In patients with XLP, a primary immunodeficiency caused by mutations in SH2D1A, EBV infection can lead to somatic reversion of the disease-causing mutation selectively in effector memory CD8 T cells; reverted CD8 cells are better able to respond to and kill EBV-infected cells.
Abstract
Patients with the primary immunodeficiency X-linked lymphoproliferative disease (XLP), which is caused by mutations in SH2D1A, are highly susceptible to Epstein-Barr virus (EBV) infection. Nonetheless, some XLP patients demonstrate less severe clinical manifestations after primary infection. SH2D1A encodes the adaptor molecule SLAM-associated protein (SAP), which is expressed in T and natural killer cells and is required for cytotoxicity against B cells, the reservoir for EBV. It is not known why the clinical presentation of XLP is so variable. In this study, we report for the first time the occurrence of somatic reversion in XLP. Reverted SAP-expressing cells resided exclusively within the CD8+ T cell subset, displayed a CD45RA−CCR7− effector memory phenotype, and were maintained at a stable level over time. Importantly, revertant CD8+ SAP+ T cells, but not SAP− cells, proliferated in response to EBV and killed EBV-infected B cells. As somatic reversion correlated with EBV infection, we propose that the virus exerts a selective pressure on the reverted cells, resulting in their expansion in vivo and host protection against ongoing infection.
X-linked lymphoproliferative disease (XLP-1; referred to hereafter as XLP) is a primary immunodeficiency (PID) resulting from loss of function mutations in SH2D1A (Coffey et al., 1998; Nichols et al., 1998; Sayos et al., 1998). SH2D1A encodes SLAM-associated protein (SAP), a cytoplasmic adaptor protein involved in intracellular signaling downstream of the SLAM family of surface receptors (Ma et al., 2007; Schwartzberg et al., 2009; Cannons et al., 2011). XLP patients exhibit exquisite sensitivity to infection with the herpes group virus EBV (Bar et al., 1974; Purtilo et al., 1975; Sumegi et al., 2000; Nichols et al., 2005b). In contrast to healthy individuals, in whom primary infection is often asymptomatic (Hislop et al., 2007), many XLP patients suffer from severe, and often-fatal, fulminant infectious mononucleosis caused by an inability to control EBV infection (Nichols et al., 2005b; Ma et al., 2007). XLP patients can also develop hypogammaglobulinemia and B-lymphoma (Sumegi et al., 2000; Nichols et al., 2005b; Ma et al., 2007).
Several immune cell defects have been identified in XLP patients and Sh2d1a-deficient mice. These include reduced cytotoxicity of CD8+ T and NK cells, abolished NKT cell development, and impaired humoral immunity caused by compromised function of SAP-deficient CD4+ T cells (Nichols et al., 2005b; Ma et al., 2007; Schwartzberg et al., 2009; Cannons et al., 2011). We recently showed that SAP-deficient CD8+ T cells selectively fail to respond to B cell targets, yet they can be activated normally by other APCs, such as monocytes, DCs, or fibroblasts (Hislop et al., 2010; Palendira et al., 2011). Because EBV persists predominantly in B cells (Hislop et al., 2007), this provides a mechanism for the molecular pathogenesis of EBV infection in XLP. However, variability in the clinical presentation of XLP and the lack of a genotype-phenotype correlation (Sumegi et al., 2000; Booth et al., 2011) suggest that other factors influence disease progression, pathogenesis, and severity. Indeed, despite the presence of persistently high EBV viral loads (Chaganti et al., 2008), some XLP patients have milder clinical features, such as isolated hypogammaglobulinemia, and some exceed the mean life expectancy of XLP by several decades (Sumegi et al., 2000; Booth et al., 2011).
Milder clinical presentations in PIDs are often associated with somatic reversion, which results from a spontaneous genetic change in a disease-causing mutation in a somatic cell (Hirschhorn, 2003; Wada and Candotti, 2008). Cells harboring somatic reversions often expand because of a growth advantage or selective pressure (Hirschhorn, 2003; Wada and Candotti, 2008). Revertant somatic mosaicisms have been reported in several PIDs, including SCID (caused by mutations in ADA [Hirschhorn et al., 1996], IL2RG [Stephan et al., 1996; Speckmann et al., 2008], RAG1 [Wada et al., 2005], and CD3ζ [Rieux-Laucat et al., 2006]), X-linked ectodermal dysplasia with immunodeficiency (XL-EDA-ID; Nishikomori et al., 2004), Wiskott-Aldrich syndrome (WAS; Ariga et al., 2001; Wada et al., 2003; Stewart et al., 2007; Trifari et al., 2010), and lymphocyte adhesion deficiency-1 (LAD-1; Tone et al., 2007; Uzel et al., 2008). Although somatic reversion is infrequent overall, it has been observed in 11%, 18%, and 35% of patients with WAS (Stewart et al., 2007), Fanconi anemia (Kalb et al., 2007), and epidermolysis bullosa (Jonkman and Pasmooij, 2009), respectively. The resulting phenotype of patients with somatic reversion/mosaicism can range from mild immune defects to a completely normal state (Hirschhorn, 2003; Wada and Candotti, 2008). In this study, we examined reversion in XLP patients who have not undergone BM transplant and present evidence demonstrating that somatic mosaicism exists in a high proportion of patients. Somatic reversion was restricted to CD8+ T cells and correlated with exposure to EBV. Thus, the extrinsic selective effect of the virus appeared to be responsible for expanding the reverted SAP+ cells. Moreover, these SAP+ CD8+ T cells displayed EBV-specific cytotoxicity, indicating that they have the potential to protect XLP patients from the severe effects of EBV infection and possible progression to lymphoma in these individuals.
RESULTS AND DISCUSSION
Detection of somatic reversion in XLP patients
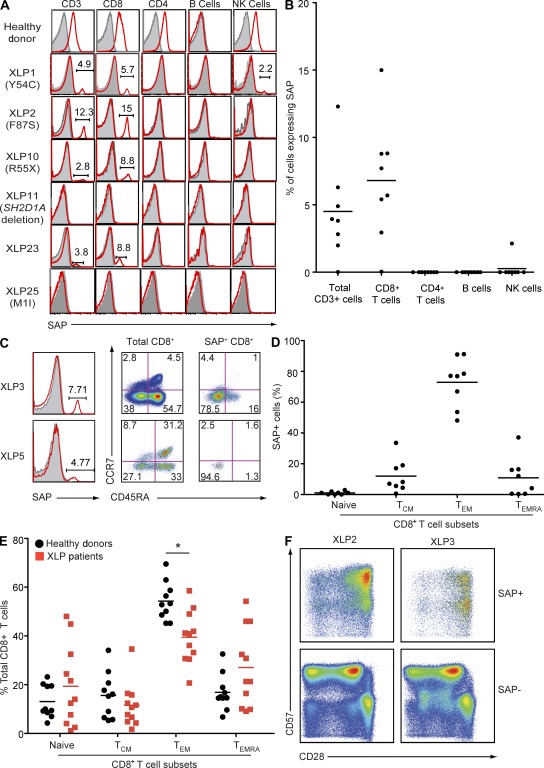
Previous Western blot analyses of SAP expression in XLP patients (Morra et al., 2001; Hare et al., 2006) demonstrated that most missense SH2D1A mutations abrogate expression. However, in some cases, residual levels of SAP were detected, which were assumed to represent expression of mutant protein. We have now reassessed SAP expression in 12 XLP patients from 10 different kindreds at the single cell level using flow cytometric–based intracellular staining (Palendira et al., 2011). The clinical features of these patients are listed in Table 1. In normal individuals, SAP is expressed in CD4+ and CD8+ T cells and NK cells but not B cells (Fig. 1 A; Palendira et al., 2011). As expected, SAP was not detected in any lymphocyte lineage from an XLP patient (XLP11) with a complete deletion of the SH2D1A locus (Fig. 1 A and Table 1). In contrast, 8/11 patients with a missense or nonsense SH2D1A mutations showed evidence of SAP expression in a small fraction (0.01–8.5%) of PBMCs (Table 1). This apparent reversion to SAP positivity occurred within the CD8+ T cell population in all cases, and in one case (XLP1), a small population (∼2%) of NK cells also expressed SAP (Fig. 1, A and B). In contrast, SAP was not detected in CD4+ T cells from any of the patients examined (Fig. 1, A and B). NKT cell development is largely abrogated by mutations in SH2D1A, i.e., percentage of NKT cells in controls, ∼0.2 ± 0.1% of CD3+ cells; XLP, 0.00095 ± 0.0002% (Nichols et al., 2005a). Although SAP was expressed by a small percentage of CD3+ T cells in the blood of XLP2 and XLP3, it appeared to be absent from the few residual NKT cells that could be detected in these patients (unpublished data). Thus, SAP is predominantly reexpressed in CD8+ T cells in XLP patients.
