Abstract
Wild-type p53 is a stress-responsive tumor suppressor and potent growth inhibitor. Genotoxic stresses (e.g. ionizing and UV radiation or chemotherapeutic drug treatment) can activate p53, but also induce mutations in the P53 gene and thus select for p53-mutated cells. Nutlin-3a (Nutlin) is pre-clinical drug that activates p53 in a non-genotoxic fashion. Nutlin occupies the p53-binding pocket of MDM2, activating p53 by blocking the p53-MDM2 interaction. Because Nutlin neither binds p53 directly nor introduces DNA damage, we hypothesized Nutlin would not induce P53 mutations and therefore not select for p53-mutated cells. To test this, populations of SJSA-1 (p53 wild-type) cancer cells were expanded that survived repeated Nutlin exposures, and individual clones were isolated. Group 1 clones were resistant to Nutlin-induced apoptosis, but still underwent growth-arrest. Surprisingly, while some Group 1 clones retained wild-type p53, others acquired a heterozygous p53 mutation. Apoptosis resistance in Group 1 clones was associated with decreased PUMA induction and decreased caspase 3/7 activation. Group 2 clones were resistant to both apoptosis and growth-arrest induced by Nutlin. Group 2 clones had acquired mutations in the p53 DNA-binding domain and expressed only mutant p53s that were induced by Nutlin treatment, but were unable to bind the P21 and PUMA gene promoters, and unable to activate transcription. These results demonstrate that non-genotoxic p53 activation (e.g. by Nutlin treatment) can lead to the acquisition of somatic mutations in p53 and select for p53-mutated cells. These findings have implications for the potential clinical use of Nutlin and other small molecule MDM2 antagonists.
Introduction
Wild-type p53 is a stress-activated tumor suppressor. P53 is normally expressed at low levels and inactive due to the action of MDM2, an E3 ubiquitin-ligase that binds p53 and promotes its degradation (Haupt et al., 1997; Kubbutat et al., 1997). However, the p53 protein is stabilized in response to stresses, such as DNA damage and inappropriate oncogene signaling, that might otherwise predispose a normal cell toward carcinogenesis (Horn and Vousden, 2007; Maki and Howley, 1997; Maltzman and Czyzyk, 1984). The stress-induced stabilization of p53 results from disruption of p53-MDM2 binding and/or inhibition of MDM2 E3 ligase activity. The majority of stabilized p53 accumulates in the nucleus where it functions as a transcription factor, activating expression of genes that cause cell cycle arrest (P21) or apoptosis (PUMA, bax, Noxa) (Brown et al., 2007). A smaller portion of p53 accumulates in mitochondria, where it interacts with pro- and anti-apoptotic members of the Bcl-2 family, resulting in release of factors from mitochondria that drive apoptosis (Mihara et al., 2003; Vaseva et al., 2009). Thus, p53 eliminates cells with potentially cancer-promoting lesions by either inhibiting their growth or causing them to die. In light of this, it is not surprising that P53 gene status often correlates with the responsiveness of cancer cells to radiation and other therapeutic agents. In several reports, p53 wild-type cancer cells respond better to DNA damaging therapeutics than p53 mutated or p53-null cancer cells, due to activation of wild-type p53 growth inhibitory pathways (McDermott et al., 2005; Wu and Ding, 2002; Wu and El-Deiry, 1996).
Inactivation of p53 is considered essential for the development of most human cancers. Approximately 50% of cancers harbor inactivating P53 gene mutations (Hollstein et al., 1991). While radiation and other DNA damaging stresses can activate p53, they also exert pressure for mutation acquisition on p53, and thus can select for p53-mutated cells. This is perhaps best illustrated in therapy-induced, secondary cancers. For example, Hodgkins disease patients treated with radiotherapy have an increased risk of developing lung cancer after treatment. These lung cancers often have acquired mutations in p53 that appear to have resulted from the direct mutagenic effects of radiotherapy (De Benedetti et al., 1996). Similarly, ovarian cancer patients treated with platinum-based drugs are at risk for secondary leukemias that often harbor p53 mutations. P53 sequence analyses in these leukemias are consistent with the mutations resulting from direct damage to the p53 gene by the platinum drugs (Leonard et al., 2002). Finally, p53 mutations are common in secondary leukemias that arise in patients treated with alkylating agents, most likely due to alkylating agent-induced mutations in the p53 gene (Christiansen et al., 2001). Thus, radiation and DNA damaging chemotherapeutics can activate p53, but can also induce mutations in the P53 gene and thus select for p53 mutated cells. A potentially adverse side effect of DNA damaging therapeutic drug treatment is the development of secondary cancers which are associated with therapy-induced mutations in p53.
Nutlin-3a (Nutlin) is a small molecule MDM2 antagonist that occupies the p53 binding pocket in MDM2, effectively blocking the p53-MDM2 interaction (Vassilev et al., 2004). This results in the stabilization and activation of p53. Thompson et al. (Thompson et al., 2004) monitored p53 phosphorylation at six key serine residues (Ser (6), Ser (15), Ser (20), Ser (37), Ser (46), and Ser (392)) in cells in which p53 was induced by either genotoxic stresses (doxorubicin or etoposide) or induced by Nutlin. P53 phosphorylations induced by genotoxic stress were not observed in cells in which p53 was induced by Nutlin. This led to the conclusion, subsequently supported by other studies (Drakos et al., 2007; Kumamoto et al., 2008), that Nutlin stabilizes p53 in a non-genotoxic fashion, as would be expected from simply blocking the binding between p53 and MDM2. The fact that Nutlin can activate the p53 pathway in a non-genotoxic fashion is attractive from a therapeutic standpoint. As mentioned above, most cancer therapeutics cause DNA damage, drawbacks being the potential for collateral damage to normal surrounding tissue and the potential for secondary malignancies. By activating p53 through a non-genotoxic fashion, the use of Nutlin as a therapeutic agent would presumably be without these potential drawbacks.
