Full text
PDF



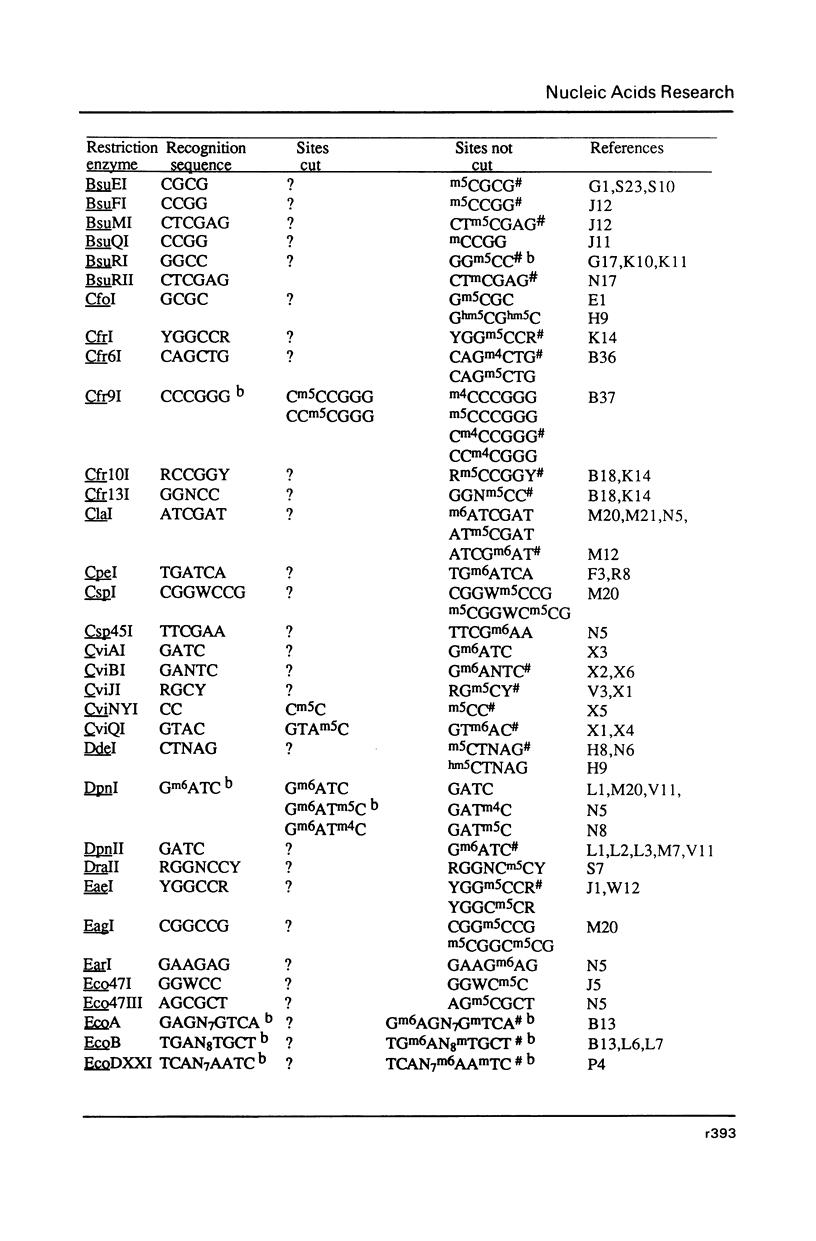
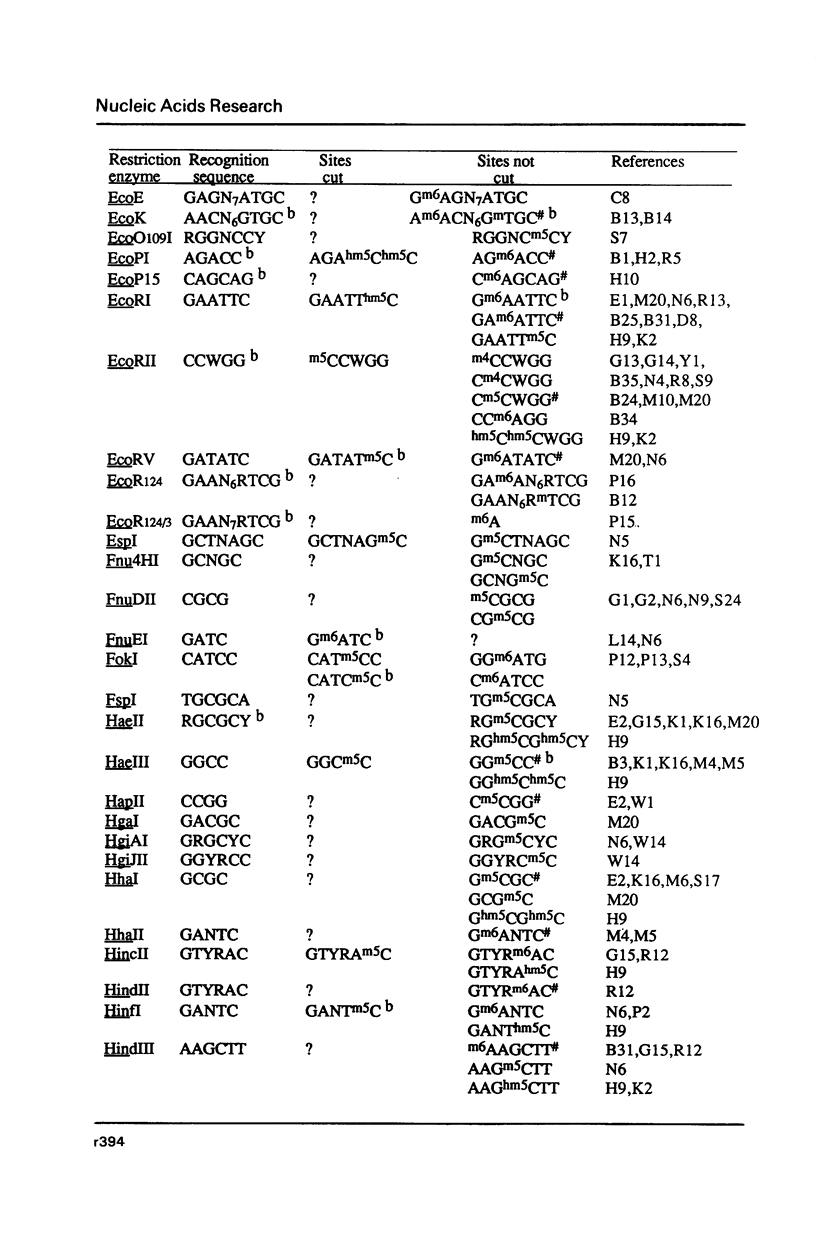
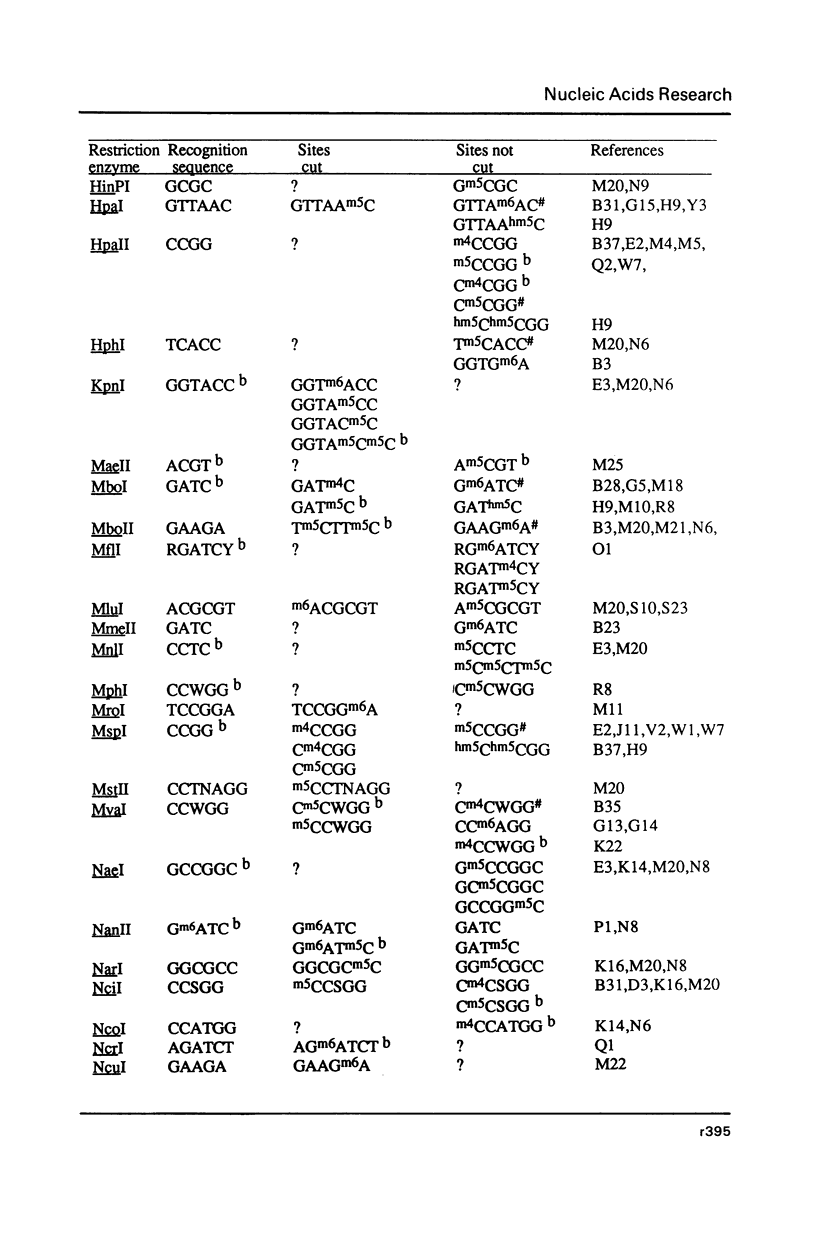
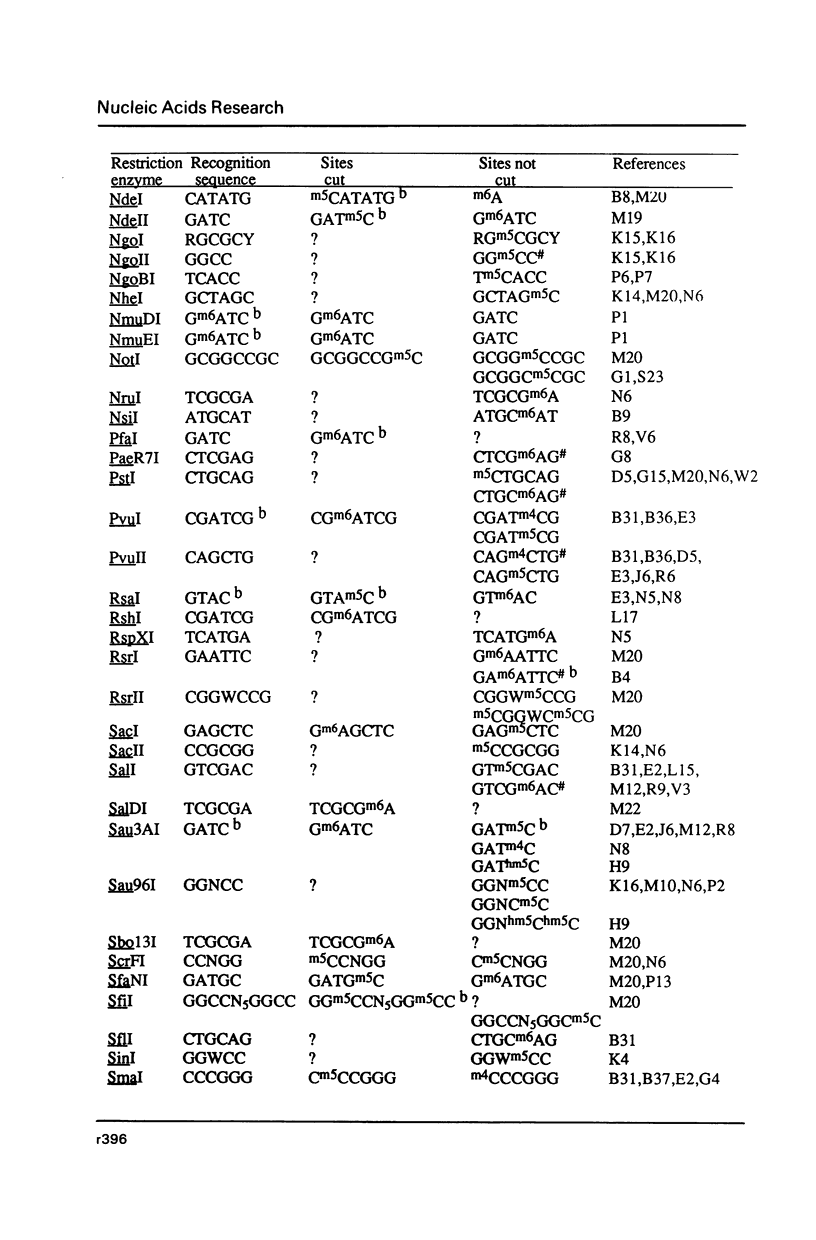

















Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K. A cautionary note on the use of certain restriction endonucleases with methylated substrates. Gene. 1980 Oct;11(1-2):169–171. doi: 10.1016/0378-1119(80)90097-9. [DOI] [PubMed] [Google Scholar]
- Barbeyron T., Kean K., Forterre P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J Bacteriol. 1984 Nov;160(2):586–590. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens B., Noyer-Weidner M., Pawlek B., Lauster R., Balganesh T. S., Trautner T. A. Organization of multispecific DNA methyltransferases encoded by temperate Bacillus subtilis phages. EMBO J. 1987 Apr;6(4):1137–1142. doi: 10.1002/j.1460-2075.1987.tb04869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham A. H., Atkinson T., Sciaky D., Roberts R. J. A specific endonuclease from Bacillus caldolyticus. Nucleic Acids Res. 1978 Oct;5(10):3457–3467. doi: 10.1093/nar/5.10.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Smith B. A. Methylated and unmethylated DNA compartments in the sea urchin genome. Cell. 1979 Aug;17(4):889–901. doi: 10.1016/0092-8674(79)90329-5. [DOI] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Bitinaité J. B., Klimasauskas S. J., Butkus V. V., Janulaitis A. A. Characterization of restriction-modification enzymes Cfr13 I from Citrobacter freundii RFL13. FEBS Lett. 1985 Mar 25;182(2):509–513. doi: 10.1016/0014-5793(85)80364-1. [DOI] [PubMed] [Google Scholar]
- Blumenthal R. M., Gregory S. A., Cooperider J. S. Cloning of a restriction-modification system from Proteus vulgaris and its use in analyzing a methylase-sensitive phenotype in Escherichia coli. J Bacteriol. 1985 Nov;164(2):501–509. doi: 10.1128/jb.164.2.501-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borck K., Beggs J. D., Brammar W. J., Hopkins A. S., Murray N. E. The construction in vitro of transducing derivatives of phage lambda. Mol Gen Genet. 1976 Jul 23;146(2):199–207. doi: 10.1007/BF00268089. [DOI] [PubMed] [Google Scholar]
- Bougueleret L., Schwarzstein M., Tsugita A., Zabeau M. Characterization of the genes coding for the Eco RV restriction and modification system of Escherichia coli. Nucleic Acids Res. 1984 Apr 25;12(8):3659–3676. doi: 10.1093/nar/12.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A. C., Charles I. G., Keyte J. W., Brammar W. J. Isolation and computer-aided characterization of MmeI, a type II restriction endonuclease from Methylophilus methylotrophus. Nucleic Acids Res. 1986 Jul 11;14(13):5255–5274. doi: 10.1093/nar/14.13.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Van Cleve M. D., Gumport R. I. The effects of base analogue substitutions on the methylation by the EcoRI modification methylase of octadeoxyribonucleotides containing modified EcoRI recognition sequences. J Biol Chem. 1986 Jun 5;261(16):7279–7286. [PubMed] [Google Scholar]
- Bron S., Murray K., Trautner T. A. Restriction and modification in B. subtilis. Purification and general properties of a restriction endonuclease from strain R. Mol Gen Genet. 1975 Dec 30;143(1):13–23. doi: 10.1007/BF00269416. [DOI] [PubMed] [Google Scholar]
- Brooks J. E., Blumenthal R. M., Gingeras T. R. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983 Feb 11;11(3):837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Hattman S. In vitro methylation of bacteriophage lambda DNA by wild type (dam+) and mutant (damh) forms of the phage T2 DNA adenine methylase. J Mol Biol. 1978 Dec 15;126(3):381–394. doi: 10.1016/0022-2836(78)90047-5. [DOI] [PubMed] [Google Scholar]
- Brooks J. E. Properties and uses of restriction endonucleases. Methods Enzymol. 1987;152:113–129. doi: 10.1016/0076-6879(87)52014-6. [DOI] [PubMed] [Google Scholar]
- Brooks J. E., Roberts R. J. Modification profiles of bacterial genomes. Nucleic Acids Res. 1982 Feb 11;10(3):913–934. doi: 10.1093/nar/10.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhk H. J., Behrens B., Tailor R., Wilke K., Prada J. J., Günthert U., Noyer-Weidner M., Jentsch S., Trautner T. A. Restriction and modification in Bacillus subtilis: nucleotide sequence, functional organization and product of the DNA methyltransferase gene of bacteriophage SPR. Gene. 1984 Jul-Aug;29(1-2):51–61. doi: 10.1016/0378-1119(84)90165-3. [DOI] [PubMed] [Google Scholar]
- Bur'ianov Ia I., Nesterenko V. F., Vagabova L. M. Prisutstvie dvukh DNK-tsitozin-metilaz v kletkakh Escherichia coli MRE 600. Dokl Akad Nauk SSSR. 1976;227(6):1472–1475. [PubMed] [Google Scholar]
- Buryanov Y. I., Bogdarina I. G., Bayev A. A. Site specificity and chromatographic properties of E. coli K12 and EcoRIIDNA-cytosine methylases. FEBS Lett. 1978 Apr 15;88(2):251–254. doi: 10.1016/0014-5793(78)80186-0. [DOI] [PubMed] [Google Scholar]
