Abstract
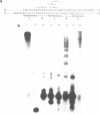
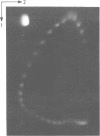
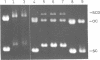
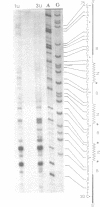
Polymorphic forms of the DNA duplex with long stretches of structural monotony are known. Several alternating purine-pyrimidine sequences have been shown to adopt left-handed Z-conformation. We report a DNA sequence d(CGCGCGATCGAT)n exhibiting alternating right-handed B and left-handed Z helical conformation after every half a turn. Further, this unusual conformation with change in handedness after every six base pairs was induced at physiological superhelical density.
Full text
PDF

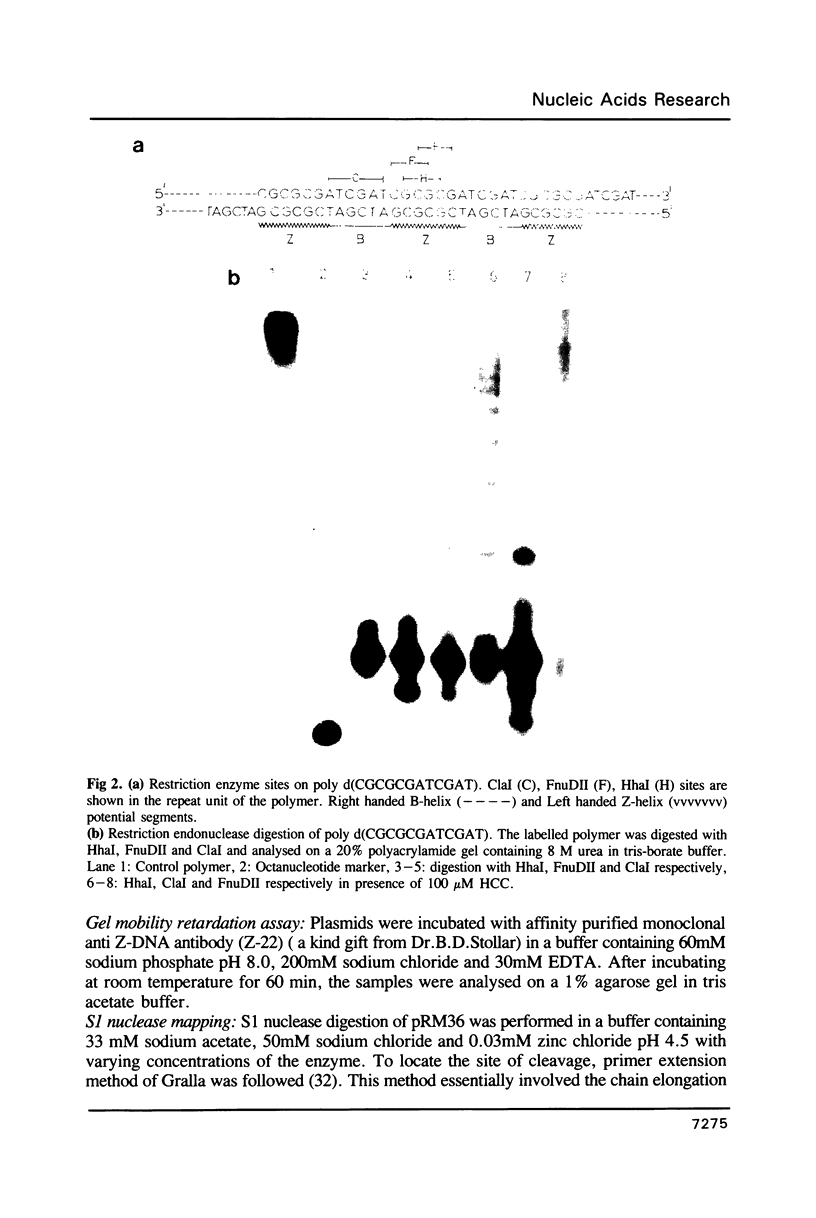

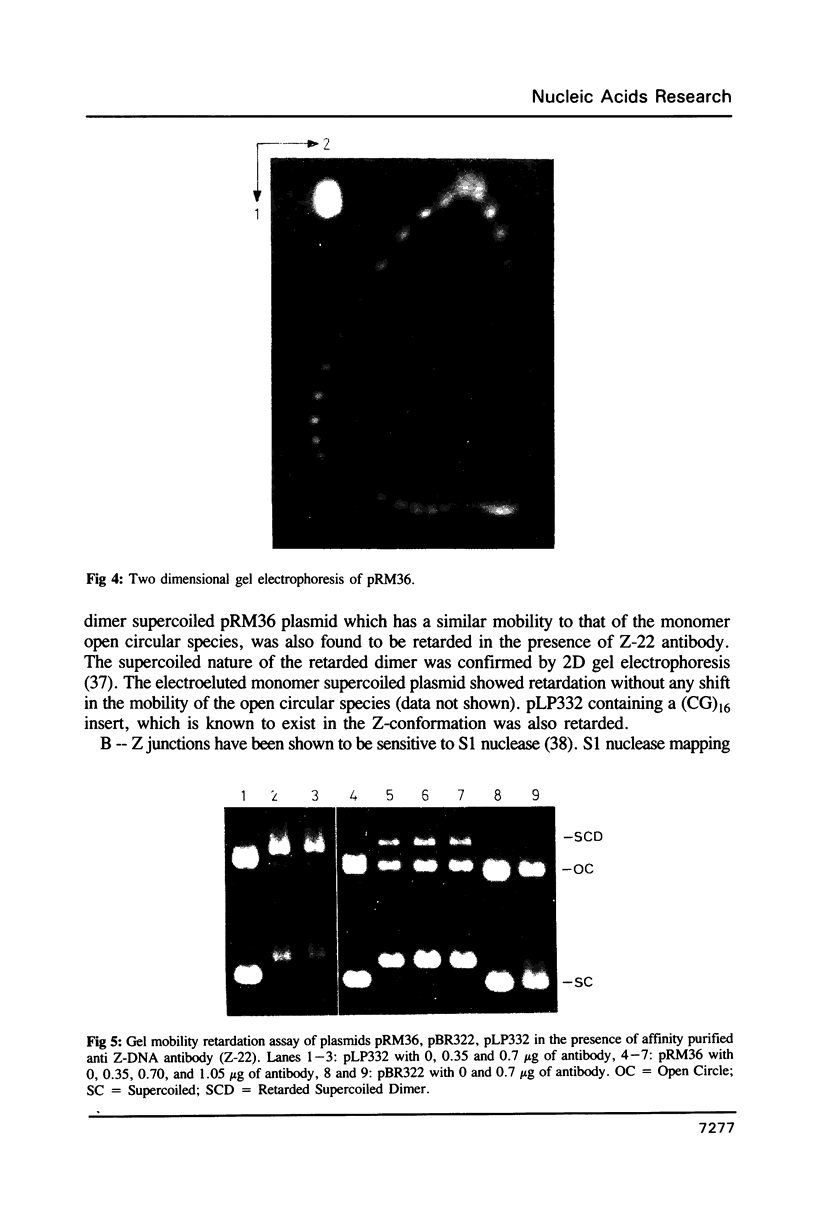




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azorin F., Hahn R., Rich A. Restriction endonucleases can be used to study B-Z junctions in supercoiled DNA. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5714–5718. doi: 10.1073/pnas.81.18.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behe M., Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B--Z transition in poly(dG-m5dC).poly(dG-m5dC). Proc Natl Acad Sci U S A. 1981 Mar;78(3):1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari S. K., Shouche Y. S., Cantor C. R., McClelland M. Sequences that adopt non-B-DNA conformation in form V DNA as probed by enzymic methylation. J Mol Biol. 1987 Jan 5;193(1):201–211. doi: 10.1016/0022-2836(87)90637-1. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Influence of DNA sequence and supercoiling on the process of cruciform formation. J Mol Biol. 1988 Jul 5;202(1):35–43. doi: 10.1016/0022-2836(88)90516-5. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Jendrisak J. J., Hager D. A., Burgess R. R. Purification and characterization of wheat germ DNA topoisomerase I (nicking-closing enzyme). J Biol Chem. 1981 Jun 10;256(11):5860–5865. [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K., Lilley D. M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985 Oct 5;185(3):461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Gupta G., Bansal M., Sasisekharan V. Conformational flexibility of DNA: polymorphism and handedness. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6486–6490. doi: 10.1073/pnas.77.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G., Bansal M., Sasisekharan V. Reversal of handedness in DNA: a stable link between RU and LZ helices. Biochem Biophys Res Commun. 1980 Dec 31;97(4):1258–1267. doi: 10.1016/s0006-291x(80)80002-7. [DOI] [PubMed] [Google Scholar]
