Abstract
Background
Phytophthora infestans (Mont.) de Bary, the causal organism of late blight, is economically the most important pathogen of potato and resistance against it has been one of the primary goals of potato breeding. Some potentially durable, broad-spectrum resistance genes against this disease have been described recently. However, to obtain durable resistance in potato cultivars more genes are needed to be identified to realize strategies such as gene pyramiding or use of genotype mixtures based on diverse genes.
Results
A major resistance gene, Rpi-rzc1, against P. infestans originating from Solanum ruiz-ceballosii was mapped to potato chromosome X using Diversity Array Technology (DArT) and sequence-specific PCR markers. The gene provided high level of resistance in both detached leaflet and tuber slice tests. It was linked, at a distance of 3.4 cM, to violet flower colour most likely controlled by the previously described F locus. The marker-trait association with the closest marker, violet flower colour, explained 87.1% and 85.7% of variance, respectively, for mean detached leaflet and tuber slice resistance. A genetic linkage map that consisted of 1,603 DArT markers and 48 reference sequence-specific PCR markers of known chromosomal localization with a total map length of 1204.8 cM was constructed.
Conclusions
The Rpi-rzc1 gene described here can be used for breeding potatoes resistant to P. infestans and the breeding process can be expedited using the molecular markers and the phenotypic marker, violet flower colour, identified in this study. Knowledge of the chromosomal localization of Rpi-rzc1 can be useful for design of gene pyramids. The genetic linkage map constructed in this study contained 1,149 newly mapped DArT markers and will be a valuable resource for future mapping projects using this technology in the Solanum genus.
Keywords: Diversity Array Technology, Mapping, Phytophthora infestans, Solanum tuberosum, Rpi-rzc1
Background
Domestication of crop plants involving selection through many generations has brought yield and quality improvement but also narrowed the gene pools of cultivated species [1]. Therefore, for the last hundred years, modern plant breeders have searched wild relatives of crop plants for valuable genes, alleles and Quantitative Trait Loci (QTL) encoding desirable phenotypic characters. In potato, more than 210 wild and cultivated species [1] provide a good chance of finding useful traits. However, till date only a limited number has been used in breeding programs. Exploitation of wild Solanum species has focused on those resistant to pests and pathogens. Among potato pathogens, Phytophthora infestans (Mont.) de Bary, the causal organism of late blight, is economically the most important. Resistance against it has been one of the primary goals of potato breeding [2]. The first introduction of major genes for resistance (R genes) to P. infestans from the wild species S. demissum early in the 20th century brought disappointment, due to the rapid spread of P. infestans strains possessing the corresponding virulence factors. Cultivars with different combinations of these genes quickly became infected [3]. However, some potentially more durable, broad-spectrum R genes have recently been described, including RB/Rpi-blb1 [4,5], Rpi-blb3 [6], Rpi-vnt1.1 [7,8], Rpi-phu1 [9] and Rpi-sto1 [10]. New strategies preventing such rapid evolution of compatible P. infestans races have been proposed. All these are based on avoiding monocultures and use of as many broad-spectrum R genes as possible. These can either be stacked within cultivars or used in mixtures of breeding lines each containing a different R gene. More simply, cultivars containing different R genes could be grown next to each other or could be changed between seasons to achieve R gene diversification [11]. Use of molecular markers and transgenesis can facilitate this process. Triple R gene transformants have been obtained stacked with three R genes against P. infestans, Rpi-sto1, Rpi-vnt1.1 and Rpi-blb3, and these plants combined resistance spectra of the individual R genes [12]. PCR markers have also been applied in selection of plants possessing both Rpi-mcd1 gene from S. microdontum [13] and RPi-ber from S. berthaultii [14-16], though these two genes provided only limited resistance [17]. To improve the likelihood that these strategies will be successful, breeders need to have many such genes available and therefore the search for these continues among wild relatives of potato.
Solanum ruiz-ceballosii Cárd. (syn. Solanum sparsipilum (Bitt.) Juz. et Buk.) is a species closely related to potato, diploid, 2 EBN (Endosperm Balance Number), originating from Peru and Bolivia. It is a polymorphic weed species of walls, fields and field borders growing at altitudes of 2,400-3,800 m, with a corolla pale to medium blue-violet [18]. It is also known as Solanum ruiz-zeballosii Cárd. and, together with S. sparsipilum (Bitt.) Juz. et Buk., is accepted as belonging to the species Solanum brevicaule Bitt. [19] or is included in the brevicaule-complex [20]. The distribution of S. brevicaule is similar to that of the species mentioned above, spanning Bolivia near the border with Peru and south to northwest Argentina, from 2,000 up to 4,180 m above sea level and its corolla colour is described as purple violet to light blue [19]. Another slightly differently spelled name, Solanum ruiz-cevallosii Cárd., was published together with the proposed species abbreviation rzc [21]. Being a diploid, 2 EBN species, S. ruiz-ceballosii can be crossed with diploid S. tuberosum, but seeds were also obtained from its cross with a tetraploid potato cultivar Aurora [22].
S. ruiz-ceballosii was previously described as highly resistant to potato late blight both in leaves and in tubers [23,24]. Quantitative trait locus for late blight resistance with major effects originating from the synonymous S. sparsipilum has previously been mapped on potato chromosome X using stem and foliage tests and explained up to 29% of variance in the stem resistance of the mapping population [25]. S. sparsipilum has been also described as a source of resistance to the nematodes Meloidogyne fallax [26] and Globodera pallida [27] as well as to Potato Virus Y (PVY) [28]. Therefore, it may become a valuable component of potato breeding programs. In the case of PVY, a dominant resistance gene Ncspl originating from S. sparsipilum has been mapped to potato chromosome IV [28]. In addition, S. sparsipilum has been proposed as a potential source of resistance against potato tuber moth, Phthorimaea operculella [29].
The goal of this study was to characterize the late blight resistance of the clone chosen from S. ruiz-ceballosii accession VIR 8664 (VIR 7370) [23,24], originating from the Vavilov Collection in Russia (VIR), and to map the underlying R gene, named Rpi-rzc1. Within our study, the Rpi-rzc1 gene was mapped to potato chromosome X, it proved to be effective in providing a high level of resistance to P. infestans both in detached leaflet and in tuber slice tests. We also noted a close linkage between Rpi-rzc1 and violet flower colour encoded by the F locus [30,31], which may be useful for selection of resistant individuals from certain crosses.
Methods
Plant material
A late blight resistant clone 99-10/36, selected from S. ruiz-ceballosii accession VIR 7370 (VIR 8664) obtained from the VIR collection in Russia and collected by Cárdenas in Horlahon, Bolivia, and a susceptible S. tuberosum dihaploid of the Polish cultivar (cv.) Balbina (dH Balbina) from IHAR-PIB O/Młochów's haploidization program, were crossed to obtain an F1 mapping population of 114 individuals. The parent dH Balbina and the progeny were propagated each year in the field, whereas clone 99-10/36 was propagated in the glasshouse. Along with the mapping population and the parental clones, standard cultivars (cvs) proposed by the Eucablight consortium [32], Alpha, Bintje (susceptible to late blight), Biogold (moderately resistant, containing an R gene), Eersteling (susceptible), Escort (resistant, with R1, R2, R3 and R10 [33]), Gloria, Robijn (moderately resistant), Sárpo Mira (very resistant) and two additional diploid hybrid clones, DG 94-15 (resistant in leaflets and tubers) and DG 94-668 (resistant in tubers) [9], were included in tests of resistance to P. infestans. From the mapping population, 13 highly resistant individuals were chosen on the basis of the laboratory tests results. These individuals were assessed for resistance in the field conditions. Standards, except cv. Biogold and diploid clones, were also included in the field test.