Because Nutlin neither binds p53 directly nor introduces DNA damage, we hypothesized Nutlin would not induce mutations and would not select for p53 mutated cells. To test this hypothesis, populations of SJSA-1 (p53 wild-type) cancer cells were expanded that survived repeated Nutlin exposures, and individual clones isolated. Group 1 clones were resistant to Nutlin-induced apoptosis, but still underwent growth arrest. Surprisingly, while some Group 1 clones retained wild-type p53, others had acquired a heterozygous mutation in the p53 DNA binding domain. Apoptosis resistance in these Group 1 clones was associated with decreased induction of the pro-apoptotic p53 target gene PUMA, and decreased caspase 3/7 activation. Other clones (Group 2) were resistant to both apoptosis and growth arrest induced by Nutlin. Group 2 clones had also acquired mutations in the p53 DNA binding domain and expressed only mutated p53. These mutant p53s were induced by Nutlin treatment, but were unable to bind the P21 and PUMA gene promoters, and unable to activate transcription. These results demonstrate that non-genotoxic stresses (e.g. Nutlin-3a treatment) can lead to the acquisition of somatic mutations in p53 and select for p53 mutated cells. These findings have implications for the potential clinical use of Nutlin and other small molecule MDM2 antagonists.
Results
Selection of Nutlin-Resistant SJSA-1 cell populations
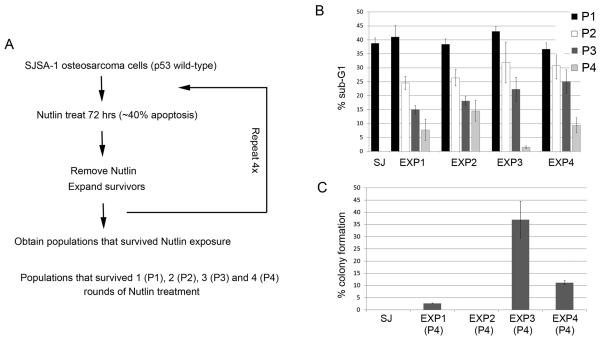
SJSA-1 is a p53 wild-type osteosarcoma cell line that undergoes apoptosis as one of its primary responses to Nutlin (Vassilev et al 2004). In initial experiments, 1×107 SJSA-1 cells were plated into 5 separate 10 cm dishes (2×106 cells per dish). The cells were cultured in the continued presence of Nutlin (10 μM) and allowed to grow for a 2-3 week period. Zero colonies formed (data not shown). This demonstrated the parental SJSA-1 population does not contain Nutlin-resistant clones. In parallel experiments, 2×106 SJSA-1 cells were treated with Nutlin (10 μM) for 3 days. At this time point, ~40% of the cells were apoptotic, determined by sub-G1 DNA content. The cells were then rinsed to remove the Nutlin, and the remaining cells were expanded in normal medium (minus Nutlin). The process was repeated four times, and populations that survived 1-4 rounds of Nutlin treatment were obtained (P1-P4, Fig 1A). We compared the extent to which SJSA-1 cells and the P1-P4 populations underwent apoptosis when treated for 3 days with Nutlin. The results indicated that the selected populations became progressively more resistant to apoptosis (Fig 1B). Thus, whereas parental SJSA-1 cells underwent apoptosis to relatively high extents (~40% apoptosis) after 3 days Nutlin treatment, the P4 populations displayed only minimal apoptosis when similarly treated (~10% apoptosis in P4 from Exp 1). We repeated these studies in 4 separate experiments and in each experiment Nutlin resistant populations were obtained (Fig 1B).
Figure 1. Selection of Nutlin-resistant SJSA-1 populations.
A. The procedure used to select SJSA-1 populations resistant to Nutlin-induced apoptosis. B. SJSA-1 (SJ) was obtained from American Type Culture Collection and grown in RPMI1640 medium (100 units/mL penicillin, 100 μg/mL streptomycin, 10% FBS). SJSA-1 and Nutlin-selected populations (P1, P2, P3 and P4) were treated with 10 μM Nutlin-3 (Nutlin, Sigma, USA) for 72 hrs. Cells were harvested, fixed in 25% ethanol, stained with propidium iodide and subjected to FACS analysis as previously described (Shen et al., 2008). Percentage of cells with sub-G1 DNA content was determined from the DNA profile histogram using FlowJo (Treestar Inc., USA). The mean of three independent experiments is shown ± S.E. (error bars). C. SJSA-1 (SJ) and Nutlin-resistant populations (P4) were plated at low density (1×102 - 1×105 cells / 10cm dish). Cells were either untreated or treated with 10 μM Nutlin and allowed to grow for a 2-3 week period. Colonies were stained with 0.5% Crystal Violet and counted. The plating efficiency for untreated sample was set at 100%. The mean of three independent experiments is shown ± S.E. (error bars).
Next, P4 populations and SJSA-1 parental cells were seeded at increasing densities (1×102-1×105 cells per 10 cm dish) and compared for their ability to grow into colonies when cultured in the continued presence of Nutlin (10 μM). While SJSA-1 cells did not form colonies, the P4 populations from each experiment were able to form colonies in the presence of Nutlin to varying extents (Fig 1C). In the presence of Nutlin, the P4 population from Experiment 1 had ~3% colony forming ability, the P4 population from Experiment 2 had 0.008% colony forming ability, the P4 population from Experiment 3 had ~35% colony forming ability, and the P4 population from Experiment 4 had ~10% colony forming ability. Taken together, the results of Figs 1B and 1C suggest the majority of cells within the P4 populations are resistant to Nutlin-induced apoptosis but unable to grow in the continued presence of Nutlin. Nonetheless, within each P4 population resides a smaller percentage of cells that appear resistant to both apoptosis and growth arrest, and these cells are able to grow and form colonies in the presence of Nutlin.