- Busslinger M., deBoer E., Wright S., Grosveld F. G., Flavell R. A. The sequence GGCmCGG is resistant to MspI cleavage. Nucleic Acids Res. 1983 Jun 11;11(11):3559–3569. doi: 10.1093/nar/11.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkus V., Klimasauskas S., Kersulyte D., Vaitkevicius D., Lebionka A., Janulaitis A. Investigation of restriction-modification enzymes from M. varians RFL19 with a new type of specificity toward modification of substrate. Nucleic Acids Res. 1985 Aug 26;13(16):5727–5746. doi: 10.1093/nar/13.16.5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkus V., Klimasauskas S., Petrauskiene L., Maneliene Z., Lebionka A., Janulaitis A. Interaction of AluI, Cfr6I and PvuII restriction-modification enzymes with substrates containing either N4-methylcytosine or 5-methylcytosine. Biochim Biophys Acta. 1987 Aug 25;909(3):201–207. doi: 10.1016/0167-4781(87)90078-9. [DOI] [PubMed] [Google Scholar]
- Butkus V., Petrauskiene L., Maneliene Z., Klimasauskas S., Laucys V., Janulaitis A. Cleavage of methylated CCCGGG sequences containing either N4-methylcytosine or 5-methylcytosine with MspI, HpaII, SmaI, XmaI and Cfr9I restriction endonucleases. Nucleic Acids Res. 1987 Sep 11;15(17):7091–7102. doi: 10.1093/nar/15.17.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caserta M., Zacharias W., Nwankwo D., Wilson G. G., Wells R. D. Cloning, sequencing, in vivo promoter mapping, and expression in Escherichia coli of the gene for the HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4770–4777. [PubMed] [Google Scholar]
- Casjens S., Hayden M., Jackson E., Deans R. Additional restriction endonuclease cleavage sites on the bacteriophage P22 genome. J Virol. 1983 Feb;45(2):864–867. doi: 10.1128/jvi.45.2.864-867.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. M., Hartley B. S. Purification, properties and specificity of the restriction endonuclease from Bacillus stearothermophilus. Biochem J. 1979 Jan 1;177(1):49–62. doi: 10.1042/bj1770049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985 May 10;13(9):3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan G. M., Gann A. A., Murray N. E. Conservation of complex DNA recognition domains between families of restriction enzymes. Cell. 1989 Jan 13;56(1):103–109. doi: 10.1016/0092-8674(89)90988-4. [DOI] [PubMed] [Google Scholar]
- Daniel A. S., Fuller-Pace F. V., Legge D. M., Murray N. E. Distribution and diversity of hsd genes in Escherichia coli and other enteric bacteria. J Bacteriol. 1988 Apr;170(4):1775–1782. doi: 10.1128/jb.170.4.1775-1782.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A. P., Dobritsa S. V. DNA protection with the DNA methylase M . BbvI from Bacillus brevis var. GB against cleavage by the restriction endonucleases PstI and PvuII. Gene. 1980 Jul;10(2):105–112. doi: 10.1016/0378-1119(80)90128-6. [DOI] [PubMed] [Google Scholar]
- Doolittle M. M., Sirotkin K. Bacteriophage T2 and T4, dam+ and damh and Eco dam+ methylation: preference at different sites. Biochim Biophys Acta. 1988 Feb 28;949(2):240–246. doi: 10.1016/0167-4781(88)90088-7. [DOI] [PubMed] [Google Scholar]
- Dreiseikelmann B., Eichenlaub R., Wackernagel W. The effect of differential methylation by Escherichia coli of plasmid DNA and phage T7 and lambda DNA on the cleavage by restriction endonuclease MboI from Moraxella bovis. Biochim Biophys Acta. 1979 May 24;562(3):418–428. doi: 10.1016/0005-2787(79)90105-9. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Hedgpeth J., Boyer H. W., Goodman H. M. Physical identity of the SV40 deoxyribonucleic acid sequence recognized by the Eco RI restriction endonuclease and modification methylase. Biochemistry. 1974 Jan 29;13(3):503–512. doi: 10.1021/bi00700a016. [DOI] [PubMed] [Google Scholar]
- Dybvig K., Swinton D., Maniloff J., Hattman S. Cytosine methylation of the sequence GATC in a mycoplasma. J Bacteriol. 1982 Sep;151(3):1420–1424. doi: 10.1128/jb.151.3.1420-1424.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Wilson G. G., Kuo K. C., Gehrke C. W. N4-methylcytosine as a minor base in bacterial DNA. J Bacteriol. 1987 Mar;169(3):939–943. doi: 10.1128/jb.169.3.939-943.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér Z., Kiss A., Venetianer P. Expression of a bacterial modification methylase gene in yeast. Nature. 1983 Mar 17;302(5905):266–268. doi: 10.1038/302266a0. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Bullas L. R., Delius H., Murray N. E. Genetic recombination can generate altered restriction specificity. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6095–6099. doi: 10.1073/pnas.81.19.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Murray N. E. Two DNA recognition domains of the specificity polypeptides of a family of type I restriction enzymes. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9368–9372. doi: 10.1073/pnas.83.24.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaido M. L., Prostko C. R., Strobl J. S. Isolation and characterization of BsuE methyltransferase, a CGCG specific DNA methyltransferase from Bacillus subtilis. J Biol Chem. 1988 Apr 5;263(10):4832–4836. [PubMed] [Google Scholar]
- Gann A. A., Campbell A. J., Collins J. F., Coulson A. F., Murray N. E. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol. 1987 Jul;1(1):13–22. doi: 10.1111/j.1365-2958.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- Gautier F., Bünemann H., Grotjahn L. Analysis of calf-thymus satellite DNA: evidence for specific methylation of cytosine in C-G sequences. Eur J Biochem. 1977 Oct 17;80(1):175–183. doi: 10.1111/j.1432-1033.1977.tb11869.x. [DOI] [PubMed] [Google Scholar]