Table 1.
Molecular features of somatic reversion and EBV infection in XLP patients
| XLP patient | Age at analysis | Clinical features | Current status | Original mutation | Reversion | Reversion mutation | % SAP+ cells in PBMCs | EBV status |
| yr | % | |||||||
| 1 | 36–47 | •Clinical EBV: severe (19 yr) | •Deceased (age 49 yr) | c.161A>G; Y54C | Yes | c.161G>A; C54Y | 4.42 | + |
| •HLH: yes (49 yr) | ||||||||
| •Lymphoma: B-cell lymphoma, large bowel (10 yr) | ||||||||
| •Hypogamma: yes (19 yr) | ||||||||
| •Other: meningitis (19 yr), lymphoproliferation of CD4+CD8+ T cells (40 yr) | ||||||||
| 2 | 38–45 | •Clinical EBV: mild (17 yr) | •Stable | c.259T>C; F87S | Yes | c.259C>T/A; S87Y; S87F | 8.45 | + |
| •HLH: nil | •On IVIg | |||||||
| •Lymphoma: nil | ||||||||
| •Hypogamma: yes (26 yr) | ||||||||
| 3 | 42–52 | •Clinical EBV: mild (20 yr) | •Stable | c.259T>C; F87S | Yes | c.259C>A; S87Y | 2.29 | + |
| •HLH: nil | •On IVIg | |||||||
| •Lymphoma: nil | ||||||||
| •Hypogamma: yes (21 yr) | ||||||||
| 5 | 11–17 | •Clinical EBV: nil | •Stable | c.251T>C; I84T | Yes | c.251C>T; T84I | 1.67 | + |
| •HLH: nil | ||||||||
| •Lymphoma: B-cell lymphoma | ||||||||
| •Hypogamma: nil | ||||||||
| 8 | 16 | •Clinical EBV: severe (10-12 yr); severe reactivation (21 yr) | •BMT (age 21 yr) | c.251T>C; I84T | Yes | c.251C>T; T84I | 1.63 | + |
| •HLH: nil | •Stable | |||||||
| •Lymphoma: nil | ||||||||
| •Hypogamma: yes (13 yr) | ||||||||
| 10 | 31 | •Clinical EBV: nil | •Deceased (age 31 yr) | c.163C>T; R55X | Yes | c.163T>A/C; X55R | 1.82 | + |
| •HLH: yes (26 yr) | ||||||||
| •Lymphoma: nil | ||||||||
| •Hypogamma: nil | ||||||||
| •Other: vasculitis | ||||||||
| 11 | 34 | •Clinical EBV: fulminant | •Stable | Deletion | No | NA | Nil | + |
| •HLH: nil | •On IVIg | |||||||
| •Lymphoma: nil | ||||||||
| •Hypogamma: nil | ||||||||
| •Other: psoriasis, chronic active hepatitis, IBD | ||||||||
| 23 | 38 | •Clinical EBV: severe (12 yr) | •Deceased (age 37 yr) | ND | Yes | ND | 1.94 | + |
| •HLH: nil | ||||||||
| •Lymphoma: unknown | ||||||||
| •Hypogamma: yes (16 yr) | ||||||||
| 24 | 3 | •Clinical EBV: fulminant (3 yr) | •Stable 3 yr after BMT | c.3G>T; M1I | No | NA | <LOD | + (acute infection) |
| •HLH: yes (3 yr) | ||||||||
| •Lymphoma: possible non-Hodgkin’s lymphoma (3 yr) | ||||||||
| •Hypogamma: unknown | ||||||||
| 25 | 17 | •Clinical EBV: nil | •Stable 2 yr after BMT | c.3G>T; M1I | No | NA | <LOD | − |
| •HLH: nil | ||||||||
| •Lymphoma: small bowel lymphoma (12 yr) | ||||||||
| •Hypogamma: yes | ||||||||
| 26 | 11 | •Clinical EBV: nil | •Stable | c.160T>C; Y54H | Yes | Below limit of sensitivity | 0.01 | + |
| •HLH: nil | •On IVIg | |||||||
| •Lymphoma: nil | ||||||||
| •Hypogamma: yes (7 yr) | ||||||||
| 27 | Cord blood | •Clinical EBV: nil | •Deceased | c.163C>T; R55X | No | NA | <LOD | − |
| •HLH: nil | ||||||||
| •Lymphoma: nil | ||||||||
| •Hypogamma: nil |
BMT, BM transplant; HLH, hemophagocytic lymphohistiocytosis; hypogamma, hypogammaglobulinemia; IVIg, intravenous Ig; LOD, limit of detection by flow cytometry; NA, not applicable. XLP2 and XLP3 are siblings; XLP5 and XLP8 are first cousins. The values in parentheses indicate age of presentation of each clinical feature. EBV load was determined by quantitative PCR.
Figure 1.
Somatic reversion in XLP patients is confined to effector memory CD8+ T cells. (A and B) PBMCs were labeled with mAbs against CD3, CD4, CD8, CD20, and CD56, fixed and permeabilized, and then incubated with isotype control (gray histograms in A) or anti-SAP mAb (red histograms in A). Expression of SAP in total CD3+ T, CD4+ T, CD8+ T, B, and NK cells was determined by gating on CD3+, CD4+, CD8+, CD20+, and CD3−CD56+ cells, respectively. The histograms in A are from a healthy donor and six unrelated XLP patients. The text in parentheses (e.g., Y54C) refers to the mutation in SH2D1A for each patient. The symbols in B depict the percentage of SAP+ cells in each indicated lymphocyte subset examined in each of the XLP patients with somatic reversion (n = 8). (C and D) PBMCs were labeled with mAbs against CD8, CD45RA, and CCR7, fixed and permeabilized, and incubated with isotype control (gray histograms in C) or anti-SAP mAb (red histograms in C). The FACS plots in C depict SAP expression in CD8+ T cells from two unrelated XLP patients (left), CD8+ T cell subsets (naive [CD45RA+CCR7+], TCM [CD45RA−CCR7+], TEM [CD45RA−CCR7−], and TEMRA [CD45RA+CCR7−]) in two unrelated XLP patients (middle), and the distribution of SAP expression in naive and memory CD8+ T cells (right). (D) Distribution of SAP+ cells in CD8+ T cell subsets in XLP patients that have undergone reversion (n = 8). Each symbol depicts the percentage of CD8+ naive, TCM, TEM, and TEMRA cells in each XLP patient with somatic reversion that expresses SAP (n = 8). (E) The frequencies of naive, TCM, TEM, and TEMRA CD8+ T cells in the peripheral blood of healthy donors (n = 10) and XLP patients (n = 11) were determined by labeling PBMCs with mAbs against CD8, CD45RA, and CCR7. *, P < 0.005. Values for XLP27 are not included as subset analysis was performed on cord blood, which contains predominantly naive cells. (B, D, and E) Horizontal bars indicate the mean. (F) PBMCs from XLP2 and XLP3 were incubated with mAbs against CD8, CD28, and CD57, fixed and permeabilized, and then incubated with anti-SAP mAb. Expression of CD28 and CD57 on SAP+ and SAP− CD8+ T cells was then determined.