- Hintermann G., Fischer H. M., Crameri R., Hütter R. Simple procedure for distinguishing CCC, OC, and L forms of plasmid DNA by agarose gel electrophoresis. Plasmid. 1981 May;5(3):371–373. doi: 10.1016/0147-619x(81)90012-3. [DOI] [PubMed] [Google Scholar]
- Jaworski A., Hsieh W. T., Blaho J. A., Larson J. E., Wells R. D. Left-handed DNA in vivo. Science. 1987 Nov 6;238(4828):773–777. doi: 10.1126/science.3313728. [DOI] [PubMed] [Google Scholar]
- Kadalayil L. P., Majumder K., Mishra R. K., Brahmachari S. K. Sequence specificity of Z-DNA formation in oligonucleotides. Biochem Int. 1988 Jul;17(1):121–131. [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klysik J., Stirdivant S. M., Singleton C. K., Zacharias W., Wells R. D. Effects of 5 cytosine methylation on the B-Z transition in DNA restriction fragments and recombinant plasmids. J Mol Biol. 1983 Jul 25;168(1):51–71. doi: 10.1016/s0022-2836(83)80322-2. [DOI] [PubMed] [Google Scholar]
- Kmiec E. B., Holloman W. K. Homologous pairing of DNA molecules by Ustilago rec1 protein is promoted by sequences of Z-DNA. Cell. 1986 Feb 28;44(4):545–554. doi: 10.1016/0092-8674(86)90264-3. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. A pH-dependent structural transition in the homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1985 Oct;3(2):327–338. doi: 10.1080/07391102.1985.10508420. [DOI] [PubMed] [Google Scholar]
- McLean M. J., Wells R. D. The role of sequence in the stabilization of left-handed DNA helices in vitro and in vivo. Biochim Biophys Acta. 1988 Sep 7;950(3):243–254. doi: 10.1016/0167-4781(88)90120-0. [DOI] [PubMed] [Google Scholar]
- Mishra R. K., Latha P. K., Brahmachari S. K. Interruptions of (CG)n sequences by GG, TG and CA need not prevent B to Z transition in solution. Nucleic Acids Res. 1988 May 25;16(10):4651–4665. doi: 10.1093/nar/16.10.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence. Cell. 1985 Jan;40(1):129–137. doi: 10.1016/0092-8674(85)90316-2. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Dinkelspiel K., Crea R. Simultaneous stability of short alternating Z and B helices in synthetic DNA concatamers. Nucleic Acids Res. 1982 Jun 25;10(12):3759–3768. doi: 10.1093/nar/10.12.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Vasser M., Dinkelspiel K., Crea R. Conformational stability of alternating d (CG) oligomers in high salt solution. Nucleic Acids Res. 1981 May 11;9(9):2195–2206. doi: 10.1093/nar/9.9.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrifoglio F., Manzini G., Yathindra N. Short oligodeoxynucleotides with d(G-C)n sequence do not assume left-handed conformation in high salt conditions. J Mol Biol. 1984 May 25;175(3):419–423. doi: 10.1016/0022-2836(84)90358-9. [DOI] [PubMed] [Google Scholar]