- Gelinas R. E., Myers P. A., Roberts R. J. Two sequence-specific endonucleases from Moraxella bovis. J Mol Biol. 1977 Jul;114(1):169–179. doi: 10.1016/0022-2836(77)90290-x. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Brooks J. E. Cloned restriction/modification system from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1983 Jan;80(2):402–406. doi: 10.1073/pnas.80.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Gupta M., Boyer H. W., Brown W. E., Rosenberg J. M. Sequence analysis of the DNA encoding the Eco RI endonuclease and methylase. J Biol Chem. 1981 Mar 10;256(5):2143–2153. [PubMed] [Google Scholar]
- Guha S. Determination of DNA sequences containing methylcytosine in Bacillus subtilis Marburg. J Bacteriol. 1985 Aug;163(2):573–579. doi: 10.1128/jb.163.2.573-579.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günthert U., Storm K., Bald R. Restriction and modification in Bacillus subtilis. Localization of the methylated nucleotide in the BsuRI recognition sequence. Eur J Biochem. 1978 Oct 16;90(3):581–583. doi: 10.1111/j.1432-1033.1978.tb12638.x. [DOI] [PubMed] [Google Scholar]
- Günthert U., Trautner T. A. DNA methyltransferases of Bacillus subtilis and its bacteriophages. Curr Top Microbiol Immunol. 1984;108:11–22. doi: 10.1007/978-3-642-69370-0_2. [DOI] [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hattman S., Keister T., Gottehrer A. Sequence specificity of DNA methylases from Bacillus amyloliquefaciens and Bacillus brevis. J Mol Biol. 1978 Oct 5;124(4):701–711. doi: 10.1016/0022-2836(78)90178-x. [DOI] [PubMed] [Google Scholar]
- Hattman S., van Ormondt H., de Waard A. Sequence specificity of the wild-type dam+) and mutant (damh) forms of bacteriophage T2 DNA adenine methylase. J Mol Biol. 1978 Mar 5;119(3):361–376. doi: 10.1016/0022-2836(78)90219-x. [DOI] [PubMed] [Google Scholar]
- Hofer B. The sensitivity of DNA cleavage by SpeI and ApaLI to methylation by M.EcoK. Nucleic Acids Res. 1988 Jun 10;16(11):5206–5206. doi: 10.1093/nar/16.11.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard K. A., Card C., Benner J. S., Callahan H. L., Maunus R., Silber K., Wilson G., Brooks J. E. Cloning the DdeI restriction-modification system using a two-step method. Nucleic Acids Res. 1986 Oct 24;14(20):7939–7951. doi: 10.1093/nar/14.20.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hümbelin M., Suri B., Rao D. N., Hornby D. P., Eberle H., Pripfl T., Kenel S., Bickle T. A. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J Mol Biol. 1988 Mar 5;200(1):23–29. doi: 10.1016/0022-2836(88)90330-0. [DOI] [PubMed] [Google Scholar]
- Ikawa S., Shibata T., Ando T., Saito H. Genetic studies on site-specific endodeoxyribonucleases in Bacillus subtilis: multiple modification and restriction systems in transformants of Bacillus subtilis 168. Mol Gen Genet. 1980 Feb;177(3):359–368. doi: 10.1007/BF00271474. [DOI] [PubMed] [Google Scholar]
- Jacobs D., Brown N. L. Isolation and characterization of the M.EaeI modification methylase. Biochem J. 1986 Sep 1;238(2):613–615. doi: 10.1042/bj2380613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janulaitis A. A., Petrusite M. A., Jaskelavicene B. P., Krayev A. S., Skryabin K. G., Bayev A. A. A new restriction endonuclease BcnI from Bacillus centrosporus RFL 1. FEBS Lett. 1982 Jan 25;137(2):178–180. doi: 10.1016/0014-5793(82)80343-8. [DOI] [PubMed] [Google Scholar]
- Janulaitis A., Klimasauskas S., Petrusyte M., Butkus V. Cytosine modification in DNA by BcnI methylase yields N4-methylcytosine. FEBS Lett. 1983 Sep 5;161(1):131–134. doi: 10.1016/0014-5793(83)80745-5. [DOI] [PubMed] [Google Scholar]
- Janulaitis A., Petrusyté M., Butkus V. Three sequence-specific endonucleases from Escherichia coli RFL47. FEBS Lett. 1983 Sep 19;161(2):213–216. doi: 10.1016/0014-5793(83)81010-2. [DOI] [PubMed] [Google Scholar]
- Jentsch S., Günthert U., Trautner T. A. DNA methyltransferases affecting the sequence 5'CCGG. Nucleic Acids Res. 1981 Jun 25;9(12):2753–2759. doi: 10.1093/nar/9.12.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. C., Choi W. S., Yoo O. J. The effects of DNA methylation by Hha I methylase on the cleavage reactions by Hae II, Aha II and Ban I endonucleases. Biochem Biophys Res Commun. 1987 May 29;145(1):482–487. doi: 10.1016/0006-291x(87)91346-5. [DOI] [PubMed] [Google Scholar]
- Kaput J., Sneider T. W. Methylation of somatic vs germ cell DNAs analyzed by restriction endonuclease digestions. Nucleic Acids Res. 1979 Dec 20;7(8):2303–2322. doi: 10.1093/nar/7.8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman C., Tandeau de Marsac N., de Waard A. Isolation of a deoxycytidylate methyl transferase capable of protecting DNA uniquely against cleavage by endonuclease R.Aqu I (isoschizomer of Ava I). Nucleic Acids Res. 1986 Jul 11;14(13):5199–5205. doi: 10.1093/nar/14.13.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreman C., de Waard A. Isolation and characterization of the modification methylase M . SinI. J Bacteriol. 1988 Jun;170(6):2533–2536. doi: 10.1128/jb.170.6.2533-2536.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S., Kaddurah-Daouk R., Smith H. O. Purification of the HhaII restriction endonuclease from an overproducer Escherichia coli clone. J Biol Chem. 1985 Dec 5;260(28):15339–15344. [PubMed] [Google Scholar]
