved in IGF-II mRNA decay (see below), but is not likely the endonuclease that is responsible for the characteristic cleavage event.
Transferrin receptor mRNA
Post-transcriptional processes that regulate the translation of ferritin, the main iron storage protein, and transferrin receptor (TfR), the primary iron transporter, control iron levels within the cell. Ferritin mRNA has a single stem-loop iron responsive element (IRE) in its 5′-UTR and TfR mRNA has 5 copies of the IRE in its 3′-UTR, and under conditions of low iron each of these is bound by the IRE-binding proteins IRP1 and 2 [23]. This results in reduced translation of ferritin mRNA and the stabilization of TfR mRNA, from ~45 min when iron is in excess to ~ 3 hr under conditions of low iron. TfR mRNA disappears rapidly when extracellular iron is elevated and Binder et al. [24] showed that its rapid loss in cells treated with hemin (ferric protoporphyrin IX) was accompanied by the appearance of a smaller form of TfR mRNA, the kinetics of which were consistent with it being a degradation intermediate. This shorter RNA lacked poly(A), and detailed mapping showed it corresponds to the body of the mRNA upstream of the fourth IRE in the TfR 3′-UTR, ending in a sequence that is required for iron-induced destabilization [25]. Detailed analysis of the cleavage site and its surrounding sequences mapped the cleavage site with single nucleotide resolution to the scissile phosphate between G3727 and A3728. This lies just 3′ to the third IRE in a region shown previously to be single stranded. There is selectivity to the cleavage process, and cleavage was lost with changes to the sequence immediately surrounding the cleavage site.
It is rather surprising that the RNA upstream of the cleavage site was not rapidly degraded by the exosome. The most likely explanation is that this region is highly structured like apo-II and IGF-II mRNA, and this structure inhibited exonuclease decay. Since sequences downstream of endonuclease cleavage are commonly seen only after knockdown of Xrn1 [26–28] it is surprising that the authors of this study were able to also identify and characterize this fragment. Importantly, they showed that the 5′ end corresponded to the site of endonuclease cleavage and the downstream fragment retained the same length poly(A) as the parent mRNA. Finally, this paper presented convincing results that, like apo-VLDL-II and IGF-II mRNA, endonuclease cleavage is either facilitated or positioned by large secondary (and presumably tertiary) structural elements, and that stabilization results from binding of IRP1 to the IREs that surround the endonuclease cleavage site.
It is unfortunate that work did not progress beyond this paper as it left many important questions unanswered, foremost among these being the identity of the endonuclease. Given the rapidity of the response this is likely a pre-existing enzyme rather than one that is induced when cells encounter excess iron. Is this enzyme in a latent form, or is it always active but complexed with something else that prevents its action on other mRNAs? It seems unlikely that TfR mRNA is the only substrate for the endonuclease. What other mRNAs are targets of this enzyme? Is the enzyme pre-positioned on TfR mRNA and only active when one or more of the IREs are no longer occupied by IRP1? How many molecules of bound IRP1 are required to block endonuclease cleavage? Why are the decay products so stable? Hopefully this interesting story will be revisited using molecular tools that were not available at the time this work was done.
α-globin mRNA and the erythroid-enriched endonuclease (ErEN)
α-globin is among the most stable mRNAs known and the the 3′-UTR sequence and structural features responsible have been extensively characterized [29]. The core is the α-complex, which includes the poly(C) binding proteins αCP1 and αCP2 (also called hnRNP E). Wang and Kiledjian [30] found that two fragments were generated when a unadenylated α-globin 3′-UTR transcript was incubated in vitro. This appeared to be stabilized by the α-complex as the production of these fragments was enhanced by addition of oligo(dC) to the reaction. Experiments with end-labeled RNAs confirmed an endonuclease was responsible for generating the decay fragments and the authors mapped cleavage to a site 63 nt downstream of the stop codon in a region shown previously to be protected by the α-complex [31]. These in vitro results were confirmed in vivo using transfected erythroid cells, and based on the greater activity detected in extracts from erythroid versus non-erythroid cells this enzyme was named erythroid enriched endonuclease (ErEN). ErEN is selective for α-globin mRNA, with β-globin mRNA targeted by a different endonuclease decay process [32]. Like TfR mRNA, the ability of ErEN to cleave α-globin 3′-UTR was abolished by mutating residues surrounding the cleavage site.
The overall picture is analogous to TfR mRNA decay, with binding of the α-complex occluding the ErEN cleavage site. As with the case of TfR mRNA many questions remain to be answered, key among these being the identity of ErEN. Because there is not a straightforward way to predict which proteins will function as mRNA endonucleases the identity of ErEN will likely require either its purification or an RNAi screen of erythroid cells to identify this interesting enzyme and study the interplay between its function and the α-complex, and determine the overall scope of its action in catalyzing endonuclease decay.
p27KIP1 mRNA
p27KIP1 regulates cyclin-dependent protein kinases, and the 5′-UTR of this mRNA contains an GU-rich binding site for HuR, a protein that functions in stabilizing mRNAs containing AU-rich elements. Zhao et al. [33] identified an endonuclease activity in HeLa cytoplasmic extract that cleaves within the HuR binding site to generate smaller product. Experiments with end-labeled transcripts confirmed this activity as an endonuclease, and showed it cleaves within the sequence GUUUUG to generate products with 3′-hydroxl and a 5′-phosphate ends. Adding recombinant HuR to the reaction reduced cleavage of p27KIP1 5′-UTR transcript in a concentration-dependent manner, but this endonuclease has not been purified and without demonstrating in vivo activity for this in conjunction with HuR binding in regulating p27KIP1 mRNA its relevance to the decay process has yet to be determined. This is one of a handful of reports of an mRNA endonuclease cleaving within the 5′-UTR.
Identified endonucleases
Polysomal ribonuclease 1 (PMR1)
As noted above estrogen activates the transcription of a number of genes in the liver of oviparous vertebrates (eg. Xenopus, chickens) but at the same time mRNAs for serum proteins disappear [34–36]. While this was initially thought to result from transcriptional repression a comparison of changes in steady state levels of albumin mRNA with albumin gene transcription showed instead that estrogen activated mRNA decay [34] in a process that involves the classical estrogen receptor ERα [37]. While the suppression of these mRNAs could have been catalyzed by deadenylation, decapping and exonuclease decay, estrogen had no impact on poly(A) tail length of a number of target mRNAs [36, 38]. This led to experiments looking for an estrogen-induced activity that might be responsible for the selective loss of these mRNAs. Cytoplasmic extracts from livers of control or estrogen-treated frogs were devoid of activity, but a salt-extractable ribonuclease that is resistant to RNase inhibitors was recovered from polysomes from estrogen-treated frogs [39]. The production in vitro of a discrete cleavage product using 5′- or 3′-end labeled transcripts of albumin mRNA provided the first indication that this was an endonuclease. Subsequent work confirmed that the estrogen stimulated ribonuclease activity sedimented with polysomes or some form of polysome-bound mRNP [40].
Dompenciel et al. [41] used the production of this cleavage product to track the purification of the polysome-associated endonuclease from livers of estrogen-treated frogs, ultimately recovering a protein doublet that ran at 60 kDa on SDS-PAGE. Separation of the doublet into multiple spots on 2D gels and a shift after phosphatase treatment indicated that some of the protein recovered from the final column was phosphorylated. When assayed for its ability to degrade mRNA in vitro the purified protein displayed substrate selectivity similar to the in vivo decay process, with the purified enzyme degrading albumin mRNA more rapidly than ferritin mRNA. As the first ribonuclease to be purified from polysomes it was named Polysomal Ribonuclease 1 (PMR1).
Secondary structure mapping and site-directed mutagenesis of the region surrounding the main cleavage site on albumin mRNA showed that the doublet product of albumin mRNA result from cleavage between UG dinucleotide of two overlapping repeats of AU/CUGA that comprise the loop of a stem-loop structural element [42]. Mutating either of these elements had no impact on the susceptibility of the other repeat as long as it was single-stranded, whereas collapsing the loop into an extended stem structure generated an RNA that was resistant to cleavage by the purified endonuclease. Thus, like the enzymes described in the first section of this review PMR1 selectively cleaves only certain sequences and these have to be in an appropriate context. The applicability of these findings to decay in vivo was demonstrated using a sensitive ligation-mediated PCR assay to capture the 3′ ends of decay intermediates present in cytoplasmic RNA from livers of estrogen-treated frogs [43]. In addition to studying albumin mRNA decay the same study also looked at the decay of c-myc mRNA and in doing so provided the first in vivo demonstration of endonuclease cleavage within the coding region determinant (CRD) that was predicted from in vitro work of Ross and coworkers [44].