P. infestans isolate
The isolate MP324 from the pathogen collection of the Plant Breeding and Acclimatization Institute-National Research Institute, Młochów, Poland was used in all resistance tests. The isolate, collected in 1997 in Poland, was of A1 mating type, highly aggressive, metalaxyl resistant and of complex race (1.2.3.4.5.6.7.8.10.11). Black's differential set, obtained from the Scottish Agricultural Science Agency, Edinburgh, UK, was used to confirm the isolate's virulence each time in parallel to the tests. Before each resistance test, the isolate was passaged through susceptible potato tissue at least twice. Isolate MP324 has frequently been used in resistance tests in our laboratory [9,34].
Late blight resistance assessment
The resistance to P. infestans of the parental clones, the mapping population and the standard cultivars was evaluated by laboratory detached leaflet and tuber slice tests as described earlier [9,34]. Detached leaflets or tuber slices were inoculated with a 30 μl droplet of sporangia/zoospore suspension (50 sporangia/μl). After incubation of 6 days at 16°C, in high humidity and, in case of leaflets, under constant light of about 1,600 lx, they were scored using a 1-9 scale, where 9 is maximum resistant [9,34]. The detached leaflet tests were replicated as follows: 3 leaflets/genotype × 2 replications × 2 dates were tested in 2007, 3 leaflets/genotype × 2 replications in 2008 and 3 leaflets/genotype × 2 replications × 2 dates in 2010. Three tuber slices/genotype × 2 replications were tested in 2006, 2007 and 2010. A genotype was considered resistant, i.e. possessing the R gene, when its mean resistance score in both detached leaflet and in tuber slice tests was ≥ 7.
In 2008 and 2009 field resistance assessment of 13 individuals from the mapping population and seven standards (Table 1) was conducted at Southeast Poland in Boguchwala, where the weather conditions were favorable for severe late blight infection. Material was planted in randomized 4-hill plots, in two replications in 2008 and in one replication in 2009. Natural infection was evaluated weekly from the first symptoms of pathogen. Values of rAUDPC (area under disease progress curve) were calculated from 6 to 8 readings of infection in scale 1-9, according Fry (1978) [35].
Table 1.
Mean rAUDPC values of field infection for 13 individuals and standard cultivars
| Genotype | rAUDPC in | |
|---|---|---|
| 2008 | 2009 | |
| 05-18/1 | 0.012 | 0.000 |
| 05-18/9 | 0.000 | 0.000 |
| 05-18/24 | 0.000 | 0.000 |
| 05-18/25 | 0.000 | 0.000 |
| 05-18/32 | 0.000 | 0.001 |
| 05-18/33 | 0.000 | 0.001 |
| 05-18/54 | 0.000 | 0.000 |
| 05-18/56 | 0.000 | 0.000 |
| 05-18/73 | 0.000 | 0.000 |
| 05-18/98 | 0.000 | 0.000 |
| 05-18/99 | 0.000 | 0.000 |
| 05-18/118 | 0.000 | 0.001 |
| 05-18/129 | 0.000 | 0.000 |
| standard cultivars | ||
| Alpha | 0.425 | 0.312 |
| Bintje | 0.532 | 0.791 |
| Gloria | 0.512 | 0.722 |
| Eerstling | 0.446 | 0.590 |
| Escort | 0.371 | 0.426 |
| Robijn | 0.307 | 0.273 |
| Sárpo Mira | 0.029 | 0.000 |
Flower colour was assessed visually; four categories were noted in the mapping population dH Balbina × 99-10/36: (1) white, (2) pale violet, (3) violet and (4) dark violet corolla. The flower colours of the parents and examples from the progeny are shown in Figure 1. Categories 2-4 were included into the general violet group for the purpose of mapping. Assessments were repeated in 2006, 2007 and 2008, but not all individuals flowered each year.
Figure 1.
Flower colour of the parents dH Balbina (a - white) × S. ruiz-ceballosii 99-10/36 (b - pale violet) and examples of the flower colours of F1 individuals: c -white; d - pale violet; e - violet; f - dark violet.
DNA isolation and sequence-specific markers
Genomic DNA was extracted from 1 g of fresh, young leaves of field grown plants with the DNeasy Plant Maxi kit (Qiagen, Hilden, Germany). All sequence-specific markers listed in Additional file 1 were amplified using the following conditions as described by Śliwka et al. (2011) [36]. The reaction mixture of 20 μl contained 2 μl of 10 × PCR buffer, the four deoxynucleotides (0.1 mM), MgCl2 (1.5 mM), primers (0.2 μM), Taq polymerase (0.05 U/μl) and 10-30 ng of template DNA. The PCR program was 94°C - 180 s; 40 cycles of: 94°C - 30 s, 55°C - 45 s, 72°C - 90 s; 72°C - 420 s; where the annealing temperature was modified depending on primers used, this is indicated in Additional file 1. The reactions of the PCR products with the corresponding restriction endonucleases, also listed in Additional file 1 were performed according to the manufacturer's recommendations.
Diversity Array Technology (DArT)
The DArT analysis was performed in Diversity Array Pty Ltd. Canberra, Australia, exactly as described for S. michoacanum [36], following the protocols previously developed for other plant species [37-40].
Statistical and linkage analyses
The normality of the distribution of the phenotypic data was checked by the Kolmogorov-Smirnov test. The reproducibility of the resistance tests between years as well as the correlations between traits were evaluated by calculating the linear Pearson's correlation coefficients. Marker--trait linkages and determination coefficients (R2) were estimated by the Student's t-test and analysis of variance, respectively. The fit of segregation to the expected ratio was checked by the χ2 test. All statistical analyses were performed using computer program STATISTICA for Windows (Stat Soft, Inc., Tulsa, OK, U.S.A.). Linkage analyses were performed using JoinMap ® 4 [41] with the following settings: CP population type (creating maternal and paternal linkage maps first and then creating a common population map), independence LOD as a grouping parameter (linkages with LOD > 3 were considered significant), regression mapping algorithm and Haldane's mapping function.