- Keshet E., Cedar H. Effect of CpG methylation on Msp I. Nucleic Acids Res. 1983 Jun 11;11(11):3571–3580. doi: 10.1093/nar/11.11.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C., Neumaier P. S., Wolf W. Recognition sequences of restriction endonucleases and methylases--a review. Gene. 1985;33(1):1–102. doi: 10.1016/0378-1119(85)90119-2. [DOI] [PubMed] [Google Scholar]
- Kiss A., Baldauf F. Molecular cloning and expression in Escherichia coli of two modification methylase genes of Bacillus subtilis. Gene. 1983 Jan-Feb;21(1-2):111–119. doi: 10.1016/0378-1119(83)90153-1. [DOI] [PubMed] [Google Scholar]
- Kiss A., Posfai G., Keller C. C., Venetianer P., Roberts R. J. Nucleotide sequence of the BsuRI restriction-modification system. Nucleic Acids Res. 1985 Sep 25;13(18):6403–6421. doi: 10.1093/nar/13.18.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita K., Hiraoka N., Oshima A., Kadonishi S., Obayashi A. AccIII, a new restriction endonuclease from Acinetobacter calcoaceticus. Nucleic Acids Res. 1985 Dec 20;13(24):8685–8694. doi: 10.1093/nar/13.24.8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleid D., Humayun Z., Jeffrey A., Ptashne M. Novel properties of a restriction endonuclease isolated from Haemophilus parahaemolyticus. Proc Natl Acad Sci U S A. 1976 Feb;73(2):293–297. doi: 10.1073/pnas.73.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C., Kiss A., Venetianer P. Biochemical characterization of the restriction-modification system of Bacillus sphaericus. Eur J Biochem. 1978 Sep 1;89(2):523–529. doi: 10.1111/j.1432-1033.1978.tb12557.x. [DOI] [PubMed] [Google Scholar]
- Korch C., Hagblom P. In-vivo-modified gonococcal plasmid pJD1. A model system for analysis of restriction enzyme sensitivity to DNA modifications. Eur J Biochem. 1986 Dec 15;161(3):519–524. doi: 10.1111/j.1432-1033.1986.tb10473.x. [DOI] [PubMed] [Google Scholar]
- Kosykh V. G., Buryanov Y. I., Bayev A. A. Molecular cloning of EcoRII endonuclease and methylase genes. Mol Gen Genet. 1980;178(3):717–718. doi: 10.1007/BF00337884. [DOI] [PubMed] [Google Scholar]
- Kosykh V. G., Glinskaite I. V., Bur'ianov Ia I., Baev A. A. Opredelenie lokalizatsii genov restriktazy i metilazy EcoRII na rekombinantnykh plazmidakh. Dokl Akad Nauk SSSR. 1982;265(3):727–730. [PubMed] [Google Scholar]
- Kosykh V. G., Solonin A. S., Buryanov YaI, Bayev A. A. Overproduction of the EcoRII endonuclease and methylase by Escherichia coli strains carrying recombinant plasmids constructed in vitro. Biochim Biophys Acta. 1981 Aug 27;655(1):102–106. doi: 10.1016/0005-2787(81)90072-1. [DOI] [PubMed] [Google Scholar]
- Kubareva E. A., Pein C. D., Gromova E. S., Kuznezova S. A., Tashlitzki V. N., Cech D., Shabarova Z. A. The role of modifications in oligonucleotides in sequence recognition by MvaI restriction endonuclease. Eur J Biochem. 1988 Aug 15;175(3):615–618. doi: 10.1111/j.1432-1033.1988.tb14236.x. [DOI] [PubMed] [Google Scholar]
- Lacks S. A., Mannarelli B. M., Springhorn S. S., Greenberg B. Genetic basis of the complementary DpnI and DpnII restriction systems of S. pneumoniae: an intercellular cassette mechanism. Cell. 1986 Sep 26;46(7):993–1000. doi: 10.1016/0092-8674(86)90698-7. [DOI] [PubMed] [Google Scholar]
- Lacks S. A., Springhorn S. S. Cloning in Streptococcus pneumoniae of the gene for DpnII DNA methylase. J Bacteriol. 1984 Mar;157(3):934–936. doi: 10.1128/jb.157.3.934-936.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer F. W. Cleavage by ApaI is inhibited by overlapping dcm methylation. Nucleic Acids Res. 1987 Nov 11;15(21):9087–9087. doi: 10.1093/nar/15.21.9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A., Kan N. C., Lackey D., Linn S., Edgell M. H., Hutchison C. A., 3rd Recognition site of Escherichia coli B restriction enzyme on phi XsB1 and simian virus 40 DNAs: an interrupted sequence. Proc Natl Acad Sci U S A. 1978 May;75(5):2271–2275. doi: 10.1073/pnas.75.5.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenberger J. A., Linn S. The deoxyribonucleic acid modification and restriction enzymes of Escherichia coli B. I. Purification, subunit structure, and catalytic properties of the modification methylase. J Biol Chem. 1972 Oct 10;247(19):6176–6182. [PubMed] [Google Scholar]
- Levy W. P., Welker N. E. Deoxyribonucleic acid modification methylase from Bacillus stearothermophilus. Biochemistry. 1981 Mar 3;20(5):1120–1127. doi: 10.1021/bi00508a012. [DOI] [PubMed] [Google Scholar]
- Lodwick D., Ross H. N., Harris J. E., Almond J. W., Grant W. D. dam methylation in the archaebacteria. J Gen Microbiol. 1986 Nov;132(11):3055–3059. doi: 10.1099/00221287-132-11-3055. [DOI] [PubMed] [Google Scholar]
- Loenen W. A., Daniel A. S., Braymer H. D., Murray N. E. Organization and sequence of the hsd genes of Escherichia coli K-12. J Mol Biol. 1987 Nov 20;198(2):159–170. doi: 10.1016/0022-2836(87)90303-2. [DOI] [PubMed] [Google Scholar]