Using activity assays and antibodies to purified PMR1 Cunningham et al. [45] showed that a portion of the enzyme sediments with polysomes even without estrogen treatment, and both this and the population of PMR1 sedimenting further up the gradient had little enzymatic activity. After estrogen the polysome-bound population underwent a 21-fold increase in unit activity but there was no change in the activity of enzyme recovered from lighter sucrose gradient fractions. The basis for the selective activation of this population of endonuclease is still unknown. Estrogen also stabilizes the large (6 kb) mRNA encoding the yolk protein precursor vitellogenin, resulting in a half-life that is so long it essentially cannot be measured [46]. A key aspect of this is the induction of vigilin [47], a 14 KH-domain protein, which binds to a portion of the vitellogenin 3′-UTR that has overlapping PMR1 cleavage sites. Vigilin binding blocks access to these sites by estrogen-activated PMR1 [48], thus stabilizing vitellogenin mRNA. While this was not the first paper to conceptualize the idea that an RNA-binding protein might stabilize mRNA by occluding an endonuclease cleavage site, it was the first to provide experimental proof of this now well-accepted notion (eg. see [49]).
Sequencing of tryptic peptides of purified PMR1 revealed an unexpected similarity to the major human peroxidases (eg. myeloperoxidase, lactoperoxidase, eosinophil peroxidase). These guided the cloning of PMR1 cDNA from a Xenopus liver cDNA library, the sequence of which confirmed PMR1 is a member of the peroxidase gene family that lacks both heme and N-linked glycosylation [50]. This protein remained an outlier until the first assembly of the Xenopus genome, where it became apparent that Xenopus PMR1 is a product of the gene for eosinophil peroxidase moonlighting as a ribonuclease. In general each of the peroxidases function as a heterotetramer with 2 heavy and 2 light chains, that are generated by proteolytic processing of a single ~80 kDa precursor protein. This larger precursor was seen by Western blotting of liver extract with antibodies to purified PMR1, and it remains to be determined if PMR1 is generated by alternative splicing or alternative processing of the eosinophil peroxidase precursor. We have determined that human PMR1 is not a product of the same gene but instead comes from one of the poorly characterized peroxidase-like genes, with evidence of exon shuffling responsible for imparting some of the critical features of PMR1 to this closely related gene (unpublished).
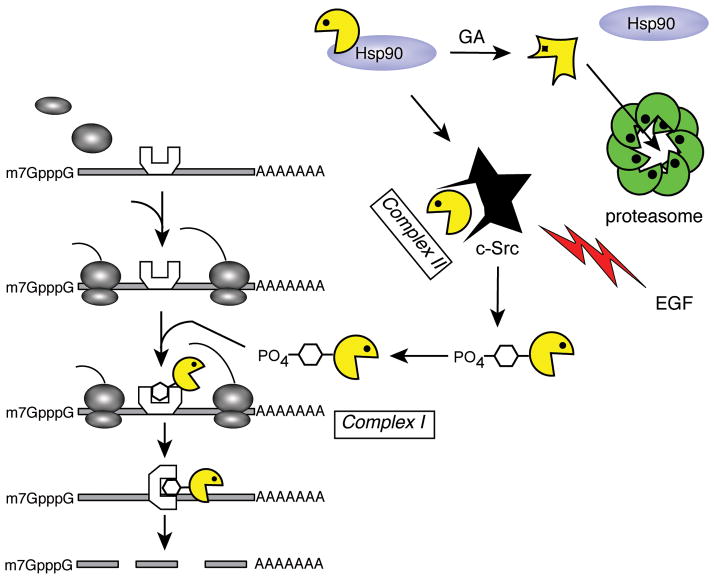
The development of cell culture models that replicated aspects of the decay process observed in cultured hepatocytes made it possible to dissect what has turned out to be complex mechanisms of PMR1 mRNA decay. In polysome profiles PMR1 sediments with polysomes and in lighter fractions of the gradient. Treating cells with puromycin prior to lysis or adding EDTA to cytoplasmic extracts released the polysome-bound protein in a complex (termed Complex I, see Fig. 2) that sediments at ~680 kDa on glycerol gradients. The ~680 kDa Complex I also contains PMR1 substrate mRNA, indicating the joining of PMR1 to this complex is the basis for substrate selectivity [51]. The population of PMR1 found in the lighter sucrose gradient fractions (termed Complex II) sediments at ~140 kDa on glycerol gradients [50] and contains PMR1 with its activating kinase (see below).
Figure 2.
PMR1 mRNA decay also requires active translation of its substrate mRNA, with decay inhibited by inserting a strong stem-loop into the 5′-UTR (Fig. 2). Deletion mutagenesis identified two domains that were critical for both targeting to polysomes and mRNA decay [51]. One lies 100–150 amino acids from the N-terminus and the other lies 50–100 amino acids from the C-terminus (see below). Deleting either of these prevented PMR1 from joining Complex I and stabilized target mRNA, indicating that decay required targeting of this endonuclease to an RNP containing its translating substrate mRNA. A motif scan identified a consensus tyrosine phosphorylation site within the C-terminal polysome targeting domain, a result consistent with the multiple spots seen on 2D gels. Western blotting of protein from Xenopus liver and transfected cells with several monoclonal antibodies phosphotyrosine (pTyr) confirmed that PMR1 is tyrosine phosphorylated in [52], and localized phosphorylation to the second of two tyrosines in the sequence FYYEQP at the core of the pTyr site. Changing this to phenylalanine blocked PMR1 binding to polysomes, its incorporation into Complex I and mRNA decay. Further proof of a requirement for tyrosine phosphorylation came from experiments with a general tyrosine kinase inhibitor, which reduced tyrosine phosphorylation of PMR1 and inhibited PMR1 mRNA decay. This finding was the first evidence linking tyrosine kinases and their associated signaling pathways to mRNA decay.
The identification of the responsible kinase was facilitated by two properties of this class of enzymes; they commonly form a complex with their substrates and they undergo autophosphorylation. Three proteins were labeled when immunoprecipitated PMR1 was incubated with γ32-P-ATP; 60 kDA PMR1, a 58 kDa protein and a 90 kDa protein. The size of the 58 kDa protein quickly narrowed the field to c-Src and the 90 kDa protein was identified as Hsp90. Experiments using geldanamycin (GA), a selective Hsp90 inhibitor, showed that PMR1 is inherently unstable and requires Hsp90 for proper folding and to prevent its degradation by the 26S proteasome (Fig. 2, upper right) [53]. Proof that c-Src was the activating kinase came from experiments using Src inhibitors and Src-deficient cells transfected with active and inactive forms of this kinase [54]. In the process these experiments also showed that the N-terminal polysome-targeting domain identified in [51] contained 2 PPXXP motifs that are bound by the c-Src SH3 domain and are responsible for joining these proteins together in the lighter Complex II. Growth factors commonly activate tyrosine kinase cascades and this was confirmed in [54] using EGF to activate c-Src phosphorylation of PMR1.
A proteomics screen using tandem affinity (TAP)-tagged PMR1 unexpectedly identified a number of actin cytoskeleton-associated proteins, key among these being the mammalian Enabled protein (Mena) and the related vasodilator stimulated phosphoprotein (VASP) [55]. Mena and VASP are multifunctional proteins that contain an N-terminal (EVH1) domain that binds to zyxin and vinculin, a C-terminal (EVH2) domain that binds to actin monomers and a central proline-rich domain that interacts with a number of signaling proteins including c-Src. The EVH1 domain binds FPPPP repeats, and in vivo interaction of PMR1 with Mena and VASP was confirmed by immunofluorescence experiments using decoy proteins containing FPPPP repeats coupled to EGFP and a mitochondrial targeting sequence. Reciprocal immunoprecipitations recovered PMR1, c-Src and Mena/VASP in a single complex, indicating that Mena/VASP likely serve as a scaffold to bring PMR1 together with its activating kinase. The recovery of Mena/VASP with PMR1 in glycerol gradient fractions of Complex I and Complex II and its co-localization with PMR1 at the leading edge of motile cells raised the possibility that the interaction with Mena/VASP also functions to localize decay to this portion of the cell. Evidence for this was seen in cell motility experiments, where expression of an active form of PMR1 stimulated motility 2-fold compared to cells expressing an inactive form or cells that were not expressing any recombinant protein. This unexpected link to cell motility suggests that some of the targets of PMR1 mRNA decay are involved in this process.
Remaining questions
Because much of the work on PMR1 used the frog protein expressed in mammalian cells the generality of this process remains to be demonstrated. Given the high degree of sequence relatedness and similar biochemical properties between the Xenopus enzyme and the presumed mammalian ortholog it seems likely this will behave similarly.