To determine which CD8+ T cells expressed SAP, XLP PBMCs were labeled with anti-CD45RA and CCR7 to identify naive (CD45RA+CCR7+), central memory (TCM; CD45RA−CCR7+), effector memory (TEM; CD45RA−CCR7−), and TEMRA (CD45RA+CCR7− effector memory) cells (Sallusto et al., 1999). Irrespective of their absolute numbers, most SAP+ cells (∼75%) lay in the TEM compartment, with only smaller proportions in the TCM or TEMRA and none in the naive subsets (Fig. 1, C and D). The total CD8+ T cell pool in the blood of XLP patients is often skewed, with most cells falling into the CD45RA+CCR7− TEMRA subset at the expense of the TEM subset (Fig. 1 E; Plunkett et al., 2005). Furthermore, the TEMRA cells showed a CD57+CD28± phenotype (Fig. 1 F) that has been linked with functional exhaustion and replicative senescence (Brenchley et al., 2003). However, the great majority of revertant SAP+ cells were not of this type; instead, they had a phenotype typical of normal TEM cells (i.e., CD45RA−CCR7−CD28+CD57+/−; Fig. 1 F). Thus, unlike XL-EDA-ID (Nishikomori et al., 2004) and some cases of LAD-1 deficiency (Tone et al., 2007; Uzel et al., 2008), where reverted CD8+ T cells are mostly exhausted (CD57+CD28−) TEMRA cells, the increased frequency of CD8+ TEMRA cells in XLP does not reflect accumulation of reverted SAP-expressing cells. Furthermore, when analyzed for TCR usage by Vβ staining, the reverted CD8+ T cells generally expressed multiple TCR Vβ chains (unpublished data), suggesting that somatic mosaicism in XLP is not caused by the occurrence of reversion in a single clone. This is similar to reverted patients with X-SCID (Stephan et al., 1996; Speckmann et al., 2008) and WAS (Trifari et al., 2010) but contrasts those with LAD-1 and XL-EDA-ID, where reverted cells express monoclonal or oligoclonal TCRs (Nishikomori et al., 2004; Wada et al., 2005; Uzel et al., 2008).
Molecular characterization of somatic reversions in SH2D1A
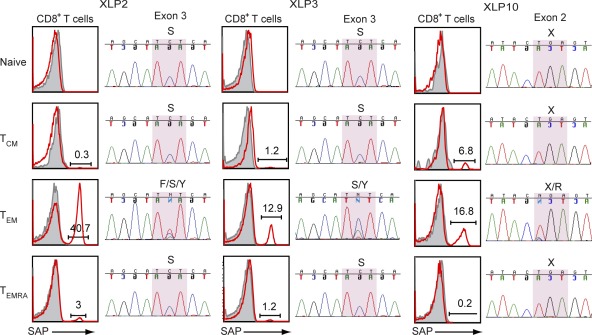
To understand the molecular basis of SAP expression in lymphocytes of XLP patients, genomic DNA was isolated from CD8+ T cell subsets, and the affected exon of SH2D1A was sequenced. This confirmed that somatic reversion within SH2D1A occurred infrequently, if at all, in naive, TCM, or TEMRA CD8+ T cells, being largely confined to CD8+ TEM cells (Fig. 2). In the patients analyzed with detectable SAP-expressing cells, we could demonstrate heterozygosity at the point of the original mutation (Fig. 2 and Table 1). With the exception of XLP3, all patients had acquired somatic reversions that resulted in expression of WT nucleotide and amino acid sequences (Table 1). The reversion in XLP3 yielded a tyrosine at position 87 (Y87), instead of a phenylalanine (F87) which occurs in the WT sequence. As tyrosine is a conserved amino acid in the SH2 domains of several proteins (Coffey et al., 1998; Sayos et al., 1998; Sumegi et al., 2000), its presence presumably allows expression of SAP in this patient’s cells (Fig. 1), whereas most other nonconserved nucleotide substitutions within SH2D1A abrogate SAP expression (Morra et al., 2001; Hare et al., 2006). We could not detect the reverted sequence in XLP26 because of the very low frequency (0.05% of CD8+ T cells) of SAP+ cells in this patient.
Figure 2.
Molecular characterization of somatic reversion in XLP. Genomic DNA was isolated from CD8+ T cell subsets (naive, TCM, TEM, and TEMRA), and the exons encoding SH2D1A were sequenced. Chromatograms of DNA sequences of affected exons from three XLP patients are shown. The highlighted section within the sequence reveals multiple histogram peaks, which represent reverted sequences. For the FACS plots shown, open red histograms represent fluorescence of cells labeled with an anti-SAP mAb; solid gray histograms represent the fluorescence of cells incubated with an isotype control mAb. See Table 1 for more details.
The SH2D1A gene in two XLP patients had undergone more than one reversion event. In XLP2, one reversion resulted in expression of the WT nucleotide (C>A) and protein (S87F), whereas another yielded the SAP87Y variant that was also present in his brother (XLP3; Fig. 2) but not their mother (Palendira et al., 2011). This latter finding suggests that the presence of SAP-expressing cells, at least in XLP2 and XLP3, was not the result of maternal engraftment. The two somatic reversions detected in XLP10 (T>A and T>C) both encode an arginine at position 55, corresponding to WT SAP. This sequencing analysis confirms the multiclonal nature of the somatic reversion, as indicated by the TCR Vβ staining.
A stable somatically reverted population of CD8+ TEM cells is maintained in vivo over time
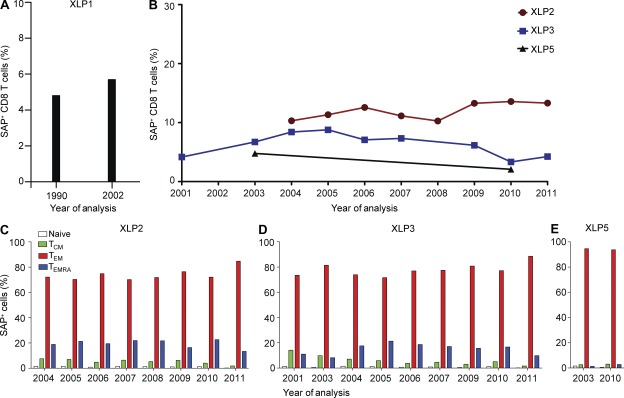
To determine whether somatically reverted cells increase in frequency in vivo over time, we enumerated the proportion of SAP+ cells in PBMCs from four XLP patients that had been sampled over a period of 7–11 yr. In all patients, SAP+ cells were present at the earliest times assessed (Fig. 3, A and B). Interestingly, the population of SAP+ CD8+ T cells was maintained at a relatively stable level (i.e., ∼5–10%; Fig. 3, A and B), with the revertants being predominantly TEM cells with a CD28+CD57low/− phenotype at all times (Fig. 3, C–E; and not depicted). Thus, reverted SAP-expressing CD8+ T cell numbers were maintained for decades, mirroring the stability of EBV loads in these same patients (Chaganti et al., 2008).