Results
Late blight resistance assessment
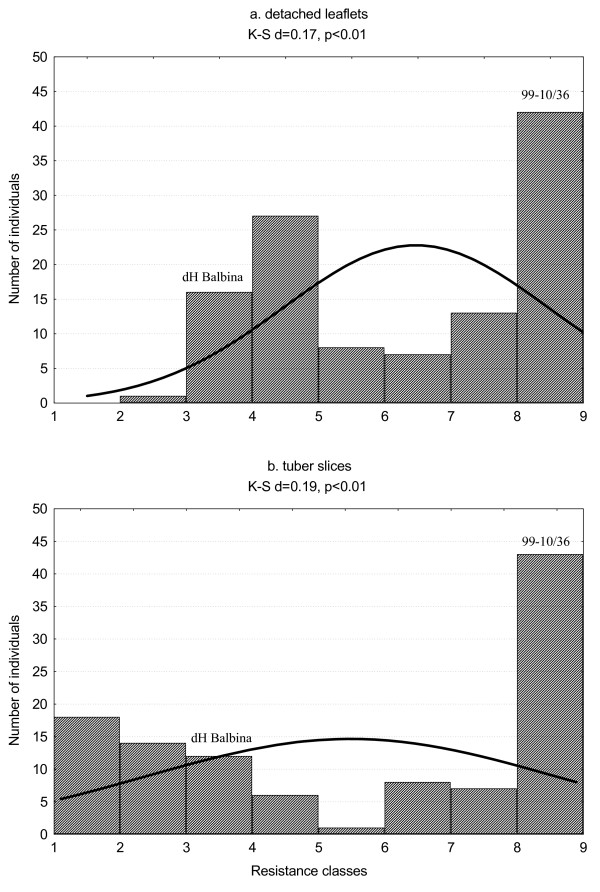
The mean resistance scores of the parental clones 99-10/36 and dH Balbina as well as of the resistance standards are presented in Table 2S. ruiz-ceballosii clone 99-10/36 was the most resistant in both detached leaflet and tuber slice tests, with mean scores 9.0 and 8.9, respectively. Most of the standards performed as expected with cv. Sárpo Mira being highly resistant and cvs Biogold and Escort and clone DG 94-15 being moderately resistant in detached leaflet tests. Cvs Gloria and Robijn and clone DG 94-668 proved more susceptible than expected. In tuber slice tests, cvs Sárpo Mira and Robijn and clone DG 94-15 showed some level of resistance to P. infestans (Table 2). The mean detached leaflet resistance results obtained for the mapping population in three subsequent years of testing were correlated with each other, with Pearson's correlation coefficients r = 0.581 (2007-2008), r = 0.664(2007-2010) and r = 0.783 (2008-2010) (p < 0.000 in all cases). Similarly, the tuber slice resistance results were correlated between years with Pearson's correlation coefficients r = 0.681 (2006-2007), r = 0.767 (2006-2010) and r = 0.742 (2007-2010); p < 0.000 in all cases. There was also a strong correlation between the mean (2007-2010) detached leaflet resistance results and the mean (2006-2010) tuber slice resistance results (Pearson's correlation coefficient r = 0.862, p < 0.000), indicating that the same genetic factor(s) was effective in both foliage and tuber resistance to P. infestans in the mapping population. Analysis of variance showed that plant genotype had the most significant effect on the resistance test results both in detached leaflet tests and in tuber slice tests. There were also significant effects of the interaction genotype × year of testing and of year of testing alone (Table 3). The distributions of the mean detached leaflet and tuber slice resistances in the mapping population were bimodal and significantly deviated from normality, which was confirmed by the Kolmogorov-Smirnov test (Figure 2). The range of the mean detached leaflet scores was 3.0-9.0 which was narrower than the case of tuber slice tests where the results ranged from 1.0 to 9.0 and covered the full assessment scale. The mean resistance ratings for the population were 6.5 and 5.5 in the leaflet and tuber slice tests, respectively, indicating less resistance in the tubers or stronger infection pressure in the tuber slice tests. When the individuals of the mapping population with mean resistance ≥ 7 were included into the resistant class, the sizes of the resistant and susceptible classes did not significantly deviate from the 1: 1 ratio expected for segregation of a single gene. This applied to the results of both the detached leaflet tests, where 52 individuals were assessed as resistant and 62 as susceptible (χ2 = 0.89, df = 1, p < 0.35), and the tuber slice tests where the resistant: susceptible ratio was 51: 63 (χ2 = 1.26, df = 1, p < 0.26). Only in 16 cases, was the leaflet resistance of a specific individual slightly above the threshold while the tuber resistance was below it, or vice versa, which we interpret as being due to test variation rather than genuine genetic difference. The majority of the individuals (98 out of 114) were either resistant both in detached leaflet and tuber slice tests or susceptible in both types of tests.
Table 2.
The late blight resistance of parental clones and standards in (a) detached leaflet and (b) tuber slice tests.
| a. Resistance of detached leaflets | b. Resistance of tuber slices | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | 2007 | 2008 | 2010 | Weighted mean: 2007, 2008 and 2010 | SD | 2006 | 2007 | 2010 | Mean: 2006, 2007 and 2010 | SD |
| 99-10/36 | 9.0 | 9.0 | 9.0 | 9.0 | 0.0 | 9.0 | 8.6 | 9.0 | 8.9 | 0.2 |
| dH Balbina | 3.8 | 4.7 | 3.4 | 3.8 | 0.7 | 4.3 | 2.5 | 4.3 | 3.7 | 1.0 |
| Alpha | 2.7 | 4.5 | 5.2 | 4.1 | 1.3 | 2.0 | 3.55 | 4.1 | 3.2 | 1.1 |
| Bintje | 2.6 | 5.1 | 4.2 | 3.7 | 1.3 | 2.1 | 2 | 2.1 | 2.1 | 0.1 |
| Biogold | 6.5 | 8.9 | 6.8 | 7.1 | 1.3 | 4.2 | 1.75 | 4.1 | 3.4 | 1.4 |
| Eersteling | 2.0 | 2.9 | 2.7 | 2.5 | 0.5 | 1.8 | 5.5 | 2.5 | 3.3 | 2.0 |
| Escort | 4.7 | 8.0 | 5.0 | 5.5 | 1.8 | 2.9 | 3.5 | 2.3 | 2.9 | 0.6 |
| Gloria | 2.8 | 5.5 | 2.9 | 3.4 | 1.5 | 1.3 | 4.2 | 1.6 | 2.4 | 1.6 |
| Robijn | 3.1 | 6.1 | 3.5 | 3.9 | 1.6 | 4.0 | 7.15 | 3.3 | 4.8 | 2.1 |
| Sárpo Mira | 8.8 | 9.0 | 8.8 | 8.8 | 0.1 | 5.9 | 7.35 | 4.7 | 6.0 | 1.3 |
| DG 94-15 | 6.1 | 6.9 | 6.4 | 6.4 | 0.4 | 7.1 | 7.25 | 2.9 | 5.8 | 2.5 |
| DG 94 -668 | 2.8 | 4.6 | 2.8 | 3.2 | 1.0 | 4.6 | 3.55 | 5.0 | 4.4 | 0.7 |
In the case of the detached leaflet tests, a weighted mean was calculated, since the numbers of tested leaflets per genotype were not equal in all years
SD standard deviation
Table 3.
Analysis of variance calculated on mean scores of each testing date in (a) detached leaflet tests across three years and (b) in tuber slice tests done in 2006, 2007 and 2010 in the mapping population dH Balbina × 99-10/36
| Factor | Dfa effect | Mean sum of squares effect | Dfa error | Mean sum of squares error | F | P | R2 (%)b |
|---|---|---|---|---|---|---|---|
| a. detached leaflet tests | |||||||
| {1}year | 2 | 79.2 | 137 | 0.4 | 194.92 | 0.000 | 4.82 |
| {2} genotype | 115 | 20.8 | 137 | 0.4 | 51.26 | 0.000 | 72.87 |
| Interaction: 1 × 2 | 162 | 3.9 | 137 | 0.4 | 9.53 | 0.000 | 19.09 |
| b. tuber slice tests | |||||||
| {1}year | 2 | 55.1 | 137 | 1.2 | 44.21 | 0.000 | 1.71 |
| {2} genotype | 115 | 43.8 | 137 | 1.2 | 35.19 | 0.000 | 78.30 |
| Interaction: 1 × 2 | 162 | 5.5 | 137 | 1.2 | 4.43 | 0.000 | 13.88 |
a - number of degrees of freedom. b - percentage of variance explained
Figure 2.