A transcriptome-wide analysis of the substrates of PMR1 mRNA decay is in progress and a proteomics analysis of the proteins of Complex I will establish the context for a particular mRNP to host this particular mechanism of mRNA decay. Since PMR1 is responsive to a number of signal transduction processes (eg. Hsp90, actin cytoskeleton, growth factors, tyrosine kinases, estrogen) it should be thought of as a post-transcriptional integrator of these processes. The link to cell motility becomes particularly interesting in this regard as it may link mRNA decay to the invasive growth of cancer cells. In stressed cells PMR1 is recruited to stress granules through direct interaction with TIA-1 [56], and this too should be investigated for its links to cell recovery from stress and cancer. The gene for the presumed mammalian PMR1 has not been knocked out, and the phenotype from this will be interesting, as will studying the impact of knocking down proteins with which it associates in the functional mRNP. Either PMR1 or a closely related protein catalyzes endonuclease cleavage of nonsense-containing β-globin mRNA in erythroid (but not in non-erythroid) cells [57]. Since this process is clearly different than the mechanisms catalyzing NMD (see SMG6 below) it will be important to learn why this happens in the context of this particular cell type and how the signal from a premature termination is codon transduced to the polysome-bound endonuclease to activate the decay process? Finally, by putting knowledge of the proteins of the polysome-bound PMR1 mRNP together with the sequence or structural determinants of its target mRNAs much will be learned about the molecular code used by cells to handle specific mRNAs or classes of mRNAs.
RasGAP SH3 binding protein (G3BP)
G3BP was originally identified as a protein that binds to RasGAP in a Ras-GTP-dependent complex. As a protein involved in signal transduction it is unusual in having two C-terminal RNA-binding motifs, an RRM domain and a downstream RGG domain. Upstream of this is a PXXP motif that serves as a ligand for SH3 domain binding. Based on similarity of the RRM to that of hnRNP C, Gallouzi et al. [58] looked at binding of G3BP expressed in baculovirus with the c-myc 3′-UTR. They unexpectedly saw a series of smaller bands that appeared in a time and temperature-dependent manner, suggestive of an endonuclease decay process. G3BP is phosphorylated on serine residues, and phosphorylation activated the catalytic activity of recombinant G3BP. In vivo support for this notion came from experiments showing significantly greater activity against the c-myc 3′-UTR of immunoprecipitated G3BP from quiescent versus dividing cells, conditions where differences in its phosphorylation had already been characterized. Given that activity was lost following phosphatase treatment these data provided evidence that phosphorylation of G3BP in quiescent cells activated endonuclease activity.
A second study from the same group presented a detailed analysis of the cleavage properties of G3BP and its relationship to growth factor stimulation. Tourriere et al. [59] showed that G3BP is quite specific in its action, cleaving between CA dinucleotides to generate an upstream product with a 3′-phosphosphate end and a downstream product with a 5′-hydroxy. This is similar to the mechanism of RNase A, and while these authors did not look for a cyclic 2′,3′ intermediate, this is likely to be the case. In vitro the recombinant protein cleaved at multiple sites within the c-myc 3′-UTR, with some cleavages stronger than others. Unexpectedly they found that immobilized G3BP lost enzymatic activity but not RNA binding activity, and using this for SELEX identified 5′-ACCCAUACGCAG-3′ as a consensus target sequence. Because this sequence is a substrate in vitro for G3BP the authors conclude that binding to this sequence juxtaposes the active site of G3BP with the scissile phosphate of the CA dinucleotide, but that remains to be determined.
G3BP is phosphorylated following serum stimulation of wild-type cells or cells heterozygous for RasGAP (RasGAP+/− cells), and while phosphorylation has little impact on the recovery of c-myc mRNA with immunoprecipitated G3BP, it has a significant impact on the turnover of c-myc mRNA, with this decaying with a half-life of 35 min in RasGAP−/− cells versus 15 min in cells expressing RasGAP. This is the first example of a post-translational modification altering the actual enzymatic activity of an mRNA endonuclease, but likely not the last. Phosphorylation also affects the subcellular distribution of G3BP, with a portion of the protein appearing in the nucleus when phosphorylated at serine 149.
Experiments with knockout mice support a role for G3BP as an integrator of signaling processes involved in cell proliferation. Loss of both copies of G3BP was lethal to most embryos, and those surviving to the neonatal stage showed extensive cell death in the central nervous system [60]. Fibroblasts from G3BP−/− embryos grow more slowly than matching cells from wild-type mice and did not show growth factor stimulation of cell growth. Microarray analysis of the transcriptome of these cells identified 5 genes whose transcripts were induced by >4-fold by loss of G3BP, the most notable of which was IGF-II (see above). IGF-II mRNA has 2 copies of the consensus G3BP binding sequence but neither of these lie in the highly structured 3′-UTR that is responsible for establishing the context for endonuclease cleavage described above. Nevertheless, the steady-state level of IGF-II mRNA in G3BP−/− fibroblasts was ~3.5-fold higher than in wild-type controls, IGF-II mRNA was recovered with G3BP from wild-type cells and by actinomycin D chase IGF-II mRNA was more stable in G3BP−/− MEFs than in wild-type MEFs. Similar results were obtained for GAS5, a non-coding RNA that was also up-regulated in G3BP knockout fibroblasts, and both this and a transcript for IGF-II 3′-UTR were rapidly degraded in vitro. Because the experiment was performed with uniformly labeled RNA it was not possible to discern the location of G3BP cleavage sites in either of these transcripts. However, in the discussion of Zekri et al. [60] the authors note that the 1.8 kb product generated by endonuclease cleavage at G2183 in the IGF-II mRNA 3′-UTR was still observed in G3BP−/− MEFs, indicating that G3BP is not the enzyme responsible for this.
While it is not directly germane to a review on endonuclease decay, G3BP is also a dominant effector of stress granule assembly [61]. Much like the TIA proteins endogenous G3BP is recruited to stress granules in arsenite-treated cells, but its overexpression also stimulates stress granule assembly through a process that involves phosphorylation at serine 149, which becomes partially dephosphorylated after arsenite stress. In keeping with a phosphomimetic (S149E) form of G3BP inhibits stress granule assembly.
Remaining questions
There is much yet to learn about the function of G3BP in endonuclease-mediated mRNA decay. Its endonuclease activity was originally linked to c-myc mRNA decay, but c-myc is not among the mRNAs that show significant increase in G3BP−/− MEFs and at this point it is unclear whether G3BP is involved in controlling c-myc in vivo. One possibility is that G3BP works in concert with APE1 (see below), but this remains to be determined. While G3BP binds IGF-II mRNA, it’s absence in G3BP−/− MEFs has no impact on endonuclease cleavage at the site described in [16, 17], which is a GA dinucleotide, not the CA site cleaved by G3BP. Given the multiple roles G3BP plays in signal transduction and stress granule assembly it is possible that G3BP has different substrates depending on signaling by different transduction processes. As with most of the enzymes described here an overriding question is how it finds its substrate mRNAs. Given that a portion of G3BP undergoes phosphorylation-dependent accumulation in the nucleus one scenario is that it forms an association with its targets in the nucleus and accompanies these into the cytoplasm where it is positioned to catalyze the decay of specific mRNAs in response to growth factor signaling. There is also the question of the ends of the decay products. Because G3BP generates products with upstream 3′-phosphate and downstream 5′-hydroxyl ends it is difficult to see how the initial products are cleared. Given this it is conceivable that the G3BP may not actually initiate decay but instead might act secondarily in the decay process.
IRE1
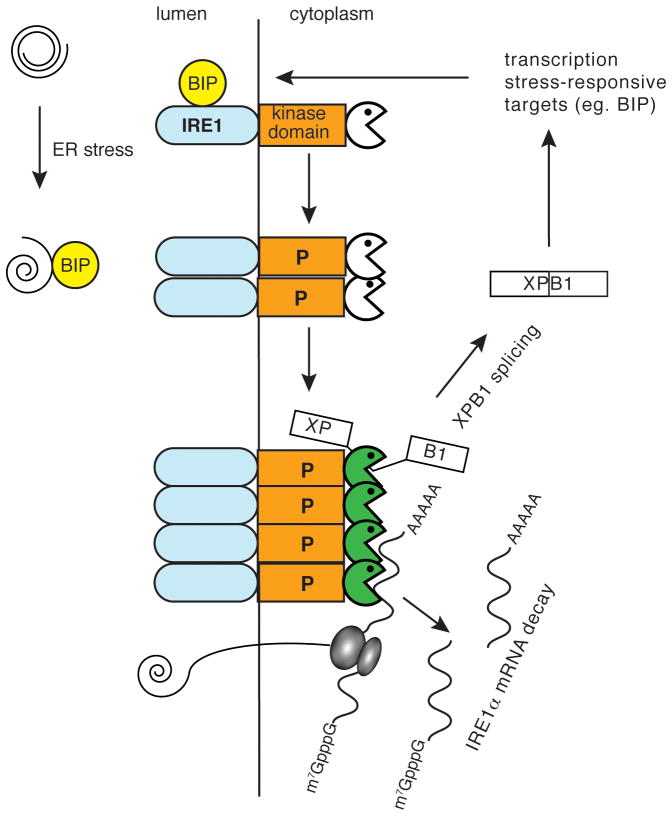
The endoplasmic reticulum in eukaryotic cells functions in the folding, post-translational modification (eg. glycoslylation) and assembly of secreted and membrane-bound proteins prior to their delivery to the plasma membrane or an intracellular organelle. Stresses that disrupt this process lead to the accumulation of misfolded or unfolded proteins, which in turn activate the unfolded protein response, or UPR, which functions to help cells adapt to these new conditions, restore the normal pattern of protein folding or induce apoptosis under cases of prolonged stress. Because UPR is the subject of an advanced review in this series only those points that are relevant to understanding its relationship to endonuclease decay are presented here and the reader is advised to look there for a more detailed treatment of this subject. Three proteins serve as major sensors for ER stress; IRE1, PERK and ATF6 (reviewed in [62]). Two of these are serine/threonine kinases (IRE1, PERK), one is a transcription factor (ATF6) and one of these (IRE1) is also a stress-induced endonuclease. IRE1 is a transmembrane protein and one of its major roles in ER stress is to catalyze the noncanoical splicing of mRNAs for stress-responsive transcription factors (HAC1 in yeast and XBP1 in mammals). In mammals this activates transcription of a number of genes including BIP, the major ER chaperone protein (Fig. 3).