Figure 3.
Reverted cells are maintained at a stable level over time in vivo. PBMCs were collected from four XLP patients at different times and labeled with mAbs against CD8, CD45RA, and CCR7, fixed and permeabilized, and then incubated with anti-SAP mAb. (A and B) The frequency of revertant (i.e., SAP+) cells within CD8+ T cells from XLP1 (A) and XLP2, XLP3, and XLP5 (B). (C–E) The distribution of SAP+ cells within the naive, TCM, TEM, and TEMRA subsets of XLP2 (C), XLP3 (D), and XLP5 (E) was determined.
In other PIDs, the expansion of reverted cells usually results from acquisition of an intrinsic survival or growth advantage. This is consistent with the finding of skewed X chromosome inactivation in female carriers of WAS, X-linked agammaglobulinemia, and X-SCID (Puck et al., 1987), where lymphocytes expressing the WT gene preferentially survive. The fact that there was no continuous expansion of reverted SAP+ cells in XLP suggested there is no growth advantage for SAP+ cells. Our findings of comparable proportions of SAP− and SAP+ cells in populations of T and NK cells in female carriers of XLP (Palendira et al., 2011) support the notion that SAP expression per se does not endow cells with a selective intrinsic growth advantage.
EBV infection is associated with somatic reversion in XLP
The finding that reverted SAP+ cells in XLP patients were predominantly CD8+ TEM cells was interesting because >50% of EBV-specific cells in healthy individuals reside within this subset (Hislop et al., 2007). This led us to investigate a potential link between the presence of somatically reverted CD8+ T cells in XLP patients and prior EBV infection. Because XLP patients receive passive Ig, the EBV infection status of most patients was determined by viral DNA detection. This approach revealed that all eight XLP patients who had undergone somatic reversion were EBV positive, whereas the two patients with missense SH2D1A mutations who did not exhibit reversion were EBV− (XLP25 and XLP27; Table 1). Moreover, XLP24, who had not undergone somatic reversion in SH2D1A and did not express detectable amounts of SAP in his lymphocytes (Table 1), was actually tested during primary EBV infection. Remarkably, three patients found to exhibit somatic reversion (XLP5, XLP10, and XLP26; Table 1) had not experienced symptomatic EBV infection, yet viral DNA was clearly detectable in their B cells (Table 1). This highlights the importance of determining EBV load in isolated B cells from XLP patients, rather than total PBMCs, to confidently infer infection history. Further evidence linking long-term EBV carriage and reversion came from the observation that the XLP patient with the highest viral load (XLP2; Chaganti et al., 2008) also had the greatest frequency of reverted cells (Table 1). A previous study has shown that, as in virus carriers, XLP patients harbor EBV only in B cells (Chaganti et al., 2008). Thus, reversion back to a functional SH2D1A gene could not have been driven by direct infection of CD8+ T cells per se. However, another possible explanation for the link between SH2D1A reversion and EBV positivity was that reverted CD8+ T cells had been selected during the host response to infection.
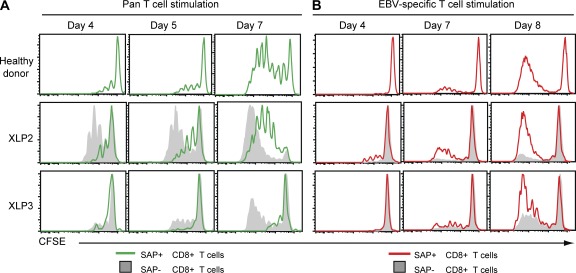
In this regard, EBV loads persist at relatively high levels in the B cell pool of XLP patients (Chaganti et al., 2008), and SAP− CD8+ T cells are impaired in their recognition of B cell targets (Hislop et al., 2010; Palendira et al., 2011). We therefore asked whether EBV infection in XLP patients was the driving force that selects for and expands reactivities from within rare revertant SAP+ CD8+ T cells. We compared the ability of reverted SAP+ and corresponding mutant SAP− cells from XLP2 and XLP3 to proliferate in vitro in response to different stimuli. Upon pan-T cell activation (anti-CD2/CD3/CD28 mAbs), SAP− CD8+ T cells underwent greater expansion than SAP+ CD8+ T cells from both XLP patients and healthy donors, as revealed by dilution of CFSE, at all times examined (Fig. 4 A, compare gray with green histograms). This is consistent with previous studies showing increased proliferation and impaired apoptosis of human and mouse SAP-deficient CD8+ T cells (Chen et al., 2005, 2007; Snow et al., 2009). In contrast, when using an EBV-specific stimulus (autologous B-lymphoblastoid cell lines [LCLs] coated with cognate EBV peptides), SAP+ CD8+ T cells from XLP patients underwent substantially more proliferation than SAP− CD8+ T cells in the same culture (Fig. 4 B, compare red with gray histograms). In fact, proliferation of reverted SAP+ CD8+ T cells in XLP patients approximated that of CD8+ T cells from normal healthy EBV-immune donors responding to the same EBV-specific stimulus (Fig. 4 B). These data demonstrate that the ability to respond to B cells presenting EBV antigen allows SAP+ CD8+ T cells to undergo preferential expansion at the expense of their mutant SAP-deficient counterparts. However, heightened proliferation of SAP− CD8+ T cells to other (non-B cell tropic) antigen challenges in vivo likely prevents SAP+ cells from dominating the CD8+ T cell subset in XLP patients and provides an explanation for the persistence of revertant cells at relatively constant levels. The acute, and often fatal, lymphoproliferation typically observed in some XLP patients in the setting of primary EBV infection (Bar et al., 1974; Purtilo et al., 1975) may reflect expansion of a functionally ineffective T cell response occurring in the absence of sufficient revertant CD8+ T cells with appropriate EBV specificity.
Figure 4.
Reverted cells expand in the presence of EBV stimulation. (A and B) CD8+ T cells from two XLP patients or a healthy control were labeled with CFSE and then cultured with anti-CD3, anti-CD28, and CD2 mAb (pan-T cell stimulation; A) or autologous LCLs pulsed with EBV peptides (EBV-specific T cell stimulation; B). EBV responses from XLP2 (HLA-B*4402 and HLA-B*5701), XLP3 (HLA-B*4402 and HLA-B*2705), and a healthy control (HLA-B*4402) were measured by stimulating cells with known EBV peptides as described in Materials and methods. Proliferation of SAP+ (green histograms in A; red in B) and SAP− cells (gray histograms) was assessed at the indicated times by determining CFSE dilution. Data are representative of two independent experiments.