Distributions of (a) mean (2007, 2008 and 2010) leaflet resistance to P. infestans and (b) mean (2006, 2007 and 2010) tuber slice resistance in the mapping population. The fitness to the normal curve: K-S - Kolmogorov-Smirnov test, d - coefficient calculated for this test, p - probability, the line indicates the normal curve. Resistance levels of parental clones are marked with their names: 99-10/36 and dH Balbina.
Based on field observations of late blight infection of highly resistant individuals selected from mapping population values of rAUDPC were calculated (Table 1). In two years all 13 tested individuals showed high field resistance to P. infestans. Mapping data confirmed the presence of Rpi-rzc1 gene in these individuals. Standard cvs, beside resistant cv. Sárpo Mira, were strongly infected, rAUDPC values of the most susceptible standard, cv. Bintje were 0.532 and 0.791, in 2008 and 2009, respectively.
Flower colour assessment
White flowers of the dH Balbina and pale violet flowers of the S. ruiz-ceballosii 99-10/36, as well as examples of the four flower colour categories noted in the progeny are shown in the Figure 1. The flower colour of 11 progeny individuals could not be assessed unambiguously due to colour discrepancies between the three years of assessment. Lack of flowering, flower fading and non-homogenous genotype could be responsible for this. Of the remaining 103 individuals, 51 had violet flowers in three different shades of violet (Figure 1) and 52 had white flowers, which is a 1: 1 segregation as confirmed by the χ2 test (χ2 = 0.01, df = 1, p < 0.92). Violet flower colour was strongly correlated with higher mean leaflet (Pearson's correlation coefficient r = 0.882, p < 0.000) and tuber resistance (Pearson's correlation coefficient r = 0.804, p < 0.000).
Linkage map
Firstly two parental linkage maps and then the common CP map were constructed using the JoinMap ® 4 software. The common map consisted of 1603 DArT markers and 48 reference sequence-specific PCR markers of known chromosomal localization (Table 4 Additional file 2). Using those PCR markers (Additional file 1 Additional file 2) all 12 potato chromosomes could be identified, although one marker, C2_At1g05055 [42], was mapped to chromosome VI, that is a different position than the expected one on chromosome IV. On average, 134 markers were located on a chromosome and this number ranged from 74 on chromosome IV to 195 on chromosome V. Total map length reached 1204.8 cM and the particular chromosomes varied in length from 79.1 cM (chromosome X) to 143.2 cm (chromosome III), with an average length of 100.4 cM. The mean interval between markers was 0.75 cM, although the markers were not distributed evenly.
Table 4.
Linkage map of the F1 population dH Balbina × S.ruiz-ceballosii 99-10/36 and its comparison to the reference Solanum phureja diploid map 2010 [42] and S. michoacanum map [36]
| Chromo-some | Number of markers | Number of reference markers | Length (cM) | Number of markers common with S. phureja map: | Number of markers common with S. michoacanum map: | ||
|---|---|---|---|---|---|---|---|
| concordant position | discordant position | concordant position | discordant position | ||||
| I | 167 | 4 | 103.5 | 36 | 1 | 13 | 2 |
| II | 168 | 4 | 113.9 | 41 | 1 | 26 | - |
| III | 183 | 4 | 143.2 | 44 | 5 | 2 | - |
| IV | 74 | 2 | 93.9 | 21 | 3 | 6 | 1 |
| V | 195 | 3 | 91.8 | 36 | 2 | 18 | 1 |
| VI | 125 | 3 + 1a | 134.8 | 11 | - | 9 | 1 |
| VII | 123 | 5 | 82.3 | 18 | - | 2 | 2 |
| VIII | 186 | 5 | 90.9 | 53 | 1 | 16 | 1 |
| IX | 84 | 4 | 107.0 | 24 | 4 | 1 | - |
| X | 75 | 4 | 79.1 | 14 | 5 | 7 | 1 |
| XI | 124 | 4 | 81.8 | 29 | 5 | 7 | 5 |
| XII | 99 | 5 | 82.6 | 5 | 2 | 15 | 1 |
| Total | 1,603 | 47 + 1a | 1204.8 | 332 | 29 | 122 | 15 |
a marker that on Tomato-EXPEN 2,000 map was located on chromosome IV [42]
The Rpi-rzc1 gene and the flower colour locus
The Rpi-rzc1 gene for resistance to potato late blight was mapped to chromosome X of the resistant parent, S. ruiz-ceballosii 99-10/36 (Figure 3). The gene was linked to the flower colour locus with a distance of 3.4 cM, and therefore violet flower colour could be treated as an additional, phenotypic marker for the presence of the Rpi-rzc1 gene. Nine markers showed statistically significant linkage with both detached leaflet and tuber slice resistance to P. infestans in each year of testing as confirmed by T-tests (Table 5). The marker-trait association with the closest marker, that is violet flower colour, explained 87.1% and 85.7% of variance, respectively in mean detached leaflet and tuber slice resistance. While, as expected, distal to the Rpi-rzc1 gene the percentages of the variance explained by the marker-trait associations decreased as the distance from the gene increased, proximal to the gene they seemed to fluctuate (Figure 3, Table 5). This could be explained by the combination of missing data both in marker and resistance scores.
Figure 3.
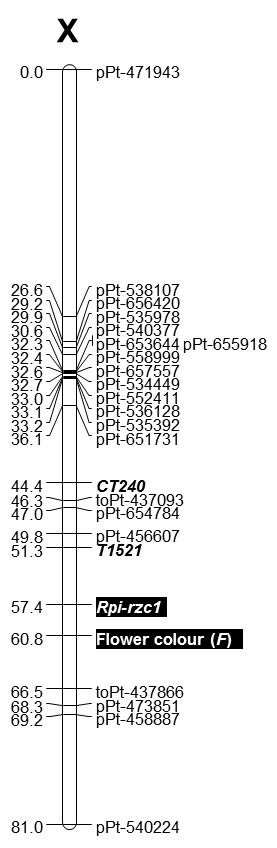
Genetic linkage map of the S. ruiz-ceballosii 99-10/36 chromosome X showing the location of the late blight resistance gene and the flower colour locus marked by black rectangles. The reference PCR markers are in bold italics. On the left, cumulative genetic distances in cM are given.
Table 5.
The percentages of variance (R2 ) in P.infestans resistance explained by the significant marker-trait linkages (T student test, p < 0.000) in (a) detached leaflet tests and (b) tuber slice tests.