Selection of Nutlin-Resistant SJSA-1 cell clones
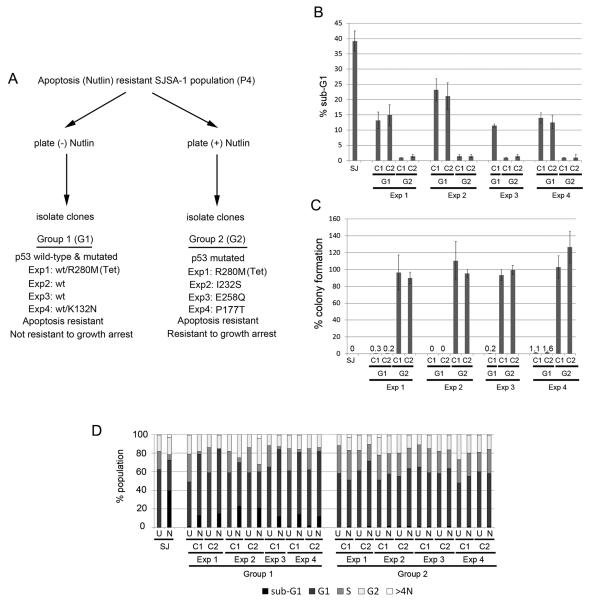
To examine this further, we isolated clones from P4 populations that were plated in absence or presence of Nutlin. We separated the clones into 2 groups (Group 1 and Group 2) based on whether the clones were isolated in the absence or presence of Nutlin (Fig 2A). Group 1 clones were isolated in the absence of Nutlin. We prepared cDNA from each clone and used it to sequence the entire p53 coding region. All Group 1 clones from Experiments 2 and 3 express wild-type p53 and appear to have normal, diploid DNA content based on flow cytometry analysis. In contrast, Group 1 clones from Experiment 1 have a heterozygous mutation in p53 (WT/R280M) and tetraploid DNA content. The majority of Group 1 clones from Experiment 4 have the p53 heterozygous mutation (WT/K132N), though one clone is p53 wild-type and another has the p53 heterozygous mutation (WT/P177T). Group 2 clones were isolated in the presence of Nutlin and express only mutant p53. The p53 mutants expressed in the Group 2 clones from each experiment are as follows: Experiment 1, p53R280M; Experiment 2, p53I232S; Experiment 3, p53E258Q; Experiment 4, four out of 5 clones p53P177T, 1 out of 5 clones p53K132N. The Group 2 clones from Experiment 1 are tetraploid, while Group 2 clones from Experiments 2, 3, and 4 are diploid. The p53 sequence of all clones isolated in each experiment is provided in Supplemental Table I.
Figure 2. Cell cycle and colony formation analysis in selected Nutlin-Resistant clones.
A. Selection of Nutlin-Resistant SJSA-1 cell clones. P4 populations from Experiments 1-4 were plated in the absence or presence of Nutlin for 14 days. Individual colonies were then isolated and expanded. Group 1 clones (isolated in the absence of Nutlin) undergo growth arrest in the presence of Nutlin but are resistant to Nutlin-induced apoptosis. Group 2 clones (isolated in the presence of Nutlin) are resistant to both apoptosis and growth arrest induced by Nutlin. The p53 status of the Group 1 and Group 2 clones from each experiment that were used in subsequent experiments was determined by cDNA sequencing and is indicated. The p53 sequence of all clones isolated in each experiment is listed in Supplemental Table 1. B, D. SJSA-1 (SJ) and Nutlin-resistant Group 1 (G1) and Group 2 (G2) clones were untreated or treated with 10 μM Nutlin for 72 hrs and subjected to FACS analysis. The percentage of cells with sub-G1 DNA content (B) and cell cycle distribution (D) were determined from DNA profile histograms using FlowJo. U: untreated; N: Nutlin treated. C. SJSA-1 (SJ) and Nutlin-resistant Group 1 and Group 2 clones were plated at low density (1×102 - 1×105 cells / 10cm dish) and either untreated or treated with 10 μM Nutlin for 14-21 days. Colony forming ability was determined as described in Figure 1.
Next, we examined individual Group 1 and 2 clones from each experiment for apoptosis and colony forming ability in the presence of Nutlin. As shown in Fig 2B, the Group 1 and 2 clones from each experiment were resistant to Nutlin-induced apoptosis compared to parental SJSA-1 cells. When treated with Nutlin for 3 days, SJSA-1 parental cells displayed ~40% apoptosis (determined by sub-G1 DNA content), Group 1 clones displayed ~10-20% apoptosis, and Group 2 clones displayed ~0-2% apoptosis. Cell cycle profiles demonstrated Group 1 clones were cell-cycle arrested when treated with Nutlin for 3 days, evidenced by increased percentage of G1-phase cells and depletion of cells in S-phase, whereas Group 2 clones were not cell cycle arrested (Fig 2D). To determine colony forming ability, between 1×102 and 1×105 cells from either SJSA-1 parental or each of the Group 1 and 2 clones were cultured in the continued presence of 10 μM Nutlin and allowed to grow over a 2-3 week period. In these experiments, SJSA-1 parental cells did not form colonies (0% colony forming ability), and individual Group 1 clones also displayed very low colony forming ability (<2%) (Fig 2C). In contrast, Group 2 clones from each experiment had ~100% colony forming ability in the presence of Nutlin (Fig 2C). Taken together, the results in Figure 2 demonstrate Group 1 clones are resistant to Nutlin-induced apoptosis but not resistant to Nutlin-induced growth arrest, while Group 2 clones are fully resistant to both apoptosis and growth arrest induced by Nutlin.