Reverted cells in XLP are functional
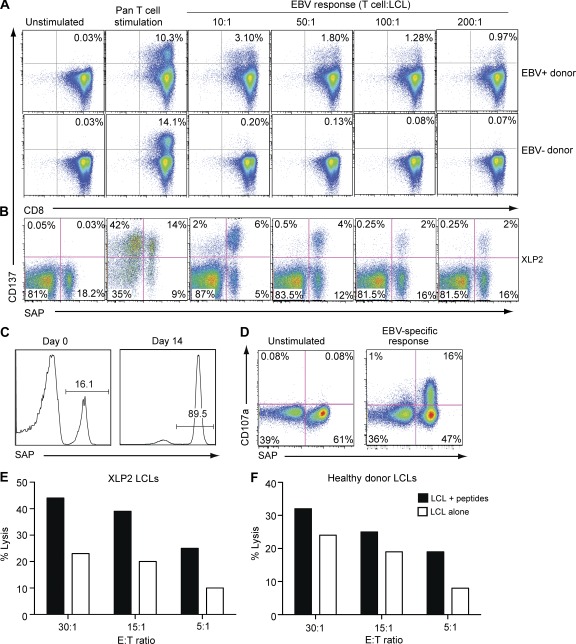
SAP-deficient CD8+ T cells have a defect in their ability to recognize EBV infection in B cells (Dupré et al., 2005; Hislop et al., 2010; Palendira et al., 2011), yet several of the patients examined here (XLP1, XLP2, XLP3, XLP10, and XLP23; Table 1) had reached an age that exceeded the median survival for XLP (4–11 yr; Table 1; Booth et al., 2011; Sumegi et al., 2000). We therefore hypothesized that some of the SAP-reverted CD8+ T cells had EBV-specific function, thereby contributing to prolonged host survival. To test this, we first examined the ability of CD8+ T cells from EBV+ XLP patients, and from EBV-seropositive or -seronegative normal donors as controls, to respond to EBV challenge in vitro. Purified CD8+ T cells were cultured with autologous EBV-transformed LCLs at different effector/stimulator ratios or with a control pan-T cell stimulus, and 48 h later, expression of the co-stimulatory molecule CD137 (41BB) was examined (Wehler et al., 2008). Pan-T cell stimulation induced detectable CD137 expression on CD8+ T cells from both EBV-seropositive and -seronegative normal donors, as well as on SAP− and reverted SAP+ CD8+ T cells from XLP patients (Fig. 5, A and B). In contrast, only CD8+ T cells from EBV-seropositive normal donors acquired CD137 after LCL stimulation, thereby demonstrating the EBV specificity of this response (Fig. 5 A). Strikingly, for the XLP patients examined, not only did most of the LCL-responding (i.e., CD137+) cells come from the reverted SAP+ population (Fig. 5 B), but at an effector/stimulator ratio of 10:1, >50% of that population responded to LCLs, suggesting that the majority of the reverted SAP+ cells are EBV specific.
Figure 5.
Reverted cells in XLP are functional. (A and B) CD8+ T cells from EBV-seropositive (EBV+) and -seronegative (EBV−) normal donors (A) and XLP2 (B) were cultured with autologous LCLs at the indicated effector/target (T cell/LCL) ratio or with staphylococcal enterotoxin B (A: normal donors) or anti-CD3, anti-CD28, and anti-CD2 mAb (B: XLP; pan-T cell stimulation); CD137 expression was determined after 48 h. (C) EBV-specific CD8+ T cell lines were generated from XLP2 by repeated stimulation with peptide-pulsed autologous LCLs as detailed in Materials and methods. The frequency of SAP+ cells was determined before and after 2 wk of in vitro expansion. (D) EBV-specific CD8+ T cell lines, containing both SAP− and SAP+ cells (as shown in C) were tested for their ability to degranulate, as measured by CD107a up-regulation, in response to antigen presented on autologous LCLs. The values in A, B, and D represent the proportion of cells in each indicated quadrant. (E and F) After 4 wk in culture, EBV-specific CD8+ T cell lines from XLP2 were tested for their ability to kill autologous (E) or HLA-matched (F) LCLs presenting endogenous antigen (open bars) or after pulsing with specific peptides (VEITPYKPTW and EENLLDFVRF; closed bars). Independent cell lines were established from XLP2 for the experiments presented in C–F.
To extend these findings to cytotoxic function, XLP CD8+ T cells were repeatedly stimulated with EBV peptide-pulsed autologous LCLs. The outcome of this was twofold. First, substantial enrichment of SAP+ cells occurred (Fig. 5 C), providing further evidence that the ability of reverted SAP+ CD8+ T cells to respond to B cell targets gives them a proliferative advantage over SAP− cells. Second, analysis of expression of CD107a, a marker of degranulation (Betts et al., 2003), to this LCL stimulus revealed that >90% of responding CD8+ T cells were SAP+ (Fig. 5 D). Moreover, in cytotoxicity assays, these expanded XLP CD8+ T cells efficiently killed autologous and MHC class I–matched LCL targets presenting endogenously processed EBV antigen, and lysis was augmented in response to the relevant peptide-pulsed targets (Fig. 5, E and F). This contrasts with the impaired cytotoxic function of EBV-specific SAP− CD8+ T cells (Dupré et al., 2005; Hislop et al., 2010; Palendira et al., 2011). Collectively, these in vitro findings demonstrate that somatically reverted CD8+ T cells in XLP are functional and provide a possible explanation of why some XLP patients may be able to control long-term EBV infection in vivo.
Overall, our data clearly demonstrate for the first time the occurrence of somatic reversion in XLP and strongly suggest that EBV plays a key role in de novo expansion of reverted cells. Somatic reversion is also largely restricted to T cells in WAS (Ariga et al., 2001; Wada et al., 2003; Trifari et al., 2010), X-SCID (Stephan et al., 1996; Speckmann et al., 2008), and LAD-1 (Tone et al., 2007; Uzel et al., 2008). In fact, reversion has not been observed in myeloid cells in any PIDs. The finding of lineage-restricted expression of a revertant disease-causing mutation in these PIDs points to the reversion event occurring in a lineage-committed precursor cell, rather than a pluripotent progenitor cell. However, it is possible that a very low frequency of reverted cells exists in lymphoid cells other than CD8+ T cells in XLP, but in the absence of any selective pressure, these cells do not undergo sufficient expansion to render them detectable by the assays used here. Our findings of a polyclonal TCR repertoire in reverted SAP+ CD8+ T cells coupled with the presence of more than one reversion in some patients suggest that the initial reversion event occurred before TCR rearrangement in developing thymocytes and that the pressure provided by EBV resulted in selective expansion of EBV-specific CD8+, but not CD4+, T cells, which then differentiated into TEM cells. Alternatively, because the reverted cells were largely confined to the CD8+ TEM subset, reversion may have occurred exclusively in naive CD8+ T cells and only became apparent once these cells became TEM cells after EBV infection and expansion. Irrespective of the mechanism, our data demonstrate that the ability of revertant SAP+ CD8+ T cells to respond to B cells, the primary reservoir of EBV, enables them to undergo preferential expansion and acquisition of effector function, which is sufficient to contain the virus and prevent overwhelming infection. This infers that lack of SAP in CD8+ T cells, but not other cells such as NK and NKT cells, is the primary factor underlying susceptibility to EBV-induced disease in XLP. Importantly, somatic reversion is unlikely to improve impaired humoral immunity in XLP, as this defect is intrinsic to SAP-deficient CD4+ T cells (Ma et al., 2005, 2007; Nichols et al., 2005b; Schwartzberg et al., 2009; Cannons et al., 2011), which did not exhibit mosaicism. Indeed, despite detection of reversion and evidence of viral control (Chaganti et al., 2008), some of the XLP patients studied continued to require Ig replacement therapy (Table 1). Based on our findings, further studies that examine the relationship between mosaicism and clinical outcome are warranted. Lastly, these observations have implications for gene therapy, inasmuch that restoring SAP in only a small subset of CD8+ T cells may be sufficient to improve anti-EBV immunity in XLP patients.