| Marker | R2 (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| a. Resistance of detached leaflets | b. Resistance of tuber slices | |||||||
| 2007 | 2008 | 2010 | Weighted mean: 2007, 2008 and 2010 | 2006 | 2007 | 2010 | Mean: 2006, 2007 and 2010 | |
| CT240 | 58.1 | 43.0 | 41.3 | 54.2 | 51.0 | 28.4 | 36.9 | 47.2 |
| toPt-437093 | 53.3 | 35.3 | 34.8 | 46.4 | 44.8 | 33.6 | 32.5 | 44.8 |
| pPt-654784 | 69.6 | 62.3 | 57.7 | 73.9 | 66.9 | 46.0 | 54.1 | 67.8 |
| pPt456607 | 81.3 | 49.4 | 45.0 | 63.9 | 63.9 | 39.6 | 44.1 | 59.8 |
| T1521 | 69.0 | 43.6 | 39.8 | 56.0 | 52.1 | 31.1 | 36.8 | 48.6 |
| Violet flower colour | 70.9 | 74.7 | 75.9 | 87.1 | 76.9 | 61.9 | 75.6 | 85.7 |
| toPt-437866 | 70.1 | 61.8 | 57.1 | 73.6 | 67.3 | 45.2 | 53.5 | 67.4 |
| pPt-473851 | 52.4 | 31.7 | 29.3 | 41.2 | 39.7 | 28.3 | 27.7 | 38.4 |
| pPt-458887 | 50.1 | 28.4 | 25.0 | 36.7 | 35.7 | 22.9 | 23.5 | 32.9 |
All the markers are located on chromosome X and are inherited from the resistant parent 99-10/36
Discussion
The Rpi-rzc1 gene from S. ruiz-ceballosii, that confers resistance to P. infestans, was mapped to potato chromosome X. The gene provides a high level of resistance in both potato foliage and tubers. The influence of field and testing conditions on results of leaflet and tuber slice tests was significant but weak (Table 3) which showed stable expression of the Rpi-rzc1 gene. Out of 130 P. infestans isolates, collected from various locations in Poland in the years 2007-2009, that were tested on S. ruiz-ceballosii plants, only 4 (3.1%) were virulent (Śliwka, unpublished), indicating the usefulness of the gene for potato breeding. Results of the field test for resistance to late blight under natural infection pressure of 13 individuals showed that gene Rpi-rzc1 confers resistance to P. infestans both in laboratory tests and in natural conditions.
The Rpi-rzc1 is a third gene for resistance to P. infestans that has been mapped to potato chromosome X. The first was an R gene originating from S. berthaultii and mapped 10.4 cM north from the marker TG403 [14]. The same gene, later named RPi-ber, was mapped more precisely to a distance of 4.6 cM from the marker TG403 [15]. The Rpi-rzc1 gene could be in a similar location since, on the common CP map of dH Balbina × 99-10/36, it was 8.1 cM north from the marker TG403 (Additional file 2). When isolates of P. infestans compatible with RPi-ber were used for inoculation, a smaller, but significant resistance effect was detected in the same map position as the R gene. This could be explained by the residual effect of the gene or/and a resistance QTL located in the same position [43]. The RPi-ber gene proved to be effective in both foliage and tubers [44], similarly to R1, R3b and Rpi-phu1 [9,45,46] and also to Rpi-rzc1 (Figure 2, Table 4). In a breeding experiment on the effects of pyramiding of the R genes, mentioned in the introduction, RPi-ber was used, together with the RPi-mcd1 gene, and both genes showed an additive effect on resistance to late blight in a field test. A larger effect was provided by the RPi-ber, which produced a three week delay in infection reaching 50% of the leaf area [17]. In the same species, S. berthaultii, two more R genes, Rpi-ber1 and Rpi-ber2, were identified and mapped to chromosome X [16]. Park et al. speculate that, on the basis of location and origin, Rpi-ber1 [16] may be identical to RPi-ber [15]. Rpi-ber2 is most likely a different gene, although located in the similar position 12 cM north from the marker TG403 [16].
Apart from R genes, a number of QTL for resistance to P. infestans have been mapped to potato chromosome X [25,34,47-49]. These QTL originate from various Solanum species, including S. sparsipilum, which is a synonym of S. ruiz-ceballosii [25]. A QTL meta-analysis that allowed more precise comparisons of different genetic maps has identified two meta-QTL for late blight resistance on potato chromosome X. The Rpi-ber1/RPi-ber is located between them and the Rpi-ber2 localization overlaps with a meta-QTL named MQTL_2_Late_blight [50]. In tomato, an incompletely dominant R gene allele, Ph-2 that originates from S. pimpinellifolium and confers resistance to P. infestans, has been mapped to chromosome X [51]. It's location in the close vicinity of the marker CT124 [51] is similar to the location of the RPi-ber gene which is 4.6 cM away from the same marker [15]. In general, on the basis of mapping data, we cannot exclude the possibility that Rpi-rzc1, Rpi-ber1/RPi-ber, Rpi-ber2, QTL for late blight resistance listed above and even the tomato Ph-2 gene are homologous and occupy the same locus on chromosome X.
An allele-specific molecular marker T1521 was located at a distance of 6.1 cM from the Rpi-rzc1 gene and this easy to score marker can be useful for marker-assisted selection (MAS) of resistant potatoes. DArT marker toPt-437866, located on the other side of the gene at a distance of 9.1 cM, can be transformed into a PCR marker and also used in MAS. The linkage of the Rpi-rzc1 gene to violet flower colour can be exploited for the selection of the resistant individuals only in certain breeding combinations. We called the colour observed in the mapping population "violet", following the botanical description of the species [18], although it may correspond to the blue or purple flower colour described in other potato germplasm [30,31]. The violet flower colour segregated in a clear 1:1 ratio in the mapping population dH Balbina × 99-10/36 indicating that this trait was controlled by a single dominant allele, heterozygous in the S. ruiz-ceballosii 99-10/36 parent. The chromosomal position of its locus suggested that most likely it was the previously mapped F locus and showed it to be involved in the flower colour expression in combination with the locus P or D [30,31]. The presence of at least a single dominant F allele and P or D allele is required to turn potato flowers from white to blue or red, respectively [30]. The presence of the both D and P alleles together with the F allele resulted in the purple flower colour [31]. We can only deduce the genotypes of the parents in the F locus as dH Balbina: ff and 99-10/36: Ff, on the basis of the segregation and chromosomal position of the flower colour locus. Within the progeny, three different shades/intensities of the violet colour were observed indicating the segregation of various numbers of D and/or P alleles, also possibly inherited from dH Balbina, in which the ff genotype would mask their presence. Flower colour could serve as a phenotypic marker of late blight resistance in other progenies with the Rpi-rzc1 gene only when the susceptible parent would have an ability to synthesize anthocyanin pigments (D and/or P alleles) but not expressed in flowers (genotype ff).
The DArT markers applied in this study are a cost-effective, quick and efficient tool for genotyping and they are being used more and more frequently for mapping studies in diverse plant species, like cotton [52], pearl millet [53], pigeon pea [54], eucalyptus [40], rye [55] and many others. The first genetic map of a potato relative constructed with the use of DArT markers was made for S. bulbocastanum (1 EBN). Twelve linkage groups consisting of 439 markers were obtained with a total map length of 403 cM. However, so far those linkage groups have not been assigned to potato chromosomes [56]. A DArT linkage map was also made for S. michoacanum (1 EBN). It consisted of 846 DArT markers and 48 sequence-specific PCR markers with known locations that allowed the identification of all particular potato chromosomes [36]. Recently, a linkage map of 2 EBN Solanum species containing, among others, DArT markers, has been published under a name: Solanum phureja diploid map 2010 [42]. It was made within the Potato Genome Sequencing Project in order to anchor and orientate physical contigs along the chromosomes [57]. A comparison of the dH Balbina × S. ruiz-ceballosii 99-10/36 map with the Solanum phureja diploid map 2010 [42] and the S. michoacanum map [36] is summarized in Table 4 and in the Additional file 2. Out of 1,603 DArT markers located on our map, only 361 (22.5%) were also present on the Solanum phureja diploid map 2010 [42], most of them (332 i.e. 92%) in similar positions on both maps (Table 4). Even fewer 137 (8.5%) DArT markers were common to both S. ruiz-ceballosii and S. michoacanum [36] linkage maps, but even so the majority of them located in concordant positions (122, i.e.89%). These results suggest that even though different DArT markers segregate in different mapping populations, there is clearly synteny within the Solanum genus as very few markers were mapped in discordant positions on the three maps. The numbers of common and similarly located markers between these three maps confirmed that indeed potatoes from the S. tuberosum × S. ruiz-ceballosii cross are more closely related to S. phureja than to the 1 EBN species S. michoacanum.
