Abstract
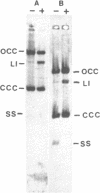
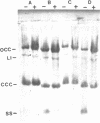
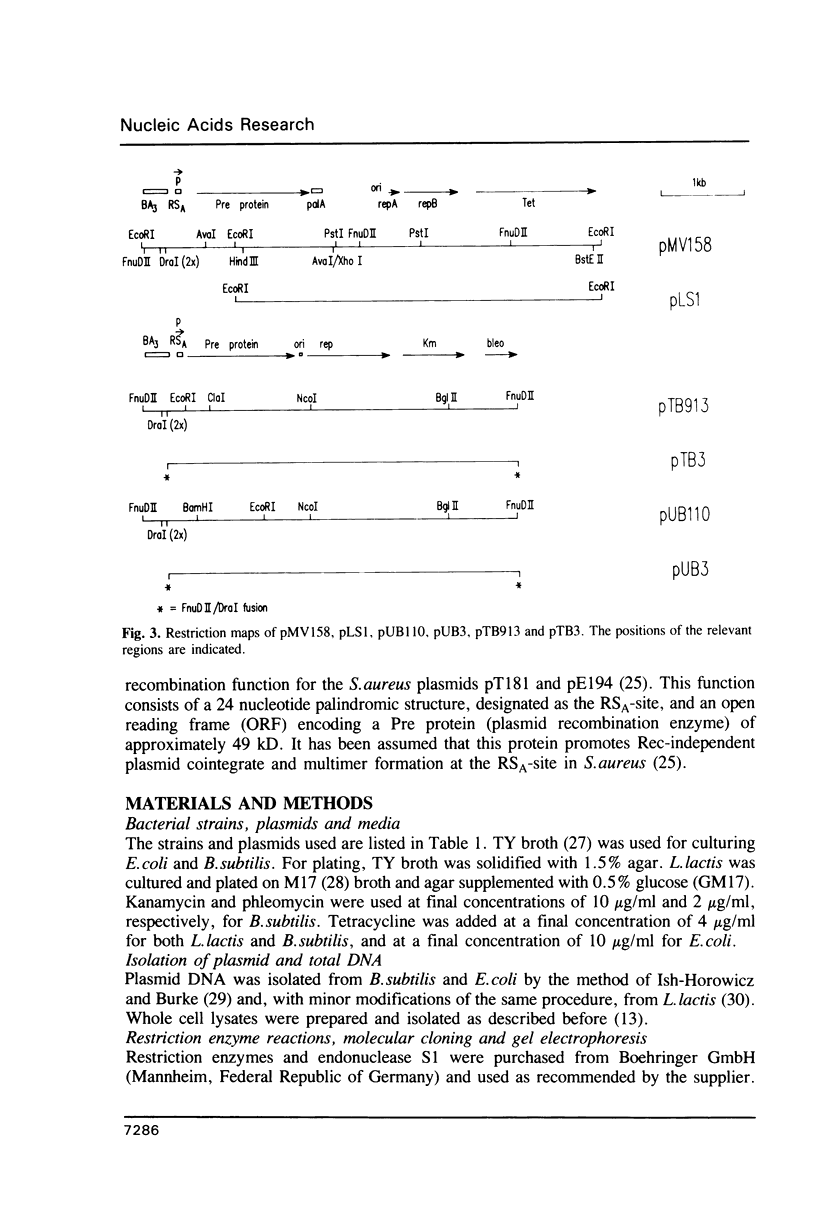
Plasmids pMV158 and pTB913, originating from Streptococcus agalactiae and a thermophilic Bacillus respectively, were sequenced to completion. Both contained a BA3-type minus origin of replication and an RSA-site, believed to constitute a site-specific recombination site. These two regions were more than 99% homologous to the corresponding regions of the Staphylococcus aureus plasmid pUB110. Deleting the BA3-type minus origin resulted in the accumulation of a considerable amount of single-stranded DNA, both in L. lactis subsp. lactis and B. subtilis, indicating that this minus origin was functional in both bacterial species. Like pUB110, both plasmids contained an open reading frame encoding a putative plasmid recombination enzyme (Pre protein), which was located downstream of the RSA-site. On the basis of sequence comparisons between pUB110, pMV158, pTB913, pT181, pE194, pNE131 and pT48 two distinct families of RSA-sites and Pre proteins could be distinguished.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bron S., Bosma P., van Belkum M., Luxen E. Stability function in the Bacillus subtilis plasmid pTA 1060. Plasmid. 1987 Jul;18(1):8–15. doi: 10.1016/0147-619x(87)90073-4. [DOI] [PubMed] [Google Scholar]
- Bron S., Luxen E., Swart P. Instability of recombinant pUB110 plasmids in Bacillus subtilis: plasmid-encoded stability function and effects of DNA inserts. Plasmid. 1988 May;19(3):231–241. doi: 10.1016/0147-619x(88)90041-8. [DOI] [PubMed] [Google Scholar]
- Bron S., Venema G. Ultraviolet inactivation and excision-repair in Bacillus subtilis. I. Construction and characterization of a transformable eightfold auxotrophic strain and two ultraviolet-sensitive derivatives. Mutat Res. 1972 May;15(1):1–10. doi: 10.1016/0027-5107(72)90086-3. [DOI] [PubMed] [Google Scholar]
- Burdett V. Identification of tetracycline-resistant R-plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother. 1980 Nov;18(5):753–760. doi: 10.1128/aac.18.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchpole I., Thomas C., Davies A., Dyke K. G. The nucleotide sequence of Staphylococcus aureus plasmid pT48 conferring inducible macrolide-lincosamide-streptogramin B resistance and comparison with similar plasmids expressing constitutive resistance. J Gen Microbiol. 1988 Mar;134(3):697–709. doi: 10.1099/00221287-134-3-697. [DOI] [PubMed] [Google Scholar]
- Chang S., Chang S. Y., Gray O. Structural and genetic analyses of a par locus that regulates plasmid partition in Bacillus subtilis. J Bacteriol. 1987 Sep;169(9):3952–3962. doi: 10.1128/jb.169.9.3952-3962.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Qasba P. K. Alkaline transfer of DNA to plastic membrane. Biochem Biophys Res Commun. 1984 Jul 18;122(1):340–344. doi: 10.1016/0006-291x(84)90480-7. [DOI] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Devine K. M., Hogan S. T., Higgins D. G., McConnell D. J. Replication and segregational stability of Bacillus plasmid pBAA1. J Bacteriol. 1989 Feb;171(2):1166–1172. doi: 10.1128/jb.171.2.1166-1172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro M. L., Kornblum J., Novick R. P. A site-specific recombination function in Staphylococcus aureus plasmids. J Bacteriol. 1987 Jun;169(6):2601–2610. doi: 10.1128/jb.169.6.2601-2610.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A. D., Ross H. F., Novick R. P. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Dubnau D. Construction and properties of chimeric plasmids in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1428–1432. doi: 10.1073/pnas.75.3.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Ano T., Fujii M., Aiba S. Two replication determinants of an antibiotic-resistance plasmid, pTB19, from a thermophilic bacillus. J Gen Microbiol. 1984 Jun;130(6):1399–1408. doi: 10.1099/00221287-130-6-1399. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aiba S. Isolation and characterization of antibiotic resistance plasmids from thermophilic bacilli and construction of deletion plasmids. J Bacteriol. 