P53 transcriptional activity and caspase activation in Group 1 and Group 2 clones
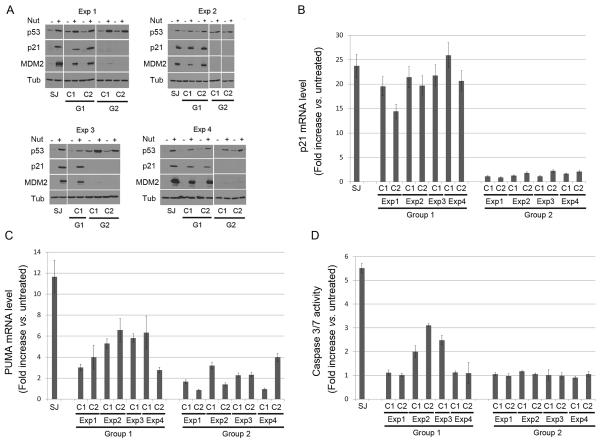
We wished to examine the mechanisms for Nutlin resistance in the Group 1 and Group 2 clones. To this end, we first tested whether expression of p53 and its downstream target genes were increased by Nutlin treatment in these clones. SJSA-1 parental cells and each of the clones were untreated or treated with Nutlin (10 μM) for 24 hrs, followed by immunoblotting for p53, p21, and MDM2 (Fig 3A). P53 was expressed at low levels in untreated cells but increased in response to Nutlin in SJSA-1 parental cells and in all the clones. The fact that p53 levels increased in response to Nutlin indicates MDM2 is degrading p53 in the clones and keeping it at a low level. In tumors, mutant p53 is usually stable, expressed at high levels, and resistant to MDM2-mediated degradation (Kubbutat et al., 1997; Midgley and Lane, 1997; Peng et al., 2001). The finding that mutant p53 increased in Nutlin-treated clones was therefore somewhat surprising. However, the notion that mutant p53 can be degraded by MDM2 is not without precedent. For example, Lozano and colleagues (Terzian et al., 2008) recently described a mutant p53 knock-in mouse in which mutant p53 was expressed at low levels (unstable) in normal tissues and some tumors unless MDM2 (or p16Ink4a) were absent. In Group 1 clones, p21 and MDM2 levels were increased with Nutlin treatment to levels comparable to or only slightly less than parental SJSA-1 cells, demonstrating p53 in these Group 1 clones is transcriptionally active. The high level of p21 induction in these clones after Nutlin treatment is consistent with them being cell cycle arrested (Fig 2D). In contrast, p21 and MDM2 levels were not increased with Nutlin treatment in Group 2 clones, suggesting the p53 in these clones is unable to activate transcription. Next, we used RT-PCR to address whether mRNA levels for p21 or the pro-apoptotic p53 target gene PUMA were increased in response to Nutlin treatment. As shown in Fig 3B, p21 mRNA levels increased after Nutlin treatment in Group 1 clones to levels only slightly less than Nutlin treated SJSA-1 parental cells, but did not increase after Nutlin treatment in Group 2 clones. PUMA mRNA levels also increased after Nutlin treatment in the Group 1 clones, though again to a lesser extent than in SJSA1 parental cells (Fig 3C). This suggests that decreased apoptosis in Group 1 clones could result, in part, from decreased expression of PUMA. In contrast, PUMA mRNA levels did not increase or were only slightly increased after Nutlin treatment in the Group 2 clones. Finally, since the execution of apoptosis results largely from caspase activation, we monitored caspase 3/7 activity in untreated and Nutlin treated SJSA-1 cells and the Group 1 and 2 clones (Fig 3D). The results show that caspases 3/7 were highly activated in Nutlin treated SJSA-1 cells, less activated or not activated in the Nutlin treated Group 1 clones, and not activated in the Nutlin treated Group 2 clones. Thus, apoptosis resistance in the Group 1 and 2 clones is associated with decreased PUMA expression and decreased activation of caspases 3/7,
Figure 3. P53 transcriptional activity and caspase activation in Nutlin-resistant clones.
SJSA-1 (SJ) and Nutlin-resistant Group 1 and Group 2 clones were untreated or treated with 10 μM Nutlin for 24 hrs. A. Cell lysates were analyzed by immunoblotting with antibodies against p53, MDM2, and p21, as previously described (Shen et al 2008). Tubulin levels were used as a loading control. Representative immunoblots are shown. B, C. Quantitative real-time PCR was performed to measure mRNA levels of p21 (B) and PUMA (C) in SJSA-1 and Nutlin-resistant clones. The complementary DNA (cDNA) was synthesized after treatment. The quantitative real-time PCR reaction was run in a 7300 Real Time PCR System (Applied Biosystems, Foster, CA) using SybrGreen PCR master mix, (Applied Biosystems, Foster City, CA) following manufacturer’s instructions. Thermocycling was done in a final volume of 20 μL containing 2 μL of cDNA and 400 nmol/L of primers (Primers are listed in Supplemental Table 2). All samples were amplified in triplicate using the following cycle scheme: 95°C for 2 minutes, 40 cycles of 95°C for 15 seconds and 55°C for 60 seconds. Fluorescence was measured in every cycle and mRNA levels were normalized using the GAPDH values in all samples. A single peak was obtained for targets, supporting the specificity of the reaction. The fold increase (compared to untreated levels) in p21 and PUMA mRNA was determined after 24hr Nutlin treatment. Data is presented as the mean fold increase ± S.E. (n=3). D. Caspase-Glo-3/7 Assay was performed with untreated and Nutlin-treated (10 μM Nutlin for 24 hrs) SJSA-1 and Nutlin-resistant clones with the Caspase-Glo-3/7 Assay (Promega Biotech, Madison, WI) per manufacture’s instruction. Results are presented as the mean (signal-to-noise ratios) ± SEM of the triplicate assays.