Conclusions
The Rpi-rzc1 gene from S. ruiz-ceballosii, that confers resistance to P. infestans, was mapped to potato chromosome X. The gene provides a high level of resistance in both foliage and tubers. The gene has already been introduced into the cultivated potato gene pool via the interspecific cross described here. Knowledge of the chromosomal localization of the Rpi-rzc1 gene can be useful for designing gene pyramids. Molecular markers identified in our study can support marker-assisted selection of individuals possessing the gene, as can also the phenotypic marker violet flower colour. We constructed a genetic linkage map for dH Balbina (S. tuberosum) × 99-10/36 (S. ruiz-ceballosii) using 1,603 DArT markers and 48 reference sequence-specific PCR markers of known chromosomal localization. This is the first DArT map for both species and one of the first within the Solanum genus. Out of these 1,603 DArT markers, 1,149 were mapped for the first time in a Solanum plant, providing a useful resource for new DArT mapping studies that will enable map comparisons, unambiguous chromosome identification and orientation, as well as validation of marker positions.
Authors' contributions
JŚ participated in the resistance testing, reference marker search, carried out statistical and linkage analyses and drafted the manuscript. HJ selected the parental clones and obtained the mapping population, assessed the flower colour and participated in the resistance testing. MC, IT and AH-S took part in search and scoring of the reference markers. AK was responsible for the DArT analyses. EZ-G participated in the design of the study and its coordination. All authors read and approved the final manuscript.
Authors' information
Professor EZ-G is a Head of Młochów Research Centre within Plant Breeding and Acclimatization Institute -National Research Institute. Doctor JŚ leads a Phytopathology Laboratory and Doctor HJ leads Genetics Laboratory, while MC, AH-S and IT are PhD students in the same institution. Doctor AK is a Director of Diversity Arrays Technology Pty Ltd.
Supplementary Material
Sequence-specific markers used for the construction of genetic maps of the S. ruiz-ceballosii clone 99-10/36 and potato dihaploid of cv. Balbina. [7,15,16,34,42,58-61].
Genetic map of the dH Balbina × S. ruiz-ceballosii 99-10/36 population constructed using JoinMap® 4 software.
Contributor Information
Jadwiga Śliwka, Email: j.sliwka@ihar.edu.pl.
Henryka Jakuczun, Email: h.jakuczun@ihar.edu.pl.
Marcin Chmielarz, Email: m.chmielarz@ihar.edu.pl.
Agnieszka Hara-Skrzypiec, Email: a.hara@ihar.edu.pl.
Iga Tomczyńska, Email: i.tomczynska@ihar.edu.pl.
Andrzej Kilian, Email: a.kilian@diversityarrays.com.
Ewa Zimnoch-Guzowska, Email: e.zimnoch-guzowska@ihar.edu.pl.
Acknowledgements
The work was financed by The National Centre for Research and Development in Poland, grant: PBZ-MNiSW-2/3/2006 and the EU Integrated Project BIOEXPLOIT: FOOD-CT-2005-513959. The authors thank Doctor Louise R. Cooke for language corrections and Doctor Jarosław Plich for help in field late blight resistance assays. The authors thank also Professor W. Marczewski for kindly sharing some of the sequence-specific markers used in this study (Additional file 1).
References
- Knapp S. Celebrating spuds. Science. 2008;321:206–207. doi: 10.1126/science.1159278. [DOI] [PubMed] [Google Scholar]
- Świeżyński KM, Zimnoch-Guzowska E. Breeding potato cultivars with tubers resistant to Phytophthora infestans. Potato Res. 2001;44:97–117. doi: 10.1007/BF02360291. [DOI] [Google Scholar]
- Malcolmson JF. Races of Phytophthora infestans occurring in Great Britain. Trans Brit Mycol Soc. 1969;53:417–423. doi: 10.1016/S0007-1536(69)80099-9. [DOI] [Google Scholar]
- Song J, Bradeen JM, Naess SK, Raasch JA, Wielgus SM, Haberlach JT, Liu J, Kuang H, Austin-Phillips S, Buell CR, Helgeson JP, Jiang J. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. PNAS. 2003;100:9128–9133. doi: 10.1073/pnas.1533501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Vossen EAG, Sikkema A, Hekkert BL, Gros J, Stevens P, Muskens M, Wouters D, Pereira A, Stiekema W, Allefs S. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003;36:867–882. doi: 10.1046/j.1365-313X.2003.01934.x. [DOI] [PubMed] [Google Scholar]
- Lokossou AA, Park T-H, van Arkel G, Arens M, Ruyter-Spira C, Morales J, Whisson SC, Birch PRJ, Visser RGF, Jacobsen E, van der Vossen EAG. Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol Plant-Microbe Interact. 2009;22:630–641. doi: 10.1094/MPMI-22-6-0630. [DOI] [PubMed] [Google Scholar]
- Foster SJ, Park T-H, Pel M, Brigneti G, Śliwka J, Jagger L, van der Vossen E, Jones JD. Rpi-vnt1.1, a Tm-22homolog from Solanum venturii, confers resistance to potato late blight. Mol Plant Microbe Interact. 2009;22:589–600. doi: 10.1094/MPMI-22-5-0589. [DOI] [PubMed] [Google Scholar]
- Pel MA, Foster SJ, Park T-H, Rietman H, van Arkel G, Jones JDG, van Eck HJ, Jacobsen E, Visser RGF, van der Vossen EAG. Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. 2009. pp. 601–615. [DOI] [PubMed]
- Śliwka J, Jakuczun H, Lebecka R, Marczewski W, Gebhardt C, Zimnoch-Guzowska E. The novel, major locus Rpi-phul for late blight resistance maps to potato chromosome IX and is not correlated with long vegetation period. Theor Appl Gen. 2006;113:685–695. doi: 10.1007/s00122-006-0336-9. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VGAA, Rietman H, Krenek P, Champouret N, Young C, Oh S-K, Wang M, Bouwmeester K, Vosman B, Visser RGF, Jacobsen E, Govers F, Kamoun S, van der Vossen EAG. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE. 2008;3:e2875. doi: 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink D, Puddephat I. Deployment of disease resistance genes by plant transformation--a 'mix and match' approach. Trends Plant Sci. 1999;4:71–75. doi: 10.1016/S1360-1385(98)01372-7. [DOI] [PubMed] [Google Scholar]
- Zhu S, Li Y, Vossen JH, Visser RGF, Jacobsen E. Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res. 2011. doi:10.1007/s11248- 011-9510-1. [DOI] [PMC free article] [PubMed]
- Tan MYA, Hutten RCB, Celis C, Park T-H, Niks RE, Visser RGF, van Eck HJ. The Rpi-mcd1 locus from Solanum microdontum involved in resistance to Phytophthora infestans, causing a delay in infection, maps on potato Chromosome 4 in a cluster of NBS-LRR genes. Mol Plant-Microbe Interact. 2008;21:909–918. doi: 10.1094/MPMI-21-7-0909. [DOI] [PubMed] [Google Scholar]