1981 Jun;146(3):1091–1097. doi: 10.1128/jb.146.3.1091-1097.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983 Nov;10(3):251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Kondo J. K., McKay L. L. Plasmid transformation of Streptococcus lactis protoplasts: optimization and use in molecular cloning. Appl Environ Microbiol. 1984 Aug;48(2):252–259. doi: 10.1128/aem.48.2.252-259.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S. A., Lopez P., Greenberg B., Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol. 1986 Dec 20;192(4):753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- Lampson B. C., Parisi J. T. Nucleotide sequence of the constitutive macrolide-lincosamide-streptogramin B resistance plasmid pNE131 from Staphylococcus epidermidis and homologies with Staphylococcus aureus plasmids pE194 and pSN2. J Bacteriol. 1986 Sep;167(3):888–892. doi: 10.1128/jb.167.3.888-892.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Katakura Y., Imanaka T., Aiba S. Enzymatic and nucleotide sequence studies of a kanamycin-inactivating enzyme encoded by a plasmid from thermophilic bacilli in comparison with that encoded by plasmid pUB110. J Bacteriol. 1984 Oct;160(1):413–420. doi: 10.1128/jb.160.1.413-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie T., Hoshino T., Tanaka T., Sueoka N. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986 Mar;15(2):93–103. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- Muller R. E., Ano T., Imanaka T., Aiba S. Complete nucleotide sequences of Bacillus plasmids pUB110dB, pRBH1 and its copy mutants. Mol Gen Genet. 1986 Jan;202(1):169–171. doi: 10.1007/BF00330534. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Iordanescu S., Surdeanu M., Edelman I. Transduction-related cointegrate formation between Staphylococcal plasmids: a new type of site-specific recombination. Plasmid. 1981 Sep;6(2):159–172. doi: 10.1016/0147-619x(81)90064-0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Puyet A., del Solar G. H., Espinosa M. Identification of the origin and direction of replication of the broad-host-range plasmid pLS1. Nucleic Acids Res. 1988 Jan 11;16(1):115–133. doi: 10.1093/nar/16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottländer E., Trautner T. A. Genetic and transfection studies with B, subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol Gen Genet. 1970;108(1):47–60. doi: 10.1007/BF00343184. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassi D. L., Lopez P., Espinosa M., Lacks S. A. Cloning of chromosomal genes in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7028–7032. doi: 10.1073/pnas.78.11.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Renz M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983 Jul 1;132(1):14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viret J. F., Alonso J. C. Generation of linear multigenome-length plasmid molecules in Bacillus subtilis. Nucleic Acids Res. 1987 Aug 25;15(16):6349–6367. doi: 10.1093/nar/15.16.6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G. H., Puyet A., Espinosa M. Initiation signals for the conversion of single stranded to double stranded DNA forms in the streptococcal plasmid pLS1. Nucleic Acids Res. 1987 Jul 24;15(14):5561–5580. doi: 10.1093/nar/15.14.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Are single-stranded circles intermediates in plasmid DNA replication? EMBO J. 1986 Mar;5(3):631–637. doi: 10.1002/j.1460-2075.1986.tb04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]
- van der Lelie D., Venema G. Bacillus subtilis generates a major specific deletion in pAM beta 1. Appl Environ Microbiol. 1987 Oct;53(10):2458–2463. doi: 10.1128/aem.53.10.2458-2463.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lelie D., van der Vossen J. M., Venema G. Effect of Plasmid Incompatibility on DNA Transfer to Streptococcus cremoris. Appl Environ Microbiol. 1988 Apr;54(4):865–871. doi: 10.1128/aem.54.4.865-871.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]