P53 mutants expressed in Group 2 clones are unable to bind DNA and activate transcription
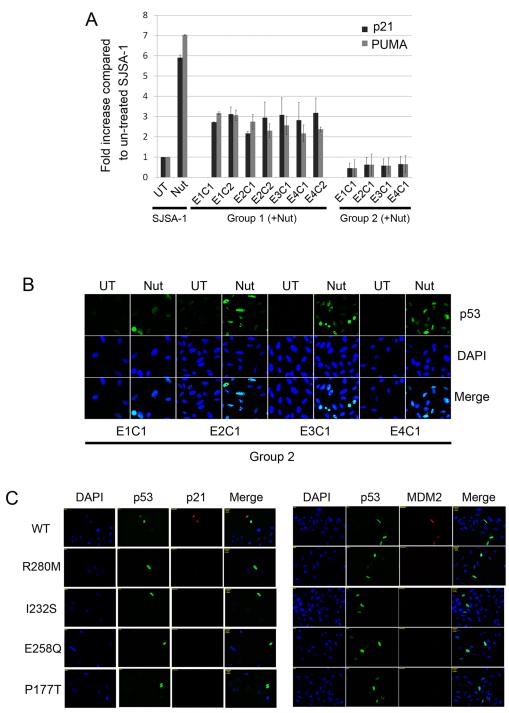
Cancer-associated mutations in p53 are found almost exclusively in the p53 DNA binding domain (Hollstein et al., 1991) and inhibit the ability of p53 to bind DNA and activate transcription. Group 2 clones express only mutant forms of p53 and, in each clone, the mutation is within the DNA binding domain. We wished to test/confirm these mutations destroyed p53 activity. First, we carried out chromatin-immunoprecipitation (ChIP) experiments to determine whether mutant p53s in group 2 clones had lost their ability to bind DNA (Fig 4A). SJSA-1 parental cells that were Nutlin treated for 24 hr showed a 5-7 fold increase in the binding of p53 to either the P21 or PUMA gene promoters. In contrast, Nutlin treated Group 2 clones showed little or no p53 binding to the P21 or PUMA gene promoters, demonstrating the mutant p53 proteins expressed in Group 2 clones are unable to bind DNA. In Group 1 clones that were Nutlin treated, p53 showed an intermediate level of binding to the P21 and PUMA gene promoters, consistent with the intermediate level of P21 and PUMA mRNA induction in these clones (Figs 3B, 3C). Next, we used immunofluorescence staining to show that the mutant p53s expressed in Group 2 clones accumulated in the nucleus after Nutlin treatment (Fig 4B). This indicates their inability to bind the P21 and PUMA gene promoters does not result from lack of nuclear accumulation. P53 in Group 1 clones also accumulated in the nucleus after Nutlin treatment (Supp Fig 1). Finally, expression vectors were generated encoding p53s that were either wild-type or contained the mutations identified in Group 2 clones (p53R280M, p53I232S, p53E258Q and p53P177T). Saos-2 cells (p53-null) were transfected with each p53 DNA and then examined by immunofluoresence co-staining with either p53 and p21, or p53 and MDM2, antibodies 48 hr after transfection. This allowed us to determine whether the p53s expressed specifically in the transfected cells could activate expression of the endogenous P21 and MDM2 genes. As shown in Fig 4C, p21 and MDM2 protein levels were increased in cells transfected with wild-type p53 but not with the various p53 mutants, indicating the mutant p53s lacked the ability to activate gene transcription. We conclude mutant p53s identified in Group 2 clones lacked the ability to bind DNA and activate transcription.
Figure 4. Mutant p53s in Nutlin-resistant clones lack DNA binding ability and transcription activity.
A. SJSA-1, Group 1 and Group 2 clones were untreated or Nutlin-treated (10 μM) for 24 hrs. Chromatin immunoprecipitation (ChIP) was performed using Human p53 ChampionChIP Antibody kit (SABiosciences/Qiagen, Frederick, MD). Briefly, formaldehyde cross-linking was performed for 10 min, and samples were sonicated to obtain DNA fragments with average size of 400–500 bp. Protein–DNA complexes were immunoprecipitated using p53 antibody (SABiosciences/Qiagen, Frederick, MD). DNAs were purified and subjected to quantitative real-time PCR amplification. The primers used are listed in Supplemental Table 2. The fold-increase ± S.E. in p53 binding to the P21 and PUMA gene promoters after 24 hrs Nutlin treatment is plotted (n=3). The level of p53 binding to each promoter in untreated SJSA-1 parental cells was given a value of 1.0, and everything else scored relative to that. B. Group 2 clones were untreated or Nutlin-treated (10 μM) for 24 hrs. Cells were fixed with 4% formaldehyde and subjected to immunofluorescence with anti-p53 antibody as described (Shen et al., 2008). Cell nuclei were counterstained with DAPI (blue). Representative images were captured at ×40 magnification and are shown. UT: untreated; Nut: Nutlin treated. C. Site directed mutations in wild-type (WT) p53 plasmid were constructed using QuikChange mutagenesis kit (Stratagene La Jolla, CA) for the mutations R280M, I232S, E258Q and P177T (Primers used for mutagenesis listed in Supplemental Table 2). Plasmids encoding wild-type p53, p53R280M, p53I232S, p53E258Q and p53P177T were transfected into Saos-2 (p53-null) cells using FuGENE-6 transfection reagent. Saos-2 cells were fixed 48 hrs after transfection and subjected to immunofluorescence with indicated antibodies.