- Lui A. C., McBride B. C., Vovis G. F., Smith M. Site specific endonuclease from Fusobacterium nucleatum. Nucleic Acids Res. 1979 Jan;6(1):1–15. doi: 10.1093/nar/6.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn S. P., Cohen L. K., Gardner J. F., Kaplan S. Characterization of a site-specific restriction endonuclease from Rhodopseudomonas sphaeroides. J Bacteriol. 1979 May;138(2):505–509. doi: 10.1128/jb.138.2.505-509.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P. M., Mosig G. Regulation of a new bacteriophage T4 gene, 69, that spans an origin of DNA replication. EMBO J. 1984 Dec 1;3(12):2863–2871. doi: 10.1002/j.1460-2075.1984.tb02221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannarelli B. M., Balganesh T. S., Greenberg B., Springhorn S. S., Lacks S. A. Nucleotide sequence of the Dpn II DNA methylase gene of Streptococcus pneumoniae and its relationship to the dam gene of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4468–4472. doi: 10.1073/pnas.82.13.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matvienko N. I., Kramarov V. M., Pachkunov D. M. Isolation and some properties of the site-specific endonuclease and methylase Bme2161 from Bacillus megaterium 216. Eur J Biochem. 1987 Jun 15;165(3):565–570. doi: 10.1111/j.1432-1033.1987.tb11477.x. [DOI] [PubMed] [Google Scholar]
- McClelland M., Kessler L. G., Bittner M. Site-specific cleavage of DNA at 8- and 10-base-pair sequences. Proc Natl Acad Sci U S A. 1984 Feb;81(4):983–987. doi: 10.1073/pnas.81.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Nelson M., Cantor C. R. Purification of Mbo II methylase (GAAGmA) from Moraxella bovis: site specific cleavage of DNA at nine and ten base pair sequences. Nucleic Acids Res. 1985 Oct 25;13(20):7171–7182. doi: 10.1093/nar/13.20.7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Nelson M. The 5'-GGATCC-3' cleavage specificity of BamHI is increased to 5'-CCGGATCCGG-3' by sequential double methylation with M.HpaII and M.BamHI. Gene. 1988 Dec 25;74(1):169–176. doi: 10.1016/0378-1119(88)90278-8. [DOI] [PubMed] [Google Scholar]
- McClelland M., Nelson M. The effect of site specific methylation on restriction endonuclease digestion. Nucleic Acids Res. 1985;13 (Suppl):r201–r207. doi: 10.1093/nar/13.suppl.r201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. Purification and characterization of two new modification methylases: MClaI from Caryophanon latum L and MTaqI from Thermus aquaticus YTI. Nucleic Acids Res. 1981 Dec 21;9(24):6795–6804. doi: 10.1093/nar/9.24.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M. The frequency and distribution of methylatable DNA sequences in leguminous plant protein coding genes. J Mol Evol. 1983;19(5):346–354. doi: 10.1007/BF02101638. [DOI] [PubMed] [Google Scholar]
- Modrich P. Structures and mechanisms of DNA restriction and modification enzymes. Q Rev Biophys. 1979 Aug;12(3):315–369. doi: 10.1017/s0033583500005461. [DOI] [PubMed] [Google Scholar]
- Molloy P. L., Watt F. Effect of cytosine methylation on cutting by the restriction enzyme MaeII. Nucleic Acids Res. 1988 Mar 25;16(5):2335–2335. doi: 10.1093/nar/16.5.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural R. J. Cleavage by the restriction endonuclease Asp718, an isoschizomer of KpnI, is sensitive to Escherichia coli Dcm methylation. Nucleic Acids Res. 1987 Nov 11;15(21):9085–9085. doi: 10.1093/nar/15.21.9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja V., Shepherd J. C., Pripfl T., Bickle T. A. Two type I restriction enzymes from Salmonella species. Purification and DNA recognition sequences. J Mol Biol. 1985 Apr 20;182(4):579–587. doi: 10.1016/0022-2836(85)90243-8. [DOI] [PubMed] [Google Scholar]
- Narva K. E., Wendell D. L., Skrdla M. P., Van Etten J. L. Molecular cloning and characterization of the gene encoding the DNA methyltransferase, M.CviBIII, from Chlorella virus NC-1A. Nucleic Acids Res. 1987 Dec 10;15(23):9807–9823. doi: 10.1093/nar/15.23.9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans D., Smith H. O. Restriction endonucleases in the analysis and restructuring of dna molecules. Annu Rev Biochem. 1975;44:273–293. doi: 10.1146/annurev.bi.44.070175.001421. [DOI] [PubMed] [Google Scholar]
- Nelson M., Christ C., Schildkraut I. Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res. 1984 Jul 11;12(13):5165–5173. doi: 10.1093/nar/12.13.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M., McClelland M. The effect of site-specific methylation on restriction-modification enzymes. Nucleic Acids Res. 1987;15 (Suppl):r219–r230. doi: 10.1093/nar/15.suppl.r219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterenko V. F., Bur'ianov Ia I., Baev A. A. Determinirovanie DNK-tsitozin-metilazy I (M.Eco MRE 600 I) plazmidoi Col A. Dokl Akad Nauk SSSR. 1980;250(5):1265–1267. [PubMed] [Google Scholar]
- Newman A. K., Rubin R. A., Kim S. H., Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J Biol Chem. 1981 Mar 10;256(5):2131–2139. [PubMed] [Google Scholar]
- Nikolskaya I. I., Lopatina N. G., Anikeicheva N. V., Debov S. S. Determination of the recognition sites of cytosine DNA-methylases from Escherichia coli SK. Nucleic Acids Res. 1979 Sep 25;7(2):517–528. doi: 10.1093/nar/7.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya I. I., Lopatina N. G., Sharkova E. V., Suchkov S. V., Somodi P., Földes I., Debov S. S. Sequence specificity of isolated DNA-adenine methylases from Mycobacterium smegmatis (butyricum) and Shigella sonnei 47 cells. Biochem Int. 1985 Mar;10(3):405–413. [PubMed] [Google Scholar]
- Nikolskaya I. I., Lopatina N. G., Suchkov S. V., Kartashova I. M., Debov S. S. Sequence specificity of isolated DNA-cytosine methylases from Shigella sonnei 47 cells. Biochem Int. 1984 Dec;9(6):771–781. [PubMed] [Google Scholar]