MATERIALS AND METHODS
XLP patients.
Blood samples collected from 12 XLP patients (11 peripheral blood and 1 cord blood) from 10 different kindreds were used in this study. Mononuclear cells were isolated using standard procedure and used either fresh or cryopreserved in liquid nitrogen. Genomic DNA was isolated from PBMCs or purified sort-purified subsets of CD8+ T cells and used for sequence analysis. In some patients, RNA was isolated and used to synthesize cDNA for sequence analysis. The primers used for amplification of the four exons of SH2D1A are as follows: Exon 1 sense, 5′-CAACATCCTGTTGTTGGGG-3′; Exon 1 antisense, 5′-CCAGGGAATGAAATCCCC-3′; Exon 2 sense, 5′-GCAATGACACCATATACG-3′; Exon 2 antisense, 5′-GAACAATTTTGGATTGGAGC-3′; Exon 3 sense, 5′-GTAAGCTCTTCTGGAATG-3′; Exon 3 antisense, 5′-CATCTACTTTCTCACTGC-3′; Exon 4 sense, 5′-CTGTGTTGTGTCATTGTG-3′; and Exon 4 antisense, 5′-GCTTCCATTACAGGACTAC-3′. Details of the mutations and the reverted sequences along with other patient information are presented in Table 1. All participants gave written informed consent, and the experiments were approved by the Human Research Ethic committees of the Sydney South West Area Health Service (Royal Prince Alfred and Concord Zones) and St. Vincent’s Hospital (Darlinghurst, Sydney).
Flow cytometric analysis.
PBMCs were stained with fluorochrome-conjugated mAbs specific for cell surface receptors. The following mAbs were used to identify different lymphocyte populations: anti-CD3, CD4, CD8 (T cells), CD56 (NK cells), and CD20 (B cells; BD). To identify naive and memory T cells, mAbs against CCR7 (R&D Systems) and CD45RA (BD; Sallusto et al., 1999) were used. For degranulation assays, mAb against CD107a (BD) was used as previously described (Betts et al., 2003; Palendira et al., 2011). Stained cells were then analyzed on either a FACSCanto I or II flow cytometer (BD), and the data were processed using FlowJo software (Tree Star).
Detection of SAP by intracellular staining.
Expression of SAP was determined as previously described (Palendira et al., 2011). Cells were first stained for surface markers and then fixed with 2% paraformaldehyde, permeabilized with 0.5% saponin, and then incubated with either Alexa Fluor 647 (Invitrogen)–conjugated isotype control or Alexa Fluor 647 anti-SAP mAb (clone 1C9; Abnova). Cells were washed and resuspended in PBS containing 1% fetal calf serum and analyzed by flow cytometry.
T cell stimulation.
The capacity of SAP+ and SAP− CD8+ T cells to respond to EBV peptides was measured using MHC class I restricted synthetic peptides. 1–2 × 106 purified CD8+ T cells were labeled with CFSE and then stimulated in vitro with autologous EBV-LCLs pulsed with irrelevant (negative control) or specific peptides (1 µg/ml) or with T cell activation and expansion (TAE) beads (anti-CD3/CD28/CD2 mAb beads; Miltenyi Biotech; positive control) for 5 d. The capacity to respond to cognate peptides was analyzed by determining the proliferation history, based on dilution of CFSE, of the CD8+ T cells after different days of culture. EBV peptides used in CFSE experiments were HLA-B*4402–restricted VEITPYKPTW (latent protein EBNA 3B), EENLLDFVRF (latent protein EBNA 3C), and HLA-B*5801–restricted VSFIEFVGW (EBNA 3B) for XLP2 and VEITPYKPTW, EENLLDFVRF, and HLA-B*2705–restricted KRPPIFIRRL (latent protein EBNA 3A) for XLP3. Control cells were stimulated with VEITPYKPTW and EENLLDFVRF peptides. Alternatively, in some experiments, CD8+ T cells were tested for their ability to degranulate in the presence of peptide-pulsed autologous EBV-LCLs by staining for CD107a (Betts et al., 2003). The capacity of CD8+ T cells to respond to EBV-LCLs was also examined ex vivo by assessing expression of CD137 (eBioscience; Wehler et al., 2008). Purified CD8+ T cells were cultured with either EBV-LCLs at different effector/target ratios or with TAE beads or Staphylococcal enterotoxin B. After 48 h of stimulation, CD137 expression by SAP− and SAP+ CD8+ T cells was analyzed by immunofluorescence and flow cytometry.
T cell lines and cytotoxicity assay.
To generate virus-specific T cell lines, CD8+ T cells were isolated from PBMCs and stimulated repeatedly over 4–8 wk with EBV peptide-pulsed autologous EBV-LCLs. Peptides used in generating EBV-specific lines were HLA-B*4402–restricted VEITPYKPTW and EENLLDFVRV. The ability of CD8+ T cell lines to respond to EBV peptides was measured by staining for the degranulation marker CD107a after stimulation with 1 µg/ml of peptide-pulsed LCLs (Betts et al., 2003). Where cytotoxicity was measured, autologous LCLs and HLA-matched control LCLs were sensitized with cognate peptide (HLA-B*4402–restricted EBV epitopes VEITPYKPTW and EENLLDFVRV) at concentrations of 1 µg/ml while loading with sodium 51chromate. After washing, T cells were incubated at different APC/T cell ratios and incubated for 5 h in standard cytotoxicity assay.
Vβ repertoire staining of cells.
Vβ repertoire staining was performed using the IOTest Beta Mark kit (Beckman Coulter). The protocol was slightly modified from the product manual because of the nature of the samples involved. Staining of PBMCs was performed as described in Flow cytometric analysis. Each tube contained mAbs to three different Vβ receptors: one conjugated to FITC, another conjugated to PE, and a third conjugated to both FITC and PE.
Measurement of EBV load.
Quantitative PCR analysis was performed to estimate viral genome levels as described previously (Junying et al., 2003). 106 B cells were sort purified from PBMCs, and genomic DNA was extracted using the UltraClean Tissue and Cells DNA isolation kit (MO BIO) in accordance with the manufacturer’s protocol.
Acknowledgments
We thank Tony Basten, Rob Brink, and Elissa Deenick for critical review of this manuscript and the XLP patients and their families for their involvement in this work.