Discussion
Wild-type p53 is a stress-activated tumor suppressor and potent inhibitor of cell growth. In most normal cells, p53 is expressed at low levels and inactive due to the action of MDM2, an E3 ubiquitin-ligase that binds p53 and promotes its degradation (Haupt et al 1997, Kubbutat et al 1997). However, the p53 protein is stabilized in response to stresses, such as DNA damage (genotoxic stress) and inappropriate oncogene signaling, that might otherwise predispose a normal cell toward carcinogenesis (Horn and Vousden 2007, Maki and Howley 1997, Maltzman and Czyzyk 1984). This stress-induced stabilization of p53 results from disruption of p53-MDM2 binding or from inhibition of MDM2 E3 ligase activity. Genotoxic stresses (e.g. ionizing and UV radiation or chemotherapeutic drug treatment) can activate p53, but can also induce mutations in the P53 gene and thus select for p53-mutated cells. Nutlin-3a (Nutlin) is a preclinical drug that binds MDM2 and prevents the interaction between MDM2 and p53, leading to the stabilization and activation of p53 (Vassilev et al., 2004). Because Nutlin neither binds p53 directly nor induces DNA damage, we hypothesized that Nutlin would not induce mutations and therefore not select for p53-mutated cells. To test this hypothesis, we expanded SJSA-1 (p53 wild-type) cell populations that survived repeated exposures to Nutlin, and then characterized individual clones from the surviving populations. Surprisingly, we found that the majority of Nutlin-selected clones had acquired inactivating mutations in the p53 DNA binding domain. The fact Nutlin-selected clones acquired mutant p53 demonstrates that Nutlin is very effective and selective against p53. Our findings demonstrate non-genotoxic p53 activation (e.g. by Nutlin treatment) can lead to the acquisition of somatic mutations in p53 and select for p53-mutated cells.
An important consideration is whether the Nutlin-resistant Group 1 and Group 2 clones were already present in the SJSA-1 parental cells, or arose during the Nutlin treatment course. Group 1 clones underwent growth arrest when Nutlin treated but were resistant to Nutlin-induced apoptosis. In two experiments the Group 1 clones expressed only wild-type p53 despite being resistant to Nutlin-induced apoptosis. It is possible these Group 1 clones were already present in the parental SJSA-1 cell population, and were simply selected for and expanded with repeated Nutlin treatments. In contrast, in two other experiments Group 1 clones acquired heterozygous mutations in p53 and expressed both wild-type and mutant forms of the protein (p53 WT/R280M for Exp 1, and p53 WT/K132N for Exp 4). The fact that different mutations were detected in two separate experiments argues that these mutated cells arose during the Nutlin treatment course and were not already present in the population. We also believe that the Group 2 clones acquired the mutations/changes that made them resistant to Nutlin during the Nutlin treatment course. We believe this for two reasons: First, Group 2 clones arose from experiments in which 2×106 cells were Nutlin treated for 3 days to induce death, surviving cells expanded, and the process repeated. The Group 2 clones must have arose from these initial 2×106 cells. In contrast, in parallel experiments 1×107 parental SJSA-1 cells were plated in the continued presence of Nutlin (2×106 cells in 5 separate dishes), and zero Nutlin-resistant colonies were obtained. If fully resistant Group 2 clones were already present in the starting SJSA-1 population, we would have expected them to grow into colonies in this experiment, but they did not. Thus, the p53 mutations that allow Group 2 clones to grow in Nutlin must have been acquired during the Nutlin-treatment course. Second, while SJSA-1 cells express wild-type p53, the Group 2 clones have inactivating mutations in the p53 DNA binding domain. Moreover, different p53 mutations were obtained in four separate experiments (Exp 1, p53R280M; Exp 2, p53I232S, Exp 3, p53E258Q, Exp 4, p53P177T). The fact that different p53 mutations were obtained in 4 separate experiments argues strongly that the p53 mutant cells were not present in the starting population but arose during the Nutlin treatment course.
Our findings invite comparison to other non-genotoxic stresses that can activate p53. At early stages of cancer development, mutations are incurred that cause constitutive activation of oncogenes. This imposes an oncogenic stress that drives hyper-proliferation, but also triggers a p53-dependent checkpoint that is mediated by p14/Arf and halts cell division (Efeyan and Serrano, 2007; Sherr and Weber, 2000). Notably, oncogenic stress does not directly cause DNA damage, but instead activates p53 by causing increased expression of p14/Arf, which then binds and inhibits MDM2. Silencing or loss of the p14/Arf locus at this early stage can eliminate p53 function, allowing cells to continue proliferation (Fulci et al., 2000; Pinyol et al., 2000). Nutlin stabilizes p53 in a non-genotoxic fashion and thus may be considered similar to oncogenic stress that activates p53 without directly damaging DNA. Our results indicate that Nutlin treatment can lead to the acquisition of somatic mutations in p53 and select for p53-mutated cells. P53 mutations have historically been considered late events in cancer development, though recent studies, particularly in ovarian cancer, suggest p53 mutations occur early and may be an “initiating” event (Ahmed et al., 2010). Our results would suggest that growth arrest/death imposed by oncogenic stress during the early stages of cancer development could be overcome not only through silencing/loss of p14/Arf, but also through the acquisition of inactivating p53 mutations. An important question is how cells acquire p53 mutations in the absence of DNA damage. One possibility is that p53 mutated cells are constantly emerging in cell culture at a low rate, perhaps due to deficiencies in DNA repair or replication fidelity. In this case, cells could acquire p53 mutations in the absence of exogenous DNA damage. Repeated exposures to Nutlin (or perhaps other growth-limiting conditions) would then select for and enrich these p53 mutated cells (Blagosklonny, 2002). A second possibility is that p53 mutations are acquired as a secondary consequence of initiating apoptosis. For example, TRAIL ligand induces apoptosis through the receptor-mediated pathway and, like Nutlin, does not directly damage DNA (Ashkenazi, 2008). In a recent study, TRAIL was found to promote mutations in cells when given at sub-lethal doses (Lovric and Hawkins, 2010). Importantly blocking the endonuclease responsible for apoptosis-associated DNA fragmentation also blocked the ability of TRAIL to induce mutations. Based on these findings a model was proposed in which cells can initiate apoptosis and fragment their DNA to some extent, but then repair the breaks in their DNA and recover. Misrepair of the DNA breaks was proposed as the mechanism that leads to mutation. Mutations in p53 arising after Nutlin treatment could also result from misrepair of DNA breaks in cells that have initiated but did not fully execute apoptosis.