- Noyer-Weidner M., Jentsch S., Kupsch J., Bergbauer M., Trautner T. A. DNA methyltransferase genes of Bacillus subtilis phages: structural relatedness and gene expression. Gene. 1985;35(1-2):143–150. doi: 10.1016/0378-1119(85)90166-0. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M., Jentsch S., Pawlek B., Günthert U., Trautner T. A. Restriction and modification in Bacillus subtilis: DNA methylation potential of the related bacteriophages Z, SPR, SP beta, phi 3T, and rho 11. J Virol. 1983 May;46(2):446–453. doi: 10.1128/jvi.46.2.446-453.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyer-Weidner M., Pawlek B., Jentsch S., Günthert U., Trautner T. A. Restriction and modification in Bacillus subtilis: gene coding for a BsuR-specific modification methyltransferase in the temperate bacteriophage phi 3T. J Virol. 1981 Jun;38(3):1077–1080. doi: 10.1128/jvi.38.3.1077-1080.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur I., Szyf M., Razin A., Glaser G., Rottem S., Razin S. Procaryotic and eucaryotic traits of DNA methylation in spiroplasmas (mycoplasmas). J Bacteriol. 1985 Oct;164(1):19–24. doi: 10.1128/jb.164.1.19-24.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwankwo D., Wilson G. Cloning of two type II methylase genes that recognise asymmetric nucleotide sequences: FokI and HgaI. Mol Gen Genet. 1987 Oct;209(3):570–574. doi: 10.1007/BF00331164. [DOI] [PubMed] [Google Scholar]
- Ono A., Ueda T. Synthesis of decadeoxyribonucleotides containing N6-methyladenine, N4-methylcytosine, and 5-methylcytosine: recognition and cleavage by restriction endonucleases (nucleosides and nucleotides part 74). Nucleic Acids Res. 1987 Jan 12;15(1):219–232. doi: 10.1093/nar/15.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Streeck R. E., Zachau H. G. Patchwork structure of a bovine satellite DNA. Cell. 1979 Nov;18(3):883–893. doi: 10.1016/0092-8674(79)90140-5. [DOI] [PubMed] [Google Scholar]
- Piekarowicz A., Goguen J. D. The DNA sequence recognized by the EcoDXX1 restriction endonuclease. Eur J Biochem. 1986 Jan 15;154(2):295–298. doi: 10.1111/j.1432-1033.1986.tb09396.x. [DOI] [PubMed] [Google Scholar]
- Piekarowicz A., Yuan R., Stein D. C. Purification and characterization of DNA methyltransferases from Neisseria gonorrhoeae. Nucleic Acids Res. 1988 Jul 11;16(13):5957–5972. doi: 10.1093/nar/16.13.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C., Shepherd J. C., Bickle T. A. DNA recognition by a new family of type I restriction enzymes: a unique relationship between two different DNA specificities. EMBO J. 1987 May;6(5):1493–1497. doi: 10.1002/j.1460-2075.1987.tb02391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Baldauf F., Erdei S., Pósfai J., Venetianer P., Kiss A. Structure of the gene coding for the sequence-specific DNA-methyltransferase of the B. subtilis phage SPR. Nucleic Acids Res. 1984 Dec 11;12(23):9039–9049. doi: 10.1093/nar/12.23.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai G., Kiss A., Erdei S., Pósfai J., Venetianer P. Structure of the Bacillus sphaericus R modification methylase gene. J Mol Biol. 1983 Nov 5;170(3):597–610. doi: 10.1016/s0022-2836(83)80123-5. [DOI] [PubMed] [Google Scholar]
- Pósfai G., Szybalski W. Increasing the Fokl cleavage specificity from 5 to 7 base pairs by two-step methylation. Nucleic Acids Res. 1988 Jul 11;16(13):6245–6245. doi: 10.1093/nar/16.13.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint A., Cedar H. In vitro methylation of DNA with Hpa II methylase. Nucleic Acids Res. 1981 Feb 11;9(3):633–646. doi: 10.1093/nar/9.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh E. A., Murray N. E., Revel H., Blumenthal R. M., Westaway D., Reith A. D., Rigby P. W., Elhai J., Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988 Feb 25;16(4):1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Urieli S., Pollack Y., Gruenbaum Y., Glaser G. Studies on the biological role of dna methylation; IV. Mode of methylation of DNA in E. coli cells. Nucleic Acids Res. 1980 Apr 25;8(8):1783–1792. doi: 10.1093/nar/8.8.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel H. R., Georgopoulos C. P. Restriction of nonglucosylated T-even bacteriophages by prophage P1. Virology. 1969 Sep;39(1):1–17. doi: 10.1016/0042-6822(69)90343-2. [DOI] [PubMed] [Google Scholar]
- Roberts R. J. Restriction enzymes and their isoschizomers. Nucleic Acids Res. 1988;16 (Suppl):r271–r313. doi: 10.1093/nar/16.suppl.r271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P. H., Smith H. O. DNA methylases of Hemophilus influenzae Rd. I. Purification and properties. J Mol Biol. 1973 Dec 25;81(4):427–444. doi: 10.1016/0022-2836(73)90515-9. [DOI] [PubMed] [Google Scholar]
- Sain B., Murray N. E. The hsd (host specificity) genes of E. coli K 12. Mol Gen Genet. 1980;180(1):35–46. doi: 10.1007/BF00267350. [DOI] [PubMed] [Google Scholar]
- Scherzer E., Auer B., Schweiger M. Identification, purification, and characterization of Escherichia coli virus T1 DNA methyltransferase. J Biol Chem. 1987 Nov 5;262(31):15225–15231. [PubMed] [Google Scholar]
- Schlagman S., Hattman S., May M. S., Berger L. In vivo methylation by Escherichia coli K-12 mec+ deoxyribonucleic acid-cytosine methylase protects against in vitro cleavage by the RII restriction endonuclease (R. Eco RII). J Bacteriol. 1976 May;126(2):990–996. doi: 10.1128/jb.126.2.990-996.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoner B., Kelly S., Smith H. O. The nucleotide sequence of the HhaII restriction and modification genes from Haemophilus haemolyticus. Gene. 1983 Oct;24(2-3):227–236. doi: 10.1016/0378-1119(83)90083-5. [DOI] [PubMed] [Google Scholar]