This project was funded by grants from the XLP Research Trust, Cancer Council New South Wales, and the Association for International Cancer Research (to S.G. Tangye), the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (to A.D. Klion), and the Medical Research Council UK and Cancer Research UK (to A.B. Rickinson). S.G. Tangye is the recipient of a Senior Research Fellowship from the National Health and Medical Research Council of Australia.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- LCL
- lymphoblastoid cell line
- PID
- primary immunodeficiency
- SAP
- SLAM-associated protein
- WAS
- Wiskott-Aldrich syndrome
- XL-EDA-ID
- X-linked ectodermal dysplasia with immunodeficiency
- XLP
- X-linked lymphoproliferative disease
References
- Ariga T., Kondoh T., Yamaguchi K., Yamada M., Sasaki S., Nelson D.L., Ikeda H., Kobayashi K., Moriuchi H., Sakiyama Y. 2001. Spontaneous in vivo reversion of an inherited mutation in the Wiskott-Aldrich syndrome. J. Immunol. 166:5245–5249 [DOI] [PubMed] [Google Scholar]
- Bar R.S., DeLor C.J., Clausen K.P., Hurtubise P., Henle W., Hewetson J.F. 1974. Fatal infectious mononucleosis in a family. N. Engl. J. Med. 290:363–367 10.1056/NEJM197402142900704 [DOI] [PubMed] [Google Scholar]
- Betts M.R., Brenchley J.M., Price D.A., De Rosa S.C., Douek D.C., Roederer M., Koup R.A. 2003. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods. 281:65–78 10.1016/S0022-1759(03)00265-5 [DOI] [PubMed] [Google Scholar]
- Booth C., Gilmour K.C., Veys P., Gennery A.R., Slatter M.A., Chapel H., Heath P.T., Steward C.G., Smith O., O’Meara A., et al. 2011. X-linked lymphoproliferative disease due to SAP/SH2D1A deficiency: a multicenter study on the manifestations, management and outcome of the disease. Blood. 117:53–62 (published erratum appears in Blood. 2011. 118:5060) 10.1182/blood-2010-06-284935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J.M., Karandikar N.J., Betts M.R., Ambrozak D.R., Hill B.J., Crotty L.E., Casazza J.P., Kuruppu J., Migueles S.A., Connors M., et al. 2003. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 101:2711–2720 10.1182/blood-2002-07-2103 [DOI] [PubMed] [Google Scholar]
- Cannons J.L., Tangye S.G., Schwartzberg P.L. 2011. SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol. 29:665–705 10.1146/annurev-immunol-030409-101302 [DOI] [PubMed] [Google Scholar]
- Chaganti S., Ma C.S., Bell A.I., Croom-Carter D., Hislop A.D., Tangye S.G., Rickinson A.B. 2008. Epstein-Barr virus persistence in the absence of conventional memory B cells: IgM+IgD+CD27+ B cells harbor the virus in X-linked lymphoproliferative disease patients. Blood. 112:672–679 10.1182/blood-2007-10-116269 [DOI] [PubMed] [Google Scholar]
- Chen G., Tai A.K., Lin M., Chang F., Terhorst C., Huber B.T. 2005. Signaling lymphocyte activation molecule-associated protein is a negative regulator of the CD8 T cell response in mice. J. Immunol. 175:2212–2218 [DOI] [PubMed] [Google Scholar]
- Chen G., Tai A.K., Lin M., Chang F., Terhorst C., Huber B.T. 2007. Increased proliferation of CD8+ T cells in SAP-deficient mice is associated with impaired activation-induced cell death. Eur. J. Immunol. 37:663–674 10.1002/eji.200636417 [DOI] [PubMed] [Google Scholar]
- Coffey A.J., Brooksbank R.A., Brandau O., Oohashi T., Howell G.R., Bye J.M., Cahn A.P., Durham J., Heath P., Wray P., et al. 1998. Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat. Genet. 20:129–135 10.1038/2424 [DOI] [PubMed] [Google Scholar]
- Dupré L., Andolfi G., Tangye S.G., Clementi R., Locatelli F., Aricò M., Aiuti A., Roncarolo M.G. 2005. SAP controls the cytolytic activity of CD8+ T cells against EBV-infected cells. Blood. 105:4383–4389 10.1182/blood-2004-08-3269 [DOI] [PubMed] [Google Scholar]
- Hare N.J., Ma C.S., Alvaro F., Nichols K.E., Tangye S.G. 2006. Missense mutations in SH2D1A identified in patients with X-linked lymphoproliferative disease differentially affect the expression and function of SAP. Int. Immunol. 18:1055–1065 10.1093/intimm/dxl039 [DOI] [PubMed] [Google Scholar]
- Hirschhorn R. 2003. In vivo reversion to normal of inherited mutations in humans. J. Med. Genet. 40:721–728 10.1136/jmg.40.10.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn R., Yang D.R., Puck J.M., Huie M.L., Jiang C.K., Kurlandsky L.E. 1996. Spontaneous in vivo reversion to normal of an inherited mutation in a patient with adenosine deaminase deficiency. Nat. Genet. 13:290–295 10.1038/ng0796-290 [DOI] [PubMed] [Google Scholar]
- Hislop A.D., Taylor G.S., Sauce D., Rickinson A.B. 2007. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu. Rev. Immunol. 25:587–617 10.1146/annurev.immunol.25.022106.141553 [DOI] [PubMed] [Google Scholar]
- Hislop A.D., Palendira U., Leese A.M., Arkwright P.D., Rohrlich P.S., Tangye S.G., Gaspar H.B., Lankester A.C., Moretta A., Rickinson A.B. 2010. Impaired Epstein-Barr virus-specific CD8+ T-cell function in X-linked lymphoproliferative disease is restricted to SLAM family-positive B-cell targets. Blood. 116:3249–3257 10.1182/blood-2009-09-238832 [DOI] [PubMed] [Google Scholar]
- Jonkman M.F., Pasmooij A.M. 2009. Revertant mosaicism—patchwork in the skin. N. Engl. J. Med. 360:1680–1682 10.1056/NEJMc0809896 [DOI] [PubMed] [Google Scholar]
- Junying J., Herrmann K., Davies G., Lissauer D., Bell A., Timms J., Reynolds G.M., Hubscher S.G., Young L.S., Niedobitek G., Murray P.G. 2003. Absence of Epstein-Barr virus DNA in the tumor cells of European hepatocellular carcinoma. Virology. 306:236–243 10.1016/S0042-6822(02)00027-2 [DOI] [PubMed] [Google Scholar]
- Kalb R., Neveling K., Hoehn H., Schneider H., Linka Y., Batish S.D., Hunt C., Berwick M., Callen E., Surralles J., et al. 2007. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am. J. Hum. Genet. 80:895–910 10.1086/517616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.S., Hare N.J., Nichols K.E., Dupré L., Andolfi G., Roncarolo M.G., Adelstein S., Hodgkin P.D., Tangye S.G. 2005. Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells. J. Clin. Invest. 115:1049–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.S., Nichols K.E., Tangye S.G. 2007. Regulation of cellular and humoral immune responses by the SLAM and SAP families of molecules. Annu. Rev. Immunol. 25:337–379 10.1146/annurev.immunol.25.022106.141651 [DOI] [PubMed] [Google Scholar]
- Morra M., Simarro-Grande M., Martin M., Chen A.S., Lanyi A., Silander O., Calpe S., Davis J., Pawson T., Eck M.J., et al. 2001. Characterization of SH2D1A missense mutations identified in X-linked lymphoproliferative disease patients. J. Biol. Chem. 276:36809–36816 10.1074/jbc.M101305200 [DOI] [PubMed] [Google Scholar]