It was interesting that certain Group 1 clones maintained wild-type p53 but were resistant to Nutlin-induced apoptosis. In previous studies, cells resistant to the growth inhibitory effects of wild-type p53 were obtained by selecting from a temperature sensitive p53 cell line clones that could grow at the non-permissive temperature (Gaitonde et al 2000, Pietenpol et al 1996). In some cases, the resistance to wild-type p53 was associated with changes in p53 protein conformation, demonstrated by altered reactivity with conformation-specific p53 antibodies (Gaitonde et al 2000, Mayelzadeh and Martinez 2007). We have used conformation-specific p53 antibodies to compare the conformation of p53 induced by Nutlin in parental SJSA-1 cells and Nutlin-selected clones. These studies suggest that p53 induced by Nutlin in Group 1 clones has an altered conformation compared to p53 induced by Nutlin in parental SJSA-1 cells (data not shown). Thus, one possibility is that apoptosis resistance in the Group 1 clones that express wild-type p53 results from a defect in p53 protein folding/conformation that diminishes its ability to activate pro-apoptotic genes (e.g. PUMA). Other studies have shown that mutant p53s can inhibit the activity of wild-type p53 when both proteins are co-expressed, and that binding between mutant p53 and wild-type p53 can drive the wild-type protein into a mutant conformation (Milner and Medcalf, 1991). One possibility is that mutant and wild-type p53s bind each other in the p53 heterozygous Group 1 clones, and that this binding alters the conformation of wild-type p53 and/or prevents it from inducing apoptosis.
Finally, our findings have implications for the potential clinical use of Nutlin or other MDM2 antagonists. The effectiveness of current cancer therapies is often limited by an acquired resistance of cancer cells to the therapeutic agent. Many DNA damaging chemotherapeutics not only kill cancer cells, but can also induce cancer-promoting mutations and in this way promote tumorigenesis (Blagosklonny, 2005). Cancer treatment schedules often include extended intervals between courses of chemotherapy, to allow recovery of normal cells and tissues. Our data show that, just as cancer cells can become resistant to conventional chemotherapeutic agents, cancer cells can also gain resistance to Nutlin through mechanisms that include acquired mutations in p53. We would suggest Nutlin or other MDM2 antagonists be used in combination with agents that can also target or limit the potential outgrowth of p53 mutated cells.
Supplementary Material
Supplemental Figure 1: p53 localization in Group 1 cells. SJSA-1 and Group 1 clones were either untreated or treated with 10 μM Nutlin for 24 hrs. Cells were fixed with 4% formaldehyde and subjected to immunofluorescence analysis with anti-p53 antibody as described in Figure 4.
Supplemental Table 1: List of Clones. Individual clones were isolated from Nutlin-selected P4 populations in the absence (Group 1) or in the continued presence of Nutlin (Group 2). The p53 status and genotype were determined by sequence analysis. Total RNA was extracted with RNeasy Mini kit (Qiagen Inc., Valencia, CA). Complimentary DNA (cDNA) was prepared by reverse transcription using SuperScript III First-Strand Synthesis SuperMix system (Invitrogen Corporation, Carlsbad, CA) following the manufacturer’s instructions. The full-length human p53 cDNA was amplified by using polymerase chain reaction (PCR) with p53 primers (forward primer: 5′- GTG ACA CGCTTC CCT GGATTG G -3′ and reverse primer: 5′- GTC AGT CTG AGT CAG GCC CTT C -3′). The amplified p53 cDNA was sequenced at DNA Services Facility (Research Resources Center, University of Illinois, Chicago, IL). The Group 1 and Group 2 clones used throughout the paper in Figures 2-4 are indicated in bold.
Acknowledgements
This work supported by NCI grant 1RO1CA137598-01A1 to CGM.
Footnotes
Conflict of Interest The authors declare no conflict of interest.
References
- Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, Sharma R, Stewart C, Fereday S, Caldas C, Defazio A, Bowtell D, Brenton JD. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A. Targeting the extrinsic apoptosis pathway in cancer. Cytokine Growth Factor Rev. 2008;19:325–31. doi: 10.1016/j.cytogfr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Oncogenic resistance to growth-limiting conditions. Nat Rev Cancer. 2002;2:221–5. doi: 10.1038/nrc743. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Carcinogenesis, cancer therapy and chemoprevention. Cell Death Differ. 2005;12:592–602. doi: 10.1038/sj.cdd.4401610. [DOI] [PubMed] [Google Scholar]
- Brown L, Boswell S, Raj L, Lee SW. Transcriptional targets of p53 that regulate cellular proliferation. Crit Rev Eukaryot Gene Expr. 2007;17:73–85. doi: 10.1615/critreveukargeneexpr.v17.i1.50. [DOI] [PubMed] [Google Scholar]
- Christiansen DH, Andersen MK, Pedersen-Bjergaard J. Mutations with loss of heterozygosity of p53 are common in therapy-related myelodysplasia and acute myeloid leukemia after exposure to alkylating agents and significantly associated with deletion or loss of 5q, a complex karyotype, and a poor prognosis. J Clin Oncol. 2001;19:1405–13. doi: 10.1200/JCO.2001.19.5.1405. [DOI] [PubMed] [Google Scholar]
- De Benedetti VM, Travis LB, Welsh JA, van Leeuwen FE, Stovall M, Clarke EA, Boice JD, Jr, Bennett WP. p53 mutations in lung cancer following radiation therapy for Hodgkin’s disease. Cancer Epidemiol Biomarkers Prev. 1996;5:93–8. [PubMed] [Google Scholar]
- Drakos E, Thomaides A, Medeiros LJ, Li J, Leventaki V, Konopleva M, Andreef M, Rassidakis GZ. Inhibition of p53-murine double minute 2 interaction by nutlin-3A stabilizes p53 and induces cell cycle arrest and apoptosis in Hodgkin lymphoma. Clin Cancer Res. 2007;13:3380–7. doi: 10.1158/1078-0432.CCR-06-2581. [DOI] [PubMed] [Google Scholar]