- Shibata T., Ikawa S., Komatsu Y., Ando T., Saito H. Introduction of host-controlled modification and restriction systems of Bacillus subtilis IAM1247 into Bacillus subtilis 168. J Bacteriol. 1979 Jul;139(1):308–310. doi: 10.1128/jb.139.1.308-310.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek T. L., Nowak J. A., Maniloff J. Mycoplasma restriction: identification of a new type of restriction specificity for DNA containing 5-methylcytosine. J Bacteriol. 1986 Jan;165(1):219–225. doi: 10.1128/jb.165.1.219-225.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatko B. E., Benner J. S., Jager-Quinton T., Moran L. S., Simcox T. G., Van Cott E. M., Wilson G. G. Cloning, sequencing and expression of the Taq I restriction-modification system. Nucleic Acids Res. 1987 Dec 10;15(23):9781–9796. doi: 10.1093/nar/15.23.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H., Rueter T., Geiger R. DNA cleavage by AatI and StuI is sensitive to Escherichia coli dcm methylation. Nucleic Acids Res. 1988 Mar 25;16(6):2718–2718. doi: 10.1093/nar/16.6.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeck R. E. Single-strand and double-strand cleavage at half-modified and fully modified recognition sites for the restriction nucleases Sau3a and Taqi. Gene. 1980 Dec;12(3-4):267–275. doi: 10.1016/0378-1119(80)90109-2. [DOI] [PubMed] [Google Scholar]
- Strobl J. S., Thompson E. B. Methylation of either cytosine in the recognition sequence CGCG inhibits ThaI cleavage of DNA. Nucleic Acids Res. 1984 Nov 12;12(21):8073–8083. doi: 10.1093/nar/12.21.8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugisaki H., Maekawa Y., Kanazawa S., Takanami M. New restriction endonucleases from Acetobacter aceti and Bacillus aneurinolyticus. Nucleic Acids Res. 1982 Oct 11;10(19):5747–5752. doi: 10.1093/nar/10.19.5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sznyter L. A., Slatko B., Moran L., O'Donnell K. H., Brooks J. E. Nucleotide sequence of the DdeI restriction-modification system and characterization of the methylase protein. Nucleic Acids Res. 1987 Oct 26;15(20):8249–8266. doi: 10.1093/nar/15.20.8249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szomolányi E., Kiss A., Venetianer P. Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene. 1980 Aug;10(3):219–225. doi: 10.1016/0378-1119(80)90051-7. [DOI] [PubMed] [Google Scholar]
- Theriault G., Roy P. H., Howard K. A., Benner J. S., Brooks J. E., Waters A. F., Gingeras T. R. Nucleotide sequence of the PaeR7 restriction/modification system and partial characterization of its protein products. Nucleic Acids Res. 1985 Dec 9;13(23):8441–8461. doi: 10.1093/nar/13.23.8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thériault G., Roy P. H. Cloning of Pseudomonas plasmid pMG7 and its restriction-modification system in Escherichia coli. Gene. 1982 Oct;19(3):355–359. doi: 10.1016/0378-1119(82)90026-9. [DOI] [PubMed] [Google Scholar]
- Tran-Betcke A., Behrens B., Noyer-Weidner M., Trautner T. A. DNA methyltransferase genes of Bacillus subtilis phages: comparison of their nucleotide sequences. Gene. 1986;42(1):89–96. doi: 10.1016/0378-1119(86)90153-8. [DOI] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Günthert U., Canosi U., Jentsch S., Freund M. Restriction and modification in Bacillus subtilis: identification of a gene in the temperate phage SP beta coding for a BsuR specific modification methyltransferase. Mol Gen Genet. 1980;180(2):361–367. doi: 10.1007/BF00425849. [DOI] [PubMed] [Google Scholar]
- Urieli-Shoval S., Gruenbaum Y., Razin A. Sequence and substrate specificity of isolated DNA methylases from Escherichia coli C. J Bacteriol. 1983 Jan;153(1):274–280. doi: 10.1128/jb.153.1.274-280.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heuverswyn H., Fiers W. Nucleotide sequence of the Hind-C fragment of simian virus 40 DNA. Comparison of the 5'-untranslated region of wild-type virus and of some deletion Mutants. Eur J Biochem. 1979 Oct;100(1):51–60. doi: 10.1111/j.1432-1033.1979.tb02032.x. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Dobritsa A. P. On the nature of the cytosine-methylated sequence in DNA of Bacillus brevis var. G.-B. Biochim Biophys Acta. 1975 Sep 12;407(1):61–72. doi: 10.1016/0005-2787(75)90023-4. [DOI] [PubMed] [Google Scholar]
- Vinogradova M. N., Gromova E. S., Uporova T. M., Nikol'skaia I. I., Shabarova Z. A. Endonukleaza restriktsii SsoII: vzaimodeistvie s modifitsirovannymi substratami. Dokl Akad Nauk SSSR. 1987;295(3):732–736. [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. MspI, an isoschizomer of hpaII which cleaves both unmethylated and methylated hpaII sites. Nucleic Acids Res. 1978 Sep;5(9):3231–3236. doi: 10.1093/nar/5.9.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R. Y., Hartley J. L., Donelson J. E., Walder J. A. Cloning and expression of the Pst I restriction-modification system in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1503–1507. doi: 10.1073/pnas.78.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R. Y., Hartley J. L., Donelson J. E., Walder J. A. Cloning of the Pst I restriction-modification system. Gene Amplif Anal. 1981;1:217–227. [PubMed] [Google Scholar]