Figure 3.
The following section focuses on stress response in metazoans because IRE1 does not function in yeast mRNA decay. The multiple domains of IRE1 are diagrammed in Fig. 3. Mammals have 2 paralogs, IRE1α which is expressed ubiquitously and IRE1β which is limited to intestinal epithelium. The N-terminal half of the protein lies in the ER lumen where it binds BIP and functions as the sensor for unfolded proteins, and the kinase and RNase domains are on the cytoplasmic face of the ER membrane. Under conditions of ER stress BIP is released from its complex with IRE1, which in turn activates trans-autophosphorylation, oligomerization of IRE1 and activation of ribonuclease activity [63]. One study also provided evidence that IRE1α cleaves its own mRNA [64], with shortened forms of IRE1α mRNA detected in cells expressing catalytically-active forms of this protein. However, the endonuclease cleavage site was not mapped, and IRE1α did not appear in subsequent studies using microarrays to identify its targets, leaving open the question of its autoregulation.
Direct evidence that IRE1α is an mRNA endonuclease came from studies using microarrays to compare the impact of knocking down IRE1 versus XBP1, ATF6 or PERK on the ER stress response of Drosophila S2 cells [65]. A cluster of transcripts for genes encoding secreted or plasma membrane-bound proteins were repressed by IRE1 but not by the others, and experiments using Actinomycin D, DRB (5,6, dichloro-1-βD-ribofuranosylbenzimidazole), and a tetracycline-regulated promoter confirmed that the effect was on mRNA decay. More relevant to the current review, these authors showed that knocking down Xrn1 resulted in the IRE1- and stress-dependent appearance of 3′ fragments of several of these mRNAs, and knocking down Ski2 led to the appearance of complementary 5′ fragments. In keeping with the nature of the targets these authors also showed that the N-terminal signal sequence is the key feature targeting these mRNAs to IRE1-catalyzed endonuclease decay.
Two reports that appeared in 2009 came to similar conclusions regarding the role of IRE1α in ER stress-induced mRNA decay and XBP-1 splicing in mammalian cells [66, 67]. Both of these studies used microarrays to identify the targets as mRNAs whose protein products transit through the ER, with one using IRE1α-deficient MEFs (IRE1−/− cells) into which the authors reintroduced wild-type and mutant forms of the protein by flippase-mediated recombination [67] and the other using a tetracycline-inducible insulin-producing line of pancreatic islet cells [66] into which they introduced various forms of IRE1α. In addition to using RNase-inactive forms of the enzyme both of these studies looked at the relationship between kinase activation and RNase activity using mutations at I642 that increase the size of the ATP pocket, creating forms of the protein that show reduced autophosphorylation. This alteration in the ATP binding pocket enables binding an ATP analog (4-amino-1-tert-butyl-3-[1′-naphthylmethyl]pyrazolo[3,4-d] pyrimidine, or 1NM-PP1), which causes an allosteric change that yields IRE1α lacking kinase activity (and consequent autophosphorylation) that nonetheless has RNase activity when cells are subjected to ER stress. Importantly, both studies showed that activation of XBP-1 splicing and mRNA decay are separable processes, and results in [66] also linked IRE1α mRNA decay to apoptosis, suggesting this is part of a feed-forward loop set up by chronic ER stress.
Remaining questions
mRNA decay by IREα is important from a therapeutic and physiological point of view as the targeting of mRNAs whose products transit through the ER may facilitate a rapid response to ER stress or in situations of chronic stress help push cells toward apoptosis. Thus the work in [66] with various kinase inhibitors might lead to the development of new treatments for diseases caused by ER stress. From the point of view of mRNA decay these findings are important as they provide a direct link between translation, in this case the signal sequence, and endonuclease decay and are among the few convincing examples for a molecular code (ie. the signal sequence) that determines the targeting of specific mRNAs to endonuclease decay. Little is known about the generality of the cleavage process associated with IRE1-mediated mRNA decay; however, a recent report identified a consensus cleavage site witihin XBP1, CD59 and 13 other mRNAs [68] between the UG dinucleotides in CUGCAG in an unpaired loop of a stem-loop structure. It is unclear at present whether is a manifestation of its processing activity or is involved in the decay process, but Chernokalskaya et al. [42] showed that PMR1 also cleaves between UG dinucleotides within a consensus element (APyrUGA) in an unpaired region of a stem-loop structure.
Zc3h12a (MCPIP)
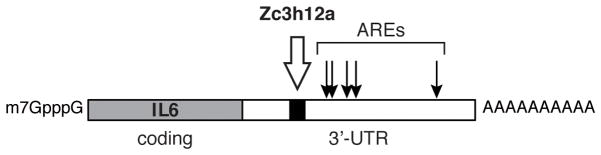
Treating macrophages with interleukin 1β (IL-1β), monocyte chemoattractant protein 1 (MCP-1), TNFα, phorbol 12-myristate (PMA) or bacterial lipopolysaccharide (LPS) induces expression of Zc3h12a, a protein that was originally thought to be a transcription factor called monocyte chemoattractant protein inducible protein (MCPIP) [69]. Zc2h12a has a single CCCH zinc finger and a PIN domain, and is one of a small family of genes that includes the similar proteins Zc3h12b, Zc3h12c and Zc3h12d, each of which is on different chromosomes but share common features. While levels of Zc3h12a mRNA are highest in lymphoid cells its expression is not limited to these as it can be induced by IL-1β in HepG2 cells and it is seen in heart and placenta [70]. Mice knocked out for Zc3h12a are anemic, showed extensive plasma cell infiltration and inflammatory changes in the lung, bile duct and pancreas and die within 3 months after birth. Zc3h12a is induced by Toll-like receptor (TLR) signaling and there was no change in the initial TLR signaling process following LPS treatment of macrophages from these mice, but interleukin 6 (IL-6) and calcitonin receptor (Calcr) mRNA were overinduced, indicating Zc3h12a might be involved in resolving the response to LPS. This was confirmed using hybrid luciferase reporters, which mapped Zc3h12a responsive sequences to the 3′-UTRs of IL-6 and Calcr mRNAs (Fig. 4). Each of these has a number of AU-rich elements, but both reports describing RNase activity of Zc3h12a showed it is a region upstream of the AREs that targets these mRNAs to Zc3h12a-mediated decay [69, 70]. Tristetraprolin (TTP, Zfp36) and related ARE-binding proteins also have CCCH zinc fingers and function to destabilize cytokine mRNAs, but their mechanism of action is quite different, activating instead the deadenylation-dependent exonuclease-catalyzed decay machinery. Interestingly, TNFα mRNA, one of the canonical targets of TTP, is not a substrate in vivo for Zc3h12a, and Zc3h12a appears to function in a feedback loop, targeting the decay of its inducer IL-1β mRNA.
Figure 4.
Remaining questions
Much remains to be learned about the mechanism of action of Zc3h12a. As might be expected from the CCCH zinc finger Zc3h12a can bind RNA and while results in [70] show substrate (IL-1β) mRNA can be recovered with this protein it has yet to be determined if this is a direct binding or secondary to binding by another protein. The precise cleavage site(s) have not been mapped, nor has it been determined if decay occurs while substrate mRNAs are being translated. The granular staining seen for a Zc3h12a-RFP fusion protein in [70] resembles that of stress granules, but overexpression of such hybrid proteins may not accurately represent the actual subcellular distribution. Much more needs to be done to clarify the relationship of Zc3h12a-mediated mRNA decay to the exonuclease-catalyzed processes. In particular, it will be important to knockdown Xrn1 and one or more exosome subunits to facilitate mapping in vivo cleavage sites. It will also be important to characterize the protein context with which Zc3h12a functions. Mizgalska et al. [70] mentioned they had identified proteins involved in mRNA decay by mass spectrometry of immunoprecipitated Zc3H12a but none of these were listed in that report.