- Ewing EE, Šimko I, Smart CD, Bonierbale MW, Mizubuti ESG, May GD, Fry WE. Genetic mapping from field tests of qualitative and quantitative resistance to Phytophthora infestans in a population derived from Solanum tuberosum and Solanum berthaultii. Mol Breeding. 2000;6:25–36. doi: 10.1023/A:1009648408198. [DOI] [Google Scholar]
- Rauscher GM, Smart CD, Šimko I, Bonierbale M, Mayton H, Greenland A, Fry WE. Characterization and mapping of RPi-ber, a novel potato late blight resistance gene from Solanum berthaultii. Theor Appl Genet. 2006;112:674–687. doi: 10.1007/s00122-005-0171-4. [DOI] [PubMed] [Google Scholar]
- Park T-H, Foster S, Brigneti G, Jones JDG. Two distinct potato late blight resistance genes from Solanum berthaultii are located on chromosome 10. Euphytica. 2009;165:269–278. doi: 10.1007/s10681-008-9784-4. [DOI] [Google Scholar]
- Tan MYA, Hutten RCB, Visser RGF, van Eck HJ. The effect of pyramiding Phytophthora infestans genes RPi-mcd1 and RPi-ber in potato. Theor Appl Genet. 2010;121:117–125. doi: 10.1007/s00122-010-1295-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes JG. The Potato, Evolution, Biodiversity and Genetic Resources. London: Belhaven Press; 1990. [Google Scholar]
- Solanaceae Source--Natural History Museum. http://www.nhm.ac.uk/research-curation/research/projects/solanaceaesource/
- Kardolus JP, van Eck HJ, van den Berg RG. The potential of AFLPs in biosystematics: a first application in Solanum taxonomy. Plant Syst Evol. 1998;210:87–103. doi: 10.1007/BF00984729. [DOI] [Google Scholar]
- Huamán Z, Ross RW. Updated listing of potato species names, abbreviations and taxonomic status. Am Potato J. 1985;62:629–641. doi: 10.1007/BF02854438. [DOI] [Google Scholar]
- Carlson-Nilsson BU, Zoteyeva NM. Crossability of wild potato species and advanced breeding lines resistant to late blight. Twelfth EuroBlight workshop, Arras (France), 3-6 May 2010. PPO-Special Report. 2010;14:209–212. [Google Scholar]
- Zoteyeva NM. Reconstruction of the potato wild species collection fromVIR and late blight research. CEEM Collaborative Project in Potato Late Blight Control, Progress Report, Cornell University; 1999. App 4. [Google Scholar]
- Zoteyeva N, Chrzanowska M, Evstratova L, Fasulati S, Yusupov T. Resistance to diseases and pests in wild potato accessions. Catalogue of VIR's Collection. St. Petersburg 88 pp. (in Russian) Issue 2004, 761.
- Danan S, Chauvin J-E, Caromel B, Moal J-D, Pellé R, Lefebvre V. Major-effect QTLs for stem and foliage resistance to late blight in the wild potato relatives Solanum sparsipilum and S. spegazzinii are mapped to chromosome X. Theor Appl Genet. 2009;119:705–719. doi: 10.1007/s00122-009-1081-7. [DOI] [PubMed] [Google Scholar]
- Bakari KA, Kerlan M-C, Caromel B, Dantec J-P, Fouville D, Manzanares-Dauleux M, Ellisèche D, Mugniéry D. A major gene mapped on chromosome XII is the main factor of a quantitatively inherited resistance to Meloidogyne fallax in Solanum sparsipilum. Theor Appl Genet. 2006;112:699–707. doi: 10.1007/s00122-005-0173-2. [DOI] [PubMed] [Google Scholar]
- Caromel B, Mugniéry D, Kerlan M-C, Andrzejewski S, Palloix A, Ellisèche D, Rousselle-Bourgeois F, Lefebvre V. Resistance quantitative trait loci originating from Solanum sparsipilum act independently on the sex ratio of Globodera pallida and together for developing a necrotic reaction. Mol Plant-Microbe Interact. 2005;18:1186–1194. doi: 10.1094/MPMI-18-1186. [DOI] [PubMed] [Google Scholar]
- Moury B, Caromel B, Johansen E, Simon V, Chauvin L, Jaquot E, Kerlan C, Lefebvre V. The helper component-proteinase cistron of Potato virus Y induces hypersensitivity and resistance in potato genotypes carrying dominant resistance genes on chromosome IV. Mol Plant-Microbe Interact. 2011;24:787–797. doi: 10.1094/MPMI-10-10-0246. [DOI] [PubMed] [Google Scholar]
- Horgan FG, Quiring DT, Lagnaoui A, Salas AR, Pelletier Y. Variations in resistance against Phthorimaea operculella in wild potato tubers. Entomol Exp Appl. 2010;137:269–279. doi: 10.1111/j.1570-7458.2010.01060.x. [DOI] [Google Scholar]
- Van Eck HJ, Jacobs JME, van Dijk J, Stiekema WJ, Jacobsen E. Identification and mapping of three flower colour loci of potato (S. tuberosum L.) by RFLP analysis. Theor Appl Genet. 1993;86:295–300. doi: 10.1007/BF00222091. [DOI] [PubMed] [Google Scholar]
- Van Eck HJ, Jacobs JME, van den Berg PMMM, Stiekema WJ, Jacobsen E. The inheritance of anthocyanin pigmentation in potato (Solanum tuberosum L.) and mapping of tuber skin colour loci using RFLPs. Heredity. 1994;73:410–421. doi: 10.1038/hdy.1994.189. [DOI] [Google Scholar]
- EUCABLIGHT Potato Late Blight Network For Europe. http://www.eucablight.org/EucaBlight.asp
- Bormann CA, Rickert AM, Castillo Ruiz RA, Paal J, Lübeck J, Strahwald J, Buhr K, Gebhardt C. Tagging quantitative trait loci for maturity-corrected late blight resistance in tetraploid potato with PCR-based candidate gene markers. Mol Plant-Microbe Interact. 2004;17:1126–1138. doi: 10.1094/MPMI.2004.17.10.1126. [DOI] [PubMed] [Google Scholar]
- Śliwka J, Jakuczun H, Lebecka R, Marczewski W, Gebhardt C, Zimnoch-Guzowska E. Tagging QTLs for late blight resistance and plant maturity from diploid wild relatives in a cultivated potato (Solanum tuberosum) background. Theor Appl Gen. 2007;115:101–112. doi: 10.1007/s00122-007-0546-9. [DOI] [PubMed] [Google Scholar]
- Fry WE. Quantification of general resistance of potato cultivars and fungicide effects for integrated control of potato late blight. Phytopathology. 1978;68:1650–1655. doi: 10.1094/Phyto-68-1650. [DOI] [Google Scholar]
- Śliwka J, Jakuczun H, Chmielarz M, Hara-Skrzypiec A, Tomczyńska I, Kilian A, Zimnoch-Guzowska E. A resistance gene against potato late blight originating from Solanum x Solanum x michoacanum maps to potato chromosome VII. Theor Appl Genet. 2012;124:397–406. doi: 10.1007/s00122-011-1715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccoud D, Peng K, Feinstein D, Kilian A. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acid Res. 2001;29:E25. doi: 10.1093/nar/29.4.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzl P, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A. Diversity arrays technology (DArT) for whole genome profiling of barley. Proc Natl Acad Sci USA. 2004;101:9915–9920. doi: 10.1073/pnas.0401076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden M, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A. Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor App Genet. 2006;113:1409–1420. doi: 10.1007/s00122-006-0365-4. [DOI] [PubMed] [Google Scholar]