- Walder R. Y., Langtimm C. J., Chatterjee R., Walder J. A. Cloning of the MspI modification enzyme. The site of modification and its effects on cleavage by MspI and HpaII. J Biol Chem. 1983 Jan 25;258(2):1235–1241. [PubMed] [Google Scholar]
- Walder R. Y., Walder J. A., Donelson J. E. The organization and complete nucleotide sequence of the PstI restriction-modification system. J Biol Chem. 1984 Jun 25;259(12):8015–8026. [PubMed] [Google Scholar]
- Wang R. Y., Zhang X. Y., Khan R., Zhou Y. W., Huang L. H., Ehrlich M. Methylated DNA-binding protein from human placenta recognizes specific methylated sites on several prokaryotic DNAs. Nucleic Acids Res. 1986 Dec 22;14(24):9843–9860. doi: 10.1093/nar/14.24.9843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil M. D., McClelland M. Enzymatic cleavage of a bacterial genome at a 10-base-pair recognition site. Proc Natl Acad Sci U S A. 1989 Jan;86(1):51–55. doi: 10.1073/pnas.86.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead P. R., Jacobs D., Brown N. L. Restriction endonucleases from Herpetosiphon giganteus: an example of the evolution of DNA recognition specificity? Nucleic Acids Res. 1986 Sep 11;14(17):7031–7045. doi: 10.1093/nar/14.17.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Levy D., Perucho M. The somatic replication of DNA methylation. Cell. 1981 Apr;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- Woodcock D. M., Crowther P. J., Diver W. P., Graham M., Bateman C., Baker D. J., Smith S. S. RglB facilitated cloning of highly methylated eukaryotic DNA: the human L1 transposon, plant DNA, and DNA methylated in vitro with human DNA methyltransferase. Nucleic Acids Res. 1988 May 25;16(10):4465–4482. doi: 10.1093/nar/16.10.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. N., Burbank D. E., Uher L., Rabussay D., Van Etten J. L. IL-3A virus infection of a Chlorella-like green alga induces a DNA restriction endonuclease with novel sequence specificity. Nucleic Acids Res. 1987 Aug 11;15(15):6075–6090. doi: 10.1093/nar/15.15.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. N., Burbank D. E., Uher L., Rabussay D., Van Etten J. L. Restriction endonuclease activity induced by PBCV-1 virus infection of a Chlorella-like green alga. Mol Cell Biol. 1986 May;6(5):1430–1439. doi: 10.1128/mcb.6.5.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. N., Morgan R., Schildkraut I., Van Etten J. L. A site-specific single strand endonuclease activity induced by NYs-1 virus infection of a Chlorella-like green alga. Nucleic Acids Res. 1988 Oct 25;16(20):9477–9487. doi: 10.1093/nar/16.20.9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. N., Narva K. E., Van Etten J. L. The cleavage site of the RsaI isoschizomer, CviII, is G decreases TAC. Nucleic Acids Res. 1987 Dec 10;15(23):10063–10063. doi: 10.1093/nar/15.23.10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y. N., Van Etten J. L. DNA methyltransferase induced by PBCV-1 virus infection of a Chlorella-like green alga. Mol Cell Biol. 1986 May;6(5):1440–1445. doi: 10.1128/mcb.6.5.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Burbank D. E., Van Etten J. L. Restriction endonuclease activity induced by NC-1A virus infection of a Chlorella-like green alga. Nucleic Acids Res. 1986 Aug 11;14(15):6017–6030. doi: 10.1093/nar/14.15.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yolov A. A., Vinogradova M. N., Gromova E. S., Rosenthal A., Cech D., Veiko V. P., Metelev V. G., Kosykh V. G., Buryanov Y. I., Bayev A. A. Interaction of EcoRII restriction and modification enzymes with synthetic DNA fragments. VI. The binding and cleavage of substrates containing nucleotide analogs. Nucleic Acids Res. 1985 Dec 20;13(24):8983–8998. doi: 10.1093/nar/13.24.8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo O. J., Agarwal K. L. Isolation and characterization of two proteins possessing Hpa II methylase activity. J Biol Chem. 1980 Jul 10;255(13):6445–6449. [PubMed] [Google Scholar]
- Yoshimori R., Roulland-Dussoix D., Boyer H. W. R factor-controlled restriction and modification of deoxyribonucleic acid: restriction mutants. J Bacteriol. 1972 Dec;112(3):1275–1279. doi: 10.1128/jb.112.3.1275-1279.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Hammer S. M., Hirsch M. S., Mulder C. Methylation of the viral genome in an in vitro model of herpes simplex virus latency. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2207–2210. doi: 10.1073/pnas.79.7.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Mulder C. Detection of methylated sequences in eukaryotic DNA with the restriction endonucleases Smai and Xmai. J Mol Biol. 1981 Jul 25;150(1):133–136. doi: 10.1016/0022-2836(81)90328-4. [DOI] [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Kilpatrick M. W., Wells R. D. HhaI methylase and restriction endonuclease as probes for B to Z DNA conformational changes in d(GCGC) sequences. Nucleic Acids Res. 1984 Oct 25;12(20):7677–7692. doi: 10.1093/nar/12.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieger M., Patillon M., Roizes G., Lerouge T., Dupret D., Jeltsch J. M. Two restriction endonucleases from Bacillus sphaericus: BspXI and BspXII. Nucleic Acids Res. 1987 May 11;15(9):3919–3919. doi: 10.1093/nar/15.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]