Most of the endonucleases described in this review are expressed constitutively and their activities are controlled by interactions with other proteins or by post-translational modifications such as phosphorylation. Zc3h12a is different as its transcription is significantly induced by IL-1 or LPS activation of the MAP kinase pathway [71]. As anticipated by the presence of a PIN domain divalent cations (in this case Mg++) are required for activity in vitro, and decay in vivo was inhibited by mutating key aspartic acid residues. The preference for Mg++ is somewhat different from PIN domains of SMG6 and Dis3/Rrp44 (see below), which show greater activity with Mn++. Recombinant Zc3h12a shows no sequence selectivity, instead degrading any RNA substrate. This raises the question of how substrate selectivity is determined, particularly given that the adjacent AREs in the targets examined to date do not appear to be involved. Moreover much more needs to be done to clarify the relationship between Zc3h12a and ARE-mediated mRNA decay. While results in [70] suggested a limited range of expression a different picture emerges in a search of the Gene Expression Omnibus. There are many cells and tissues where Zc3h12a may not be as robustly expressed as in lymphoid cells but is nonetheless present and perhaps capable of targeting a broader range of substrates, the nature of which may be defined by associated RNA binding proteins. There is also the question of the function of the other members of this group of enzymes, Zc3h12b, Zc3h12c and Zc3h12d.
SMG6
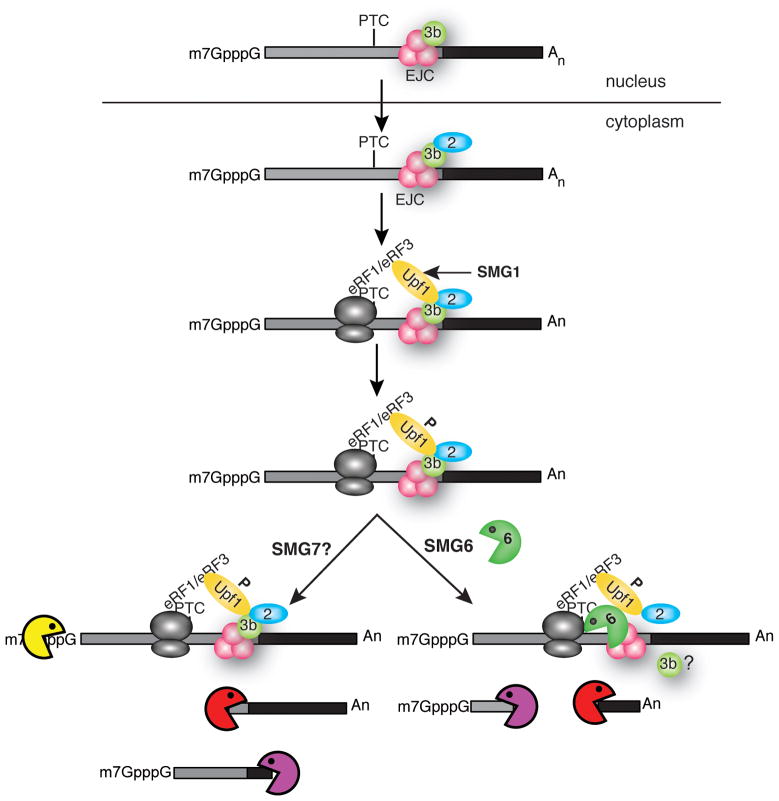
mRNA surveillance or nonsense-mediated mRNA decay, is a quality control process that is conserved throughout all eukaryotes (reviewed in [1, 72, 73]). Because NMD is the subject of several advanced reviews in this series I will summarize salient points relevant to SMG6 and recommend the reader seek out more detailed information there. NMD is a translation-dependent process in which ribosome stalling at a premature termination activates mRNA decay. In mammals NMD acts in conjunction with alternative splicing to control gene expression, and splicing determines the context for whether a stop codon is premature through deposition of the exon junction complex 24 nucleotides upstream of each splice junction. Upf3b, the first of the surveillance proteins, joins the EJC by binding to the Y14/Magoh heterodimer. These proteins accompany the mRNA into the cytoplasm where Upf3b is joined by Upf2 and the mRNA is scanned during a ‘primary round’ of translation while still bearing a number of the proteins with which it was associated in the nucleus (reviewed in [74]). Ribosome stalling at a premature termination codon (PTC) >50 nucleotides upstream of an EJC recruits the DEAD-box helicase Upf1, which binds to Upf2 and is phosphorylated by SMG1, a PI3 kinase-related kinase. This in turn recruitments SMG5, SMG6 and SMG7, each of which can bind to phosphorylated Upf1 (Fig 5).
Figure 5.
Several reports showed that the assembly of the surveillance complex activates deadenylation, decapping and decay by exonucleases acting at either end of the mRNA (Fig. 5, bottom left) [75, 76]. However, in Drosophila S2 cells knockdown of Xrn1 resulted in the accumulation of fragments transcripts downstream of the PTC which Gatfield et al. [26] confirmed to result from endonuclease cleavage. Because of differences in the way nonsense codons are detected in mammals and Drosophila [77] the endonuclease decay of PTC-containing mRNA was initially thought to be limited to this organism [78].
This began to change with the elucidation of the structure of the PIN domains of human SMG5 and SMG6 [79]. Overall their structures are similar, but differences were found in the active site. The PIN domain uses 3 conserved aspartic acid residues to coordinate a divalent cation, which is believed to activate H2O for nucleophilic attack, and while all 3 of these are present in SMG6 only 1 of these is present in SMG5. The SMG6 PIN domain is active in vitro but SMG5 is not, and the SMG6 PIN domain showed an unanticipated preference for Mn++ versus Mg++. SMG6 is highly conserved and a luciferase reporter mRNA was destabilized by tethered SMG6 or its PIN domain, but not by tethered SMG5 or a tethered SMG6 PIN domain with mutations in the critical aspartic acid residues. Finally, the dominant negative inhibition of NMD by an inactive form of Drosophila SMG6 directly linked this endonuclease to the decay process.
The role of SMG6 as an endonuclease important for NMD was confirmed in two papers that appeared in 2008 and 2009. In the first Huntzinger et al. [27] used a complementation assay to restore endonuclease cleavage of PTC-containing mRNA in cells in which SMG6 was depleted by siRNA knockdown. Loss of SMG6 was associated with loss of NMD, and NMD was restored by expression of wild-type SMG6 but not with SMG6 in which 2 of the active site aspartates were changed to asparagine. They also showed that the PIN domain from Nob1, an enzyme involved in rRNA processing, could substitute for the PIN domain of SMG6, and using the approach in [77] of knocking down Xrn1 to stabilize the downstream cleavage product confirmed that NMD was initiated by SMG6 endonuclease cleavage. Finally a similar complementation approach demonstrated that SMG6 catalyzes endonuclease cleavage of PTC-containing mRNA in human (HeLa) cells.
Eberle et al. [28] came to similar conclusions using a somewhat different approach. They started with HeLa and HEK293 cells stably expressing the same wild-type and nonsense-containing reporter mRNAs as Huntzinger et al. [27], but their approach used Northern blots to examine at the impact of knocking down Xrn1, Xrn2, Upf1, and Dcp2 alone or in pairs on each of these mRNAs. Similar to results in [77] knocking down Xrn1 resulted in the appearance of a shortened form of each nonsense-containing mRNA, and they confirmed these were polyadenylated and downstream of the PTC. This study was less successful in identifying upstream cleavage products, but that is not surprising as in most cases these are difficult to detect even with knockdown of exosome subunits. Using 5′-RACE these authors mapped sites of endonuclease cleavage to portions of target mRNAs that were both upstream and downstream of the PTC, implying that SMG6 does not cleave at a particular site but instead cleaves in the general vicinity of the PTC (Fig. 5, bottom right). A recent report by Kashima et al. [80] showed that the molecular basis for the clustering of cleavage sites and for target specificity results from binding of SMG6 to the EJC in much the same way as Upf3b. Direct binding of this effector endonuclease to the EJC solves the targeting problem and insures that SMG6 only acts on those mRNAs that are its appropriate substrates. The concept of specificity through the mRNP is likely to be a recurring theme as more is learned about the molecular code that specifies particular mRNAs for endonuclease-mediated decay.
Remaining questions
The role of endonuclease cleavage by SMG6 lies at the core of understanding NMD. Now that the context for selecting the location of cleavage sites has been solved the relative contribution of SMG6 versus exonuclease-catalyzed decay becomes the major unresolved question.. It appears that SMG6, and perhaps all PIN domain enzymes cleave nonspecifically, with specificity being determined by its positioning by interacting proteins. This makes SMG6 an ideal effector of NMD as its binding to the EJC juxtaposes it to the sensor for the nonsense codon. This is similar in concept to IREα-mediated mRNA decay where the signal sequence juxtaposes substrates to the catalytic core of the enzyme on the cytoplasmic face of the ER. It will be important now to determine whether SMG6 is regulated by the cycle of phosphorylation/dephosphorylation involved in activating and inactivating NMD and to determine the interactions that direct it’s interactions with other proteins that share the binding motif identified in [80].