- Sansaloni CP, Petroli CD, Carling J, Hudson CJ, Steane DA, Myburg AA, Grattapaglia D, Vaillancourt RE, Kilian A. A high-density Diversity Arrays Technology (DArT) microarray for genome-wide genotyping in Eucalyptus. Plant Methods. 2010;6:16. doi: 10.1186/1746-4811-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. JoinMap ® 4, Software for the Calculation of the Genetic Linkage Maps in Experimental Populations. Wageningen: Kyazma B.V.; 2006. [Google Scholar]
- SOL Genomics Network (SGN) Database. http://solgenomics.net/
- Rauscher G, Simko I, Mayton H, Bonierbale M, Smart CD, Grünwald NJ, Greenland A, Fry WE. Quantitative resistance to late blight from Solanum berthaultii cosegregates with R Pi-ber: insights in stability through isolates and environment. Theor Appl Genet. 2010;121:1553–1567. doi: 10.1007/s00122-010-1410-x. [DOI] [PubMed] [Google Scholar]
- Mayton H, Rauscher G, Simko I, Fry W. Evaluation of the RPi-ber late blight resistance gene for tuber resistance in the field and in laboratory. Plant Breed. 2011;130:464–468. doi: 10.1111/j.1439-0523.2010.01836.x. [DOI] [Google Scholar]
- Lapwood DH, McKee RK. Reaction of tubers of R-gene potato clones to inoculation with specialized races of Phytophthora infestans. Eur Potato J. 1961;4:3–14. doi: 10.1007/BF02364996. [DOI] [Google Scholar]
- Park T-H, Gros J, Sikkema A, Vleeshouwers VGAA, Muskens M, Allefs S, Jacobsen E, Visser RGF, van der Vossen EAG. The late blight resistance locus Rpi-blb3 from Solanum bulbocastanum belongs to a major late blight gene cluster on chromosome 4 of potato. Mol Plant-Microbe Interact. 2005;18:722–729. doi: 10.1094/MPMI-18-0722. [DOI] [PubMed] [Google Scholar]
- Oberhagemann P, Chatot-Balandras C, Schäfer-Pregl R, Wegener D, Palomino C, Salamini F, Bonnel E, Gebhardt C. A genetic analysis of quantitative resistance to late blight in potato: towards marker-assisted selection. Mol Breeding. 1999;5:399–415. doi: 10.1023/A:1009623212180. [DOI] [Google Scholar]
- Villamon FG, Spooner DM, Orrillo M, Mihovilovich E, Perez W, Bonierbale M. Late blight resistance linkages in a novel cross of the wild potato species Solanum paucissectum (series Piurana) Theor Appl Genet. 2005;111:1201–1214. doi: 10.1007/s00122-005-0053-9. [DOI] [PubMed] [Google Scholar]
- Simko I, Costanzo S, Ramanjulu V, Christ BJ, Haynes KG. Mapping polygenes for tuber resistance to late blight in a diploid Solanum phureja x S. stenotomum hybrid population. Plant Breeding. 2006;125:385–389. doi: 10.1111/j.1439-0523.2006.01232.x. [DOI] [Google Scholar]
- Danan S, Veyrieras J-B, Lefebvre V. Construction of a potato consensus map and QTL meta-analysis offer new insights into the genetic architecture of late blight resistance and plant maturity traits. BMC Plant Biology. 2011;11:16. doi: 10.1186/1471-2229-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P, Thoquet P, Olivier J, Laterrot H, Grimsley N. Genetic mapping of Ph-2, a single locus controlling partial resistance to Phytophthora infestans in tomato. Mol Plant-Microbe Interact. 1998;11:259–269. doi: 10.1094/MPMI.1998.11.4.259. [DOI] [Google Scholar]
- Reddy UK, Rong JK, Nimmakayala P, Vajja G, Rahman MA, Yu J, Soliman KM, Heller-Uszynska K, Kilian A, Paterson AH. Use of diversity arrays technology markers for integration into a cotton reference map and anchoring to a recombinant inbred line map. Genome. 2011;54:349–359. doi: 10.1139/g11-001. [DOI] [PubMed] [Google Scholar]
- Supriya A, Senthilvel S, Nepolean T, Eshwar K, Rajaram V, Shaw R, Hash CT, Kilian A, Yadav RC, Narasu ML. Development of a molecular linkage map of pearl millet integrating DArT and SSR markers. Theor Appl Genet. 2011;123:239–250. doi: 10.1007/s00122-011-1580-1. [DOI] [PubMed] [Google Scholar]
- Varshney RK, Penmetsa RV, Dutta S, Kulwal PL, Saxena RK, Datta S, Sharma TR, Rosen B, Carrasquilla-Garcia N, Farmer AD, Dubey A, Saxena KB, Gao J, Fakrudin B, Singh MN, Singh BP, Wanjari KB, Yuan M, Srivastava RK, Kilian A, Upadhyaya HD, Mallikarjuna N, Town CD, Bruening GE, He G, May GD, McCombie R, Jackson SA, Singh NK, Cook DR. Pigeonpea genomics initiative (PGI): an international effort to improve crop productivity of pigeonpea (Cajanus cajan L.) Mol Breed. 2010;26:393–408. doi: 10.1007/s11032-009-9327-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolibok-Brągoszewska H, Heller-Uszyńska K, Wenzl P, Uszyński G, Kilian A, Rakoczy-Trojanowska M. DArT markers for the rye genome-genetic diversity and mapping. BMC Genomics. 2009;10:578. doi: 10.1186/1471-2164-10-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H, Iorizzo M, Gao L, D'Agostino N, Carputo D, Chiusano ML, Bradeen JM. In: Genetics, Genomics and Breeding of Crop Plants: Potato. Bradeen JM, Kole C, editor. Enfield: Science Publishers and CRC Press; 2011. Molecular linkage maps: strategies, resources and achievements; pp. 68–89. [Google Scholar]
- The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189–195. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- Rickert AM, Kim JH, Meyer S, Nagel A, Ballvora A, Oefner PJ, Gebhardt C. First-generation SNP/InDel markers tagging loci for pathogen resistance in the potato genome. Plant Biotechnology J. 2003;1:399–410. doi: 10.1046/j.1467-7652.2003.00036.x. [DOI] [PubMed] [Google Scholar]
- Van der Vossen Gros JE, Sikkema A, Muskens M, Wouters D, Wolters P, Pereira A, Allefs S. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 2005;44:208–222. doi: 10.1111/j.1365-313X.2005.02527.x. [DOI] [PubMed] [Google Scholar]
- Marczewski W, Hennig J, Gebhardt C. The Potato virus S resistance gene Ns maps to potato chromosome VIII. Theor Appl Genet. 2002;105:564–567. doi: 10.1007/s00122-002-0976-3. [DOI] [PubMed] [Google Scholar]
- Huang S, Vleeshouwers VGAA, Werij JS, Hutten RCB, van Eck HJ, Visser RGF, Jacobsen E. The R3 resistance to Phytophthora infestans in potato is conferred by two closely linked R genes with distinct specificities. Mol Plant-Microbe Interact. 2004;17:428–435. doi: 10.1094/MPMI.2004.17.4.428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence-specific markers used for the construction of genetic maps of the S. ruiz-ceballosii clone 99-10/36 and potato dihaploid of cv. Balbina. [7,15,16,34,42,58-61].
Genetic map of the dH Balbina × S. ruiz-ceballosii 99-10/36 population constructed using JoinMap® 4 software.