- Ramesh N., Brahmachari S. K. Critical cation balance in B leads to Z transition: role of Li+. FEBS Lett. 1983 Nov 28;164(1):33–37. doi: 10.1016/0014-5793(83)80013-1. [DOI] [PubMed] [Google Scholar]
- Ramesh N., Brahmachari S. K. Dynamic nature of B to Z transition: role of DNA supercoiling and Z-DNA binding protein. Indian J Biochem Biophys. 1988 Dec;25(6):542–547. [PubMed] [Google Scholar]
- Ramesh N., Shouche Y. S., Brahmachari S. K. Recognition of B and Z forms of DNA by Escherichia coli DNA polymerase I. J Mol Biol. 1986 Aug 20;190(4):635–638. doi: 10.1016/0022-2836(86)90248-2. [DOI] [PubMed] [Google Scholar]
- Rich A., Nordheim A., Wang A. H. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- Rodley G. A., Scobie R. S., Bates R. H., Lewitt R. M. A possible conformation for double-stranded polynucleotides. Proc Natl Acad Sci U S A. 1976 Sep;73(9):2959–2963. doi: 10.1073/pnas.73.9.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasisekharan V., Pattabiraman N., Gupta G. Some implications of an alternative structure for DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4092–4096. doi: 10.1073/pnas.75.9.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton C. K., Kilpatrick M. W., Wells R. D. S1 nuclease recognizes DNA conformational junctions between left-handed helical (dT-dG n. dC-dA)n and contiguous right-handed sequences. J Biol Chem. 1984 Feb 10;259(3):1963–1967. [PubMed] [Google Scholar]
- Vardimon L., Rich A. In Z-DNA the sequence G-C-G-C is neither methylated by Hha I methyltransferase nor cleaved by Hha I restriction endonuclease. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3268–3272. doi: 10.1073/pnas.81.11.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wells R. D. Unusual DNA structures. J Biol Chem. 1988 Jan 25;263(3):1095–1098. [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Kilpatrick M. W., Wells R. D. HhaI methylase and restriction endonuclease as probes for B to Z DNA conformational changes in d(GCGC) sequences. Nucleic Acids Res. 1984 Oct 25;12(20):7677–7692. doi: 10.1093/nar/12.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sande J. H., Ramsing N. B., Germann M. W., Elhorst W., Kalisch B. W., von Kitzing E., Pon R. T., Clegg R. C., Jovin T. M. Parallel stranded DNA. Science. 1988 Jul 29;241(4865):551–557. doi: 10.1126/science.3399890. [DOI] [PubMed] [Google Scholar]