Apurinic/apyrimidinic endonuclease 1 (APE1)
The turnover of c-myc mRNA has been the subject of investigation since the 1980s when it was found to be more stable in embryonic and rapidly dividing cells than in normal cells. In fact, c-myc mRNA was the target of some of the earliest experiments utilizing decay in vitro to identify factors involved in mRNA turnover [81]. Early work identified 2 major determinants of c-myc mRNA decay; an AU-rich element in the 3′-UTR and a coding region determinant (CRD) that when placed in frame could impart instability to a heterologous mRNA [82, 83]. Prokipcak et al. [84] identified and purified a polysome-associated KH-domain protein (CRD-BP) that stabilized the c-myc CRD to cleavage in vitro by a 35 kDa endonuclease activity that was first identified in a high salt extract of rat liver polysomes [44].
Unlike the complicated RNA structural elements described above that facilitate or position a particular site for endonuclease decay it is the presence of rare codons in the c-myc CRD that is responsible for its endonuclease cleavage. A grouping of rare codons causes ribosomes to pause at CRD, and somewhat like No-Go decay (see below) it is this pausing that makes this region susceptible to endonuclease cleavage [85]. CRD-BP, which is elevated under conditions when c-myc is most highly expressed (ie. during embryogenesis, in liver regeneration and in cancer) [86] stabilizes c-myc mRNA by occluding the endonuclease cleavage site. On a side note recent work showed that CRD-BP functions similarly to stabilize betaTrCP1 mRNA by occluding a coding region site for miR-183 [49].
In 2009 the 35 kDa endonuclease from earlier studies was identified as apurinic/apyrimidinic endonuclease 1 (APE1) [87]. APE1 has a number of activities that complicated efforts to definitively link this to c-myc mRNA decay. As inferred from its name APE1 is best known for its role in DNA repair [88]. It is overexpressed in cancer, its expression is repressed by wild-type but not mutant p53 [89], and it functions in redox-dependent regulation of transcription [90]. Complicating matters further APE1 also has a 3′-5′ exonuclease and RNase H activity [91]. Nevertheless, its knockdown stabilized c-myc mRNA, thus providing direct evidence for its involvement in the decay process.
Remaining questions
Much remains to be learned about the role of APE1 in regulating c-myc mRNA. Earlier work showing CRD-BP blocks cleavage by unpurified or semi-purified protein should be repeated using each of the individual components. While much will be learned by this reductionist approach the key questions revolve around how APE1 functions in vivo in the context of ribosomes stalled at the CRD. If APE1 functions in the decay of c-myc mRNA why are both elevated in cancer? Is it simply a matter of elevated CRD-BP blocking cleavage by APE1, or is something else going on? Are there other mRNA targets for APE1? Do they behave similarly to c-myc, undergoing endonuclease cleavage at sites of ribosome pausing? This takes on added importance in light of the transcriptome-wide prevalence of microRNA and Drosha-independent endonuclease cleavages described in [2].
Additional cases of endonuclease decay
This review would be incomplete without at least mentioning two other well-studied examples of endonuclease decay; No-go decay, and endonuclease cleavage by Dis3/Rrp44, the catalytic engine of the exosome. These are considered separately because none of these are native to metazoan mRNA decay.
No-go decay
In yeast endonuclease cleavage can be induced by the presence of rare codons or a stem-loop structure that is sufficiently strong to inhibit ribosome scanning [92]. This requires two proteins, Dom34, a protein related to eRF1, and Hbs1, a protein related to eRF3. Neither of these proteins has endonuclease activity [93] and the entity responsible for the cleavage event is currently unknown. The reader is directed to a review on No-go decay in this series.
Dis3/Rrp44
The exosome is covered in depth in other advanced reviews in this series, but I would be remiss not to note the importance of endonuclease activity in Dis3 (also called Rrp44). The catalytic activity of the exosome is imparted by two associated nucleases; Rrp6 (also called PM/Scl-100 in metazoans), and Dis3/Rrp44. Dis3/Rrp44 has an RNB-type 3′-5′ exonuclease domain and 3 separate groups [94–97] also showed that an N-terminal PIN domain is an endonuclease that imparts dual functionality to this enzyme. Like other PIN domains it requires divalent cations for activity in vitro with Mn++ preferred over Mg++. The endonuclease activity of Dis3/Rrp44 is less robust than the activity of the RNB domain exonuclease, but to some extent the activities are redundant at least in terms of lethality as yeast will grow with mutations that inactivate either but not both. The relative contributions of the PIN and RNB domains of Dis3/Rrp44 to different aspects of RNA processing and decay are an area of active investigation and the reader is directed to reviews on this and nonstop decay in this series.
Conclusion
Even though endonuclease cleavage was one of the first identified mechanisms of mRNA decay this was until recently considered a minor process that acted on a specific part of the transcriptome or functioned in parallel with the more prevalent processes of deadenylation and exonuclease-catalyzed decay. Within the past several years this view has changed significantly. Some of this is due to the discovery of microRNAs as post-transcriptional regulators of much of the transcriptome, but as pointed out in [2] microRNAs and to a lesser extent Drosha only target some of the transcriptome. This raises the major question how many mRNA endonucleases are there? If one looks at the scope of the enzymes described here only proteins with a PIN domain share similarities that classify them as endonucleases. Others like PMR1 and G3BP, and perhaps even APE1 would never have been identified as mRNA endonucleases if they had been selected based on sequence alone. It seems likely that a genome-wide RNAi screen coupled to transcriptome-wide profiling of 5′ ends, perhaps in Xrn1 knockdown cells, will be needed to get a comprehensive answer to this. Related to this is the issue of contaminating nucleases. It is not sufficient to simply characterize an enzyme as an endonuclease because it demonstrates endonuclease activity when recovered from cell extracts or activity is lost when it is knocked down, since in each of these cases the protein in question could instead associate with an endonuclease in vivo or bind nonspecifically during cell fractionation. The ultimate biochemical proof of endonuclease also requires expression of recombinant protein, inactivation by mutating active site residues and reconstitution of activity after purification under denaturing conditions.
A second major question is how an mRNA endonuclease finds its target. In the case of RISC this is Ago-bound microRNA, for IRE1α this is likely proximity to the ER membrane, and for SMG6 this is the assembly of the surveillance complex at the premature termination codon and its binding to the EJC. For the other enzymes described in this review and endonucleases that are yet to be identified this remains an open question. The answer will likely involve recruitment of the endonuclease to a complex containing its substrate mRNA, the determinants of which will be higher order RNA structures and their cognate RNA-binding proteins. It is also possible that the same enzyme might have different substrates in different cells.
A third major question is how endonuclease decay is regulated. Links to signal transduction are a repeating theme throughout the literature. There already is a substantial body of evidence linking phosphorylation and other signaling events to the activation of a number of the enzymes described here, and with the exception of Zc3h12a, most endonucleases appear to be present prior to their activating stimulus. The coordination of signal transduction with mRNA decay will likely be an important area for the foreseeable future.
Finally, while links between endonuclease and exonuclease decay are standard in bacterial mRNA decay there are only a few reports linking these processes in mammals and other metazoans (see [97] for an example). It is clear that exonucleases degrade the products generated by endonuclease cleavage, but it isn’t known whether in the same cell the same mRNA is handled by both decay mechanisms, and whether there is a balance between these. Given consistent evidence of signaling events activating endonuclease decay it seems reasonable that both processes act on any given mRNA with endonuclease decay coming into play after some form of molecular switch that targets an mRNA to this process. In summary, while there has been progress in elucidating a few mechanisms of endonuclease decay much work remains to be done to understand the breadth of this process, how it is regulated, its relationship to physiological processes and disease, and the interplay between the various mechanisms of mRNA decay.
Acknowledgments
The work on endonuclease decay in my lab is supported by PHS grants GM038277 and GM079707 from the National Institute of General Medical Science.