- Nichols K.E., Harkin D.P., Levitz S., Krainer M., Kolquist K.A., Genovese C., Bernard A., Ferguson M., Zuo L., Snyder E., et al. 1998. Inactivating mutations in an SH2 domain-encoding gene in X-linked lymphoproliferative syndrome. Proc. Natl. Acad. Sci. USA. 95:13765–13770 10.1073/pnas.95.23.13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols K.E., Hom J., Gong S.Y., Ganguly A., Ma C.S., Cannons J.L., Tangye S.G., Schwartzberg P.L., Koretzky G.A., Stein P.L. 2005a. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat. Med. 11:340–345 10.1038/nm1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols K.E., Ma C.S., Cannons J.L., Schwartzberg P.L., Tangye S.G. 2005b. Molecular and cellular pathogenesis of X-linked lymphoproliferative disease. Immunol. Rev. 203:180–199 10.1111/j.0105-2896.2005.00230.x [DOI] [PubMed] [Google Scholar]
- Nishikomori R., Akutagawa H., Maruyama K., Nakata-Hizume M., Ohmori K., Mizuno K., Yachie A., Yasumi T., Kusunoki T., Heike T., Nakahata T. 2004. X-linked ectodermal dysplasia and immunodeficiency caused by reversion mosaicism of NEMO reveals a critical role for NEMO in human T-cell development and/or survival. Blood. 103:4565–4572 10.1182/blood-2003-10-3655 [DOI] [PubMed] [Google Scholar]
- Palendira U., Low C., Chan A., Hislop A.D., Ho E., Phan T.G., Deenick E.K., Cook M.C., Riminton D.S., Choo S., et al. 2011. Molecular pathogenesis of EBV susceptibility in XLP as revealed by analysis of female carriers with heterozygous expression of SAP. PLoS Biol. 9:e1001187 10.1371/journal.pbio.1001187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett F.J., Franzese O., Belaramani L.L., Fletcher J.M., Gilmour K.C., Sharifi R., Khan N., Hislop A.D., Cara A., Salmon M., et al. 2005. The impact of telomere erosion on memory CD8+ T cells in patients with X-linked lymphoproliferative syndrome. Mech. Ageing Dev. 126:855–865 10.1016/j.mad.2005.03.006 [DOI] [PubMed] [Google Scholar]
- Puck J.M., Nussbaum R.L., Conley M.E. 1987. Carrier detection in X-linked severe combined immunodeficiency based on patterns of X chromosome inactivation. J. Clin. Invest. 79:1395–1400 10.1172/JCI112967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purtilo D.T., Cassel C.K., Yang J.P., Harper R. 1975. X-linked recessive progressive combined variable immunodeficiency (Duncan’s disease). Lancet. 305:935–941 10.1016/S0140-6736(75)92004-8 [DOI] [PubMed] [Google Scholar]
- Rieux-Laucat F., Hivroz C., Lim A., Mateo V., Pellier I., Selz F., Fischer A., Le Deist F. 2006. Inherited and somatic CD3zeta mutations in a patient with T-cell deficiency. N. Engl. J. Med. 354:1913–1921 10.1056/NEJMoa053750 [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lenig D., Förster R., Lipp M., Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Sayos J., Wu C., Morra M., Wang N., Zhang X., Allen D., van Schaik S., Notarangelo L., Geha R., Roncarolo M.G., et al. 1998. The X-linked lymphoproliferative-disease gene product SAP regulates signals induced through the co-receptor SLAM. Nature. 395:462–469 10.1038/26683 [DOI] [PubMed] [Google Scholar]
- Schwartzberg P.L., Mueller K.L., Qi H., Cannons J.L. 2009. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat. Rev. Immunol. 9:39–46 10.1038/nri2456 [DOI] [PubMed] [Google Scholar]
- Snow A.L., Marsh R.A., Krummey S.M., Roehrs P., Young L.R., Zhang K., van Hoff J., Dhar D., Nichols K.E., Filipovich A.H., et al. 2009. Restimulation-induced apoptosis of T cells is impaired in patients with X-linked lymphoproliferative disease caused by SAP deficiency. J. Clin. Invest. 119:2976–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speckmann C., Pannicke U., Wiech E., Schwarz K., Fisch P., Friedrich W., Niehues T., Gilmour K., Buiting K., Schlesier M., et al. 2008. Clinical and immunologic consequences of a somatic reversion in a patient with X-linked severe combined immunodeficiency. Blood. 112:4090–4097 10.1182/blood-2008-04-153361 [DOI] [PubMed] [Google Scholar]
- Stephan V., Wahn V., Le Deist F., Dirksen U., Broker B., Müller-Fleckenstein I., Horneff G., Schroten H., Fischer A., de Saint Basile G. 1996. Atypical X-linked severe combined immunodeficiency due to possible spontaneous reversion of the genetic defect in T cells. N. Engl. J. Med. 335:1563–1567 10.1056/NEJM199611213352104 [DOI] [PubMed] [Google Scholar]
- Stewart D.M., Candotti F., Nelson D.L. 2007. The phenomenon of spontaneous genetic reversions in the Wiskott-Aldrich syndrome: a report of the workshop of the ESID Genetics Working Party at the XIIth Meeting of the European Society for Immunodeficiencies (ESID). Budapest, Hungary October 4-7, 2006. J. Clin. Immunol. 27:634–639 10.1007/s10875-007-9121-z [DOI] [PubMed] [Google Scholar]
- Sumegi J., Huang D., Lanyi A., Davis J.D., Seemayer T.A., Maeda A., Klein G., Seri M., Wakiguchi H., Purtilo D.T., Gross T.G. 2000. Correlation of mutations of the SH2D1A gene and Epstein-Barr virus infection with clinical phenotype and outcome in X-linked lymphoproliferative disease. Blood. 96:3118–3125 [PubMed] [Google Scholar]
- Tone Y., Wada T., Shibata F., Toma T., Hashida Y., Kasahara Y., Koizumi S., Yachie A. 2007. Somatic revertant mosaicism in a patient with leukocyte adhesion deficiency type 1. Blood. 109:1182–1184 10.1182/blood-2007-08-039057 [DOI] [PubMed] [Google Scholar]
- Trifari S., Scaramuzza S., Catucci M., Ponzoni M., Mollica L., Chiesa R., Cattaneo F., Lafouresse F., Calvez R., Vermi W., et al. 2010. Revertant T lymphocytes in a patient with Wiskott-Aldrich syndrome: analysis of function and distribution in lymphoid organs. J. Allergy Clin. Immunol. 125:439–448: e8 10.1016/j.jaci.2009.11.034 [DOI] [PubMed] [Google Scholar]
- Uzel G., Tng E., Rosenzweig S.D., Hsu A.P., Shaw J.M., Horwitz M.E., Linton G.F., Anderson S.M., Kirby M.R., Oliveira J.B., et al. 2008. Reversion mutations in patients with leukocyte adhesion deficiency type-1 (LAD-1). Blood. 111:209–218 10.1182/blood-2007-04-082552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Candotti F. 2008. Somatic mosaicism in primary immune deficiencies. Curr. Opin. Allergy Clin. Immunol. 8:510–514 10.1097/ACI.0b013e328314b651 [DOI] [PubMed] [Google Scholar]
- Wada T., Konno A., Schurman S.H., Garabedian E.K., Anderson S.M., Kirby M., Nelson D.L., Candotti F. 2003. Second-site mutation in the Wiskott-Aldrich syndrome (WAS) protein gene causes somatic mosaicism in two WAS siblings. J. Clin. Invest. 111:1389–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Toma T., Okamoto H., Kasahara Y., Koizumi S., Agematsu K., Kimura H., Shimada A., Hayashi Y., Kato M., Yachie A. 2005. Oligoclonal expansion of T lymphocytes with multiple second-site mutations leads to Omenn syndrome in a patient with RAG1-deficient severe combined immunodeficiency. Blood. 106:2099–2101 10.1182/blood-2005-03-0936 [DOI] [PubMed] [Google Scholar]
- Wehler T.C., Karg M., Distler E., Konur A., Nonn M., Meyer R.G., Huber C., Hartwig U.F., Herr W. 2008. Rapid identification and sorting of viable virus-reactive CD4(+) and CD8(+) T cells based on antigen-triggered CD137 expression. J. Immunol. Methods. 339:23–37 10.1016/j.jim.2008.07.017 [DOI] [PubMed] [Google Scholar]