- Efeyan A, Serrano M. p53: guardian of the genome and policeman of the oncogenes. Cell Cycle. 2007;6:1006–10. doi: 10.4161/cc.6.9.4211. [DOI] [PubMed] [Google Scholar]
- Fulci G, Labuhn M, Maier D, Lachat Y, Hausmann O, Hegi ME, Janzer RC, Merlo A, Van Meir EG. p53 gene mutation and ink4a-arf deletion appear to be two mutually exclusive events in human glioblastoma. Oncogene. 2000;19:3816–22. doi: 10.1038/sj.onc.1203700. [DOI] [PubMed] [Google Scholar]
- Gaitonde SV, Riley JR, Qiao D, Martinez JD. Conformational phenotype of p53 is linked to nuclear translocation. Oncogene. 2000;19:4042–9. doi: 10.1038/sj.onc.1203756. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–16. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagahima M, Takenoshita S, Yokota J, Harris CC. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard DG, Travis LB, Addya K, Dores GM, Holowaty EJ, Bergfeldt K, Malkin D, Kohler BA, Lynch CF, Wiklund T, Stovall M, Hall P, Pukkala E, Slater DJ, Felix CA. p53 mutations in leukemia and myelodysplastic syndrome after ovarian cancer. Clin Cancer Res. 2002;8:973–85. [PubMed] [Google Scholar]
- Lovric MM, Hawkins CJ. TRAIL treatment provokes mutations in surviving cells. Oncogene. 2010;29:5048–60. doi: 10.1038/onc.2010.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki CG, Howley PM. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol. 1997;17:355–63. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol. 1984;4:1689–94. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayelzadeh F, Martinez JD. DNA binding and selective gene induction by different forms of the p53 protein. Oncogene. 2007;26:2955–63. doi: 10.1038/sj.onc.1210110. [DOI] [PubMed] [Google Scholar]
- McDermott U, Longley DB, Galligan L, Allen W, Wilson T, Johnston PG. Effect of p53 status and STAT1 on chemotherapy-induced, Fas-mediated apoptosis in colorectal cancer. Cancer Res. 2005;65:8951–60. doi: 10.1158/0008-5472.CAN-05-0961. [DOI] [PubMed] [Google Scholar]
- Midgley CA, Lane DP. p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene. 1997;15:1179–89. doi: 10.1038/sj.onc.1201459. [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–90. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Milner J, Medcalf EA. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell. 1991;65:765–74. doi: 10.1016/0092-8674(91)90384-b. [DOI] [PubMed] [Google Scholar]
- Peng Y, Chen L, Li C, Lu W, Chen J. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J Biol Chem. 2001;276:40583–90. doi: 10.1074/jbc.M102817200. [DOI] [PubMed] [Google Scholar]
- Pietenpol JA, Lengauer C, Jordan J, Kinzler KW, Vogelstein B. Mammalian cells resistant to tumor suppressor genes. Proc Natl Acad Sci. 1996;93:8390–4. doi: 10.1073/pnas.93.16.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyol M, Hernandez L, Martinez A, Cobo F, Hernandez S, Bea S, Lopez-Guillermo A, Nayach I, Palacin A, Nadal A, Fernandez PL, Montserrat E, Cardesa A, Campo E. INK4a/ARF locus alterations in human non-Hodgkin’s lymphomas mainly occur in tumors with wild-type p53 gene. Am J Pathol. 2000;156:1987–96. doi: 10.1016/S0002-9440(10)65071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moran DM, Maki CG. Transient nutlin-3a treatment promotes endoreduplication and the generation of therapy-resistant tetraploid cells. Cancer Res. 2008;68:8260–8268. doi: 10.1158/0008-5472.CAN-08-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Weber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–9. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–44. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson T, Tovar C, Yang H, Carvajal D, Vu BT, Xu Q, Wahl GM, Heimbrook DC, Vassilev LT. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J Biol Chem. 2004;279:53015–22. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- Vaseva AV, Marchenko ND, Moll UM. The transcription-independent mitochondrial p53 program is a major contributor to nutlin-induced apoptosis in tumor cells. Cell Cycle. 2009;8:1711–9. doi: 10.4161/cc.8.11.8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Wu GS, Ding Z. Caspase 9 is required for p53-dependent apoptosis and chemosensitivity in a human ovarian cancer cell line. Oncogene. 2002;21:1–8. doi: 10.1038/sj.onc.1205020. [DOI] [PubMed] [Google Scholar]
- Wu GS, El-Deiry WS. Apoptotic death of tumor cells correlates with chemosensitivity, independent of p53 or bcl-2. Clin Cancer Res. 1996;2:623–33. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: p53 localization in Group 1 cells. SJSA-1 and Group 1 clones were either untreated or treated with 10 μM Nutlin for 24 hrs. Cells were fixed with 4% formaldehyde and subjected to immunofluorescence analysis with anti-p53 antibody as described in Figure 4.
Supplemental Table 1: List of Clones. Individual clones were isolated from Nutlin-selected P4 populations in the absence (Group 1) or in the continued presence of Nutlin (Group 2). The p53 status and genotype were determined by sequence analysis. Total RNA was extracted with RNeasy Mini kit (Qiagen Inc., Valencia, CA). Complimentary DNA (cDNA) was prepared by reverse transcription using SuperScript III First-Strand Synthesis SuperMix system (Invitrogen Corporation, Carlsbad, CA) following the manufacturer’s instructions. The full-length human p53 cDNA was amplified by using polymerase chain reaction (PCR) with p53 primers (forward primer: 5′- GTG ACA CGCTTC CCT GGATTG G -3′ and reverse primer: 5′- GTC AGT CTG AGT CAG GCC CTT C -3′). The amplified p53 cDNA was sequenced at DNA Services Facility (Research Resources Center, University of Illinois, Chicago, IL). The Group 1 and Group 2 clones used throughout the paper in Figures 2-4 are indicated in bold.