References
- 1.Shyu AB, Wilkinson MF, van Hoof A. Messenger RNA regulation: to translate or to degrade. EMBO J. 2008;27:471–481. doi: 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karginov FV, Cheloufi S, Chong MMW, Start A, Smith AD, Hannon GJ. Diverse endonucleolytic cleavage sits in the mammalian trenscriptome depend on microRNAs. Drosha and additional nucleases. Mol Cell. 38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: Functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shyu AB, Greenberg ME, Belasco JG. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 5.Wiskocil R, Bensky P, Dower W, Goldberger RF, Gordon JI, Deeley RG. Coordinate regulation of two estrogen-dependent genes in avian liver. Proc Natl Acad Sci USA. 1980;77:4474–4478. doi: 10.1073/pnas.77.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon DA, Shelness GS, Nicosia M, Williams DL. Estrogen-induced destabilization of yolk precursor protein mRNAs in avian liver. J Biol Chem. 1988;263:2625–2631. [PubMed] [Google Scholar]
- 7.Binder R, Hwang SP, Ratnasabapathy R, Williams DL. Degradation of apolipoprotein II mRNA occurs via endonucleolytic cleavage at 5′-AAU-3′/5′-UAA-3′ elements in single-stranded loop domains of the 3′-noncoding region. J Biol Chem. 1989;264:16910–16918. [PubMed] [Google Scholar]
- 8.Cochrane A, Deeley RG. Detection and characterization of degradative intermediates of avian apo very low density lipoprotein II mRNA present in estrogen-treated birds and following destabilization by hormone withdrawal. J Biol Chem. 1989;264:6495–6503. [PubMed] [Google Scholar]
- 9.Shelness GS, Williams DL. Secondary structure analysis of apolipoprotein II mRNA using enzymatic probes and reverse transcriptase. J Biol Chem. 1985;260:8637–8646. [PubMed] [Google Scholar]
- 10.Hwang SP, Eisenberg M, Binder R, Shelness GS, Williams DL. Predicted structures of apolipoprotein II mRNA constrained by nuclease and dimethyl sulfate reactivity: stable secondary structures occur predominantly in local domains via intraexonic base pairing. J Biol Chem. 1989;264:8410–8418. [PubMed] [Google Scholar]
- 11.Ratnasabapathy R, Hwang SP, Williams DL. The 3′-untranslated region of apolipoprotein II mRNA contains two independent domains that bind distinct cytosolic factors. J Biol Chem. 1990;265:14050–14055. [PubMed] [Google Scholar]
- 12.Brown BD, Harland RM. Endonucleolytic cleavage of a maternal homeo box mRNA in Xenopus oocytes. Genes Dev. 1990;4:1925–1935. doi: 10.1101/gad.4.11.1925. [DOI] [PubMed] [Google Scholar]
- 13.Brown BD, Zipkin ID, Harland RM. Sequence-specific endonucleolytic cleavage and protection of messenger RNA in Xenopus and Drosophila. Genes Dev. 1993;7:1620–1631. doi: 10.1101/gad.7.8.1620. [DOI] [PubMed] [Google Scholar]
- 14.Stoeckle MY, Hanafusa H. Processing of 9E3 mRNA and regulation of its stability in normal and Rous sarcoma virus-transformed cells. Mol Cell Biol. 1989;9:4738–4745. doi: 10.1128/mcb.9.11.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meinsma D, Holthuizen PE, Van dBJL, Sussenbach JS. Specific endonucleolytic cleavage of IGF-II mRNAs. Biochem Biophys Res Commun. 1991;179:1509–1516. doi: 10.1016/0006-291x(91)91743-v. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen FC, Christiansen J. Endonucleolysis in the turnover of insulin-like growth factor-II messenger RNA. J Biol Chem. 1992;267:19404–19411. [PubMed] [Google Scholar]
- 17.Meinsma D, Scheper W, Holthuizen PE, Van dBJL, Sussenbach JS. Site-specific cleavage of IGF-II mRNAs requires sequence elements from two distinct regions of the IGF-II gene. Nucleic Acids Res. 1992;20:5003–5009. doi: 10.1093/nar/20.19.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christiansen J, Kofod M, Nielsen FC. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res. 1994;22:5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheper W, Holthuizen PE, Sussenbach JS. The cis-acting elements involved in endonucleolytic cleavage of the 3′ UTR of human IGF-II mRNA bind a 50 kDa protei. Nucleic Acids Res. 1996;24:1000–1007. doi: 10.1093/nar/24.6.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Dijk EL, Sussenbach JS, Holthuizen PE. Distinct RNA structural domains cooperate to maintain a specific cleavage site in the 3′-UTR of IGF-II mRNAs. J Mol Biol. 2000;300:449–467. doi: 10.1006/jmbi.2000.3856. [DOI] [PubMed] [Google Scholar]
- 21.Scheper W, Holthuizen PE, Sussenbach JS. Growth-condition-dependent regulation of insulin-like growth factor II mRNA stability. Biochem J. 1996;318:195–201. doi: 10.1042/bj3180195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Dijk EL, Sussenbach JS, Holthuizen PE. Identification of RNA sequences and structures involved in site-specific cleavage of IGF-II mRNAs. RNA. 1998;4:1632–1635. doi: 10.1017/s1355838298981316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 24.Binder R, Horowitz JA, Basilion JP, Koeller DM, Klausner RD, Harford JB. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casey JL, Koeller DM, Ramin VC, Klausner RD, Harford JB. Iron regulation of transferrin receptor mRNA levels requires iron- responsive elements and a rapid turnover determinant in the 3′ untranslated region of the mRNA. EMBO J. 1989;8:3693–3699. doi: 10.1002/j.1460-2075.1989.tb08544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 27.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 29.Kong J, Sumaroka M, Eastmond DL, Liebhaber SA. Shared stabilization functions of pyrimidine-rich determinants in the erythroid 15-lipoxygenase and alpha-globin mRNAs. Mol Cell Biol. 2006;26:5603–5614. doi: 10.1128/MCB.01845-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Kiledjian M. Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. EMBO J. 2000;19:295–305. doi: 10.1093/emboj/19.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holick M, Liebhaber SA. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc Natl Acad Sci USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens A, Wang Y, Bremer K, Zhang J, Hoepfner R, Antoniou M, Schoenberg DR, Maquat LE. Beta-globin mRNA decay in erythroid cells: UG site-preferred endonucleolytic cleavage that is augmented by a premature termination codon. Proc Natl Acad Sci USA. 2002;99:12741–12746. doi: 10.1073/pnas.192442399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Z, Chang FC, Furneaux HM. The identification of an endonuclease that cleaves within an HuR binding site in mRNA. Nucleic Acids Res. 2000;28:2695–2701. doi: 10.1093/nar/28.14.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riegel AT, Martin MB, Schoenberg DR. Transcriptional and post-transcriptional inhibition of albumin gene expression by estrogen in Xenopus liver. Mol Cell Endocrinol. 1986;44:201–209. doi: 10.1016/0303-7207(86)90125-5. [DOI] [PubMed] [Google Scholar]
- 35.Pastori RL, Moskaitis JE, Buzek SW, Schoenberg DR. Coordinate estrogen-regulated instability of serum protein- coding messenger RNAs in Xenopus laevis. Mol Endocrinol. 1991;5:461–468. doi: 10.1210/mend-5-4-461. [DOI] [PubMed] [Google Scholar]
- 36.Schoenberg DR, Moskaitis JE, Smith JH, Jr, Pastori RL. Extranuclear estrogen-regulated destabilization of Xenopus laevis serum albumin mRNA. Mol Endocrinol. 1989;3:805–814. doi: 10.1210/mend-3-5-805. [DOI] [PubMed] [Google Scholar]
- 37.Riegel AT, Aitken SC, Martin MB, Schoenberg DR. Posttranscriptional regulation of albumin gene expression in Xenopus liver: Evidence for an estrogen receptor-dependent mechanism. Mol Endocrinol. 1987;1:160–167. doi: 10.1210/mend-1-2-160. [DOI] [PubMed] [Google Scholar]
- 38.Pastori RL, Moskaitis JE, Buzek SW, Schoenberg DR. Differential regulation and polyadenylation of transferrin mRNA in Xenopus liver and oviduct. J Steroid Biochem Mol Biol. 1992;42:649–657. doi: 10.1016/0960-0760(92)90105-r. [DOI] [PubMed] [Google Scholar]
- 39.Pastori RL, Moskaitis JE, Schoenberg DR. Estrogen-induced ribonuclease activity in Xenopus liver. Biochemistry. 1991;30:10490–10498. doi: 10.1021/bi00107a018. [DOI] [PubMed] [Google Scholar]
- 40.Pastori RL, Schoenberg DR. The nuclease that selectively degrades albumin messenger RNA in vitro associates with Xenopus liver polysomes through the 80S ribosome complex. Arch Biochem Biophys. 1993;305:313–319. doi: 10.1006/abbi.1993.1428. [DOI] [PubMed] [Google Scholar]
- 41.Dompenciel RE, Garnepudi VR, Schoenberg DR. Purification and characterization of an estrogen-regulated Xenopus liver polysomal nuclease involved in the selective destabilization of albumin mRNA. J Biol Chem. 1995;270:6108–6118. doi: 10.1074/jbc.270.11.6108. [DOI] [PubMed] [Google Scholar]
- 42.Chernokalskaya E, Dompenciel RE, Schoenberg DR. Cleavage properties of a polysomal ribonuclease involved in the estrogen-regulated destabilization of albumin mRNA. Nucleic Acids Res. 1997;25:735–742. doi: 10.1093/nar/25.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanson MN, Schoenberg DR. Identification of in vivo mRNA decay intermediates corresponding to sites of in vitro cleavage by polysomal ribonuclease 1. J Biol Chem. 2001;276:12331–12337. doi: 10.1074/jbc.M010483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee CH, Leeds P, Ross J. Purification and characterization of a polysome-associated endoribonuclease that degrades c-myc mRNA _in vitro_. J Biol Chem. 1998;273:25261–25271. doi: 10.1074/jbc.273.39.25261. [DOI] [PubMed] [Google Scholar]
- 45.Cunningham KS, Hanson MN, Schoenberg DR. Polysomal ribonuclease 1 exists in a latent form on polysomes prior to estrogen activation of mRNA decay. Nucleic Acids Res. 2001;29:1156–1162. doi: 10.1093/nar/29.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brock ML, Shapiro DJ. Estrogen stabilizes vitellogenin mRNA against cytoplasmic degradation. Cell. 1983;34:207–214. doi: 10.1016/0092-8674(83)90151-4. [DOI] [PubMed] [Google Scholar]
- 47.Dodson RE, Shapiro DJ. Vigilin, an ubiquitous protein with 14 KH domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region binding protein. J Biol Chem. 1997;272:12249–12252. doi: 10.1074/jbc.272.19.12249. [DOI] [PubMed] [Google Scholar]
- 48.Cunningham KS, Dodson RE, Nagel MA, Shapiro DJ, Schoenberg DR. Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′-UTR by the mRNA endonuclease PMR-1. Proc Natl Acad Sci USA. 2000;97:12498–12502. doi: 10.1073/pnas.220425497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell. 2009;35:240–246. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chernokalskaya E, DuBell AN, Cunningham KS, Hanson MN, Dompenciel RE, Schoenberg DR. A polysomal ribonuclease involved in the destabilization of albumin mRNA is a novel member of the peroxidase gene family. RNA. 1998;4:1537–1548. doi: 10.1017/s1355838298980451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang F, Schoenberg DR. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol Cell. 2004;14:435–445. doi: 10.1016/j.molcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Yang F, Peng Y, Schoenberg DR. Endonuclease-mediated mRNA decay requires tyrosine phosphorylation of polysomal ribonuclease 1 (PMR1) for the targeting and degradation of polyribosome-bound substrate mRNA. J Biol Chem. 2004;279:48993–49002. doi: 10.1074/jbc.M409776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng Y, Liu X, Schoenberg DR. The 90-kDa heat shock protein stabilizes the polysomal ribonuclease 1 mRNA endonuclease to degradation by the 26S proteasome. Mol Biol Cell. 2008;19:546–552. doi: 10.1091/mbc.E07-08-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng Y, Schoenberg DR. c-Src activates endonuclease-mediated mRNA decay. Mol Cell. 2007;25:779–787. doi: 10.1016/j.molcel.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng Y, Murray EL, Sarkar M, Liu X, Schoenberg DR. The cytoskeleton-associated Ena/VASP proteins are unanticipated partners of the PMR1 mRNA endonuclease. RNA. 2009;15:576–587. doi: 10.1261/rna.1206209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang F, Peng Y, Murray EL, Otsuka Y, Kedersha N, Schoenberg DR. Polysome-bound endonuclease PMR1 Is targeted to stress granules via stress-specific binding to TIA-1. Mol Cell Biol. 2006;26:8803–8813. doi: 10.1128/MCB.00090-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bremer KA, Stevens A, Schoenberg DR. An endonuclease activity similar to Xenopus PMR1 catalyzes the degradation of normal and nonsense-containing human beta-globin mRNA in erythroid cells. RNA. 2003;9:1157–1167. doi: 10.1261/rna.5720303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gallouzi IE, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, Capony JP, Tocque B, Tazi J. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: A potential link between signal transduction and RNA stability. Mol Cell Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tourriere H, Gallouzi IE, Chebli K, Capony JP, Mouaikel J, van der Geer P, Tazi J. RasGAP-associated endoribonuclease G3Bp: selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol. 2001;21:7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zekri L, Chebli K, Tourriere H, Nielsen FC, Hansen TVO, Rami A, Tazi J. Control of fetal growth and neonatal survival by the RasGAP-associated endoribonuclease G3BP. Mol Cell Biol. 2005;25:8703–8716. doi: 10.1128/MCB.25.19.8703-8716.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Kohno K. Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. J Biochem. 2010;147:27–33. doi: 10.1093/jb/mvp196. [DOI] [PubMed] [Google Scholar]
- 63.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 66.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, Hagen A, Backes BJ, Oakes SA, Papa FR. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated IRE1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oikawa D, Tokuda M, Hosoda A, Iwawaki T. Identification of a consensus element recognized and cleaved by IRE1α. Nucl Acids Res. 2010;38:6265–6273. doi: 10.1093/nar/gkq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsushita K, Takeuchi O, Standley DM, Kumagai Y, Kawagoe T, Miyake T, Satoh T, Kato H, Tsujimura T, Nakamura H, Akira S. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 70.Mizgalska D, Wegrzyn P, Murzyn K, Kasza A, Koj A, Jura J, Jarzab B, Jura J. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FEBS J. 2009;276:7386–7399. doi: 10.1111/j.1742-4658.2009.07452.x. [DOI] [PubMed] [Google Scholar]
- 71.Kasza A, Wyrzykowska P, Horwacik I, Tymoszuk P, Mizgalska D, Palmer K, Rokita H, Sharrocks AD, Jura J. Transcription factors Elk-1 and SRF are engaged in IL1-dependent regulation of ZC3H12A expression. BMC Mol Biol. 2010;11:14. doi: 10.1186/1471-2199-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 73.Rehwinkel J, Raes J, Izaurralde E. Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem Sci. 2006;31:639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 74.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 76.Couttet P, Grange T. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res. 2004;32:488–494. doi: 10.1093/nar/gkh218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Valencia-Sanchez MA, Maquat LE. An enemy within: fly reconnaissance deploys an endonuclease to destroy nonsense-containing mRNA. Trends Cell Biol. 2004;14:594–597. doi: 10.1016/j.tcb.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 79.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kashima I, Jonas S, Jayachandran U, Buchwald G, Conti E, Lupas AN, Izaurralde E. SMG6 interacts with the exon junction complex via two conserved EJC-binding motifs (EBMs) required for nonsense-mediated mRNA decay. Genes Dev. 2010;24:2440–2550. doi: 10.1101/gad.604610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brewer G, Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989;9:1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bernstein PL, Herrick DJ, Prokipcak RD, Ross J. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 1992;6:642–654. doi: 10.1101/gad.6.4.642. [DOI] [PubMed] [Google Scholar]
- 83.Herrick DJ, Ross J. The half-life of c-myc mrna in growing and serum-stimulated cells - influence of the coding and 3′ untranslated regions and role of ribosome translocation. Mol Cell Biol. 1994;14:2119–2128. doi: 10.1128/mcb.14.3.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prokipcak RD, Herrick DJ, Ross J. Purification and properties of a protein that binds to the C- terminal coding region of human c-myc mRNA. J Biol Chem. 1994;269:9261–9269. [PubMed] [Google Scholar]
- 85.Lemm I, Ross J. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol Cell Biol. 2002;22:3959–3969. doi: 10.1128/MCB.22.12.3959-3969.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ross J, Lemm I, Berberet B. Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene. 2001;20:6544–6550. doi: 10.1038/sj.onc.1204838. [DOI] [PubMed] [Google Scholar]
- 87.Barnes T, Kim WC, Mantha AK, Kim SE, Izumi T, Mitra S, Lee CH. Identification of Apurinic/apyrimidinic endonuclease 1 (APE1) as the endoribonuclease that cleaves c-myc mRNA. Nucleic Acids Res. 2009;37:3946–3958. doi: 10.1093/nar/gkp275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zaky A, Busso C, Izumi T, Chattopadhyay R, Bassiouny A, Mitra S, Bhakat KK. Regulation of the human AP-endonuclease (APE1/Ref-1) expression by the tumor suppressor p53 in response to DNA damage. Nucleic Acids Res. 2008;36:1555–1566. doi: 10.1093/nar/gkm1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid Redox Signal. 2009;11:621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yoshida A, Ueda T. Human AP endonuclease possesses a significant activity as major 3′-5′ exonuclease in human leukemia cells. Biochem Biophys Res Commun. 2003;310:522–528. doi: 10.1016/j.bbrc.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 92.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Passos DO, Doma MK, Shoemaker CJ, Muhlrad D, Green R, Weissman J, Hollien J, Parker R. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell. 2009;20:3025–3032. doi: 10.1091/mbc.E09-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 95.Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schneider C, Leung E, Brown J, Tollervey D. The N-terminal PIN domain of the exosome subunit Rrp44 harbors endonuclease activity and tethers Rrp44 to the yeast core exosome. Nucleic Acids Res. 2009;37:1127–1140. doi: 10.1093/nar/gkn1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nechama M, Peng Y, Bell O, Briata P, Gherzi R, Schoenberg DR, Naveh-Many T. KSRP-PMR1-exosome association determines parathyroid hormone mRNA levels and stability in transfected cells. BMC Cell Biol. 2009;10:70. doi: 10.1186/1471-2121-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Murray EL, Schoenberg DR. A+U-rich instability elements differentially activate 5′-3′ and 3′-5′ mRNA decay. Mol Cell Biol. 2007;27:2791–2799. doi: 10.1128/MCB.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]