Chikungunya virus induces autophagy by triggering ER and oxidative stress, and this autophagy restricts apoptosis and viral propagation.
Abstract
Autophagy is an important survival pathway and can participate in the host response to infection. Studying Chikungunya virus (CHIKV), the causative agent of a major epidemic in India, Southeast Asia, and southern Europe, we reveal a novel mechanism by which autophagy limits cell death and mortality after infection. We use biochemical studies and single cell multispectral assays to demonstrate that direct infection triggers both apoptosis and autophagy. CHIKV-induced autophagy is mediated by the independent induction of endoplasmic reticulum and oxidative stress pathways. These cellular responses delay apoptotic cell death by inducing the IRE1α–XBP-1 pathway in conjunction with ROS-mediated mTOR inhibition. Silencing of autophagy genes resulted in enhanced intrinsic and extrinsic apoptosis, favoring viral propagation in cultured cells. Providing in vivo evidence for the relevance of our findings, Atg16LHM mice, which display reduced levels of autophagy, exhibited increased lethality and showed a higher sensitivity to CHIKV-induced apoptosis. Based on kinetic studies and the observation that features of apoptosis and autophagy were mutually exclusive, we conclude that autophagy inhibits caspase-dependent cell death but is ultimately overwhelmed by viral replication. Our study suggests that inducers of autophagy may limit the pathogenesis of acute Chikungunya disease.
Chikungunya virus (CHIKV) is the causative agent for Chikungunya fever, an arboviral disease transmitted by mosquitoes. CHIKV was first isolated in 1953 during an epidemic in Tanzania, East Africa (Mason and Haddow, 1957) and has recently emerged in islands of the Indian Ocean in 2005 (Enserink, 2006). La Reunion, an island in the Indian Ocean with a population of almost 785,000, was the most affected region, with an estimation of 300,000 cumulative cases in 2005–2006 (Schuffenecker et al., 2006; Simon et al., 2006; Gérardin et al., 2008). The epidemic involved India, where estimates approach six million infected people (Watson, 2007). It has also emerged in Italy, southern France, and Australia, and ongoing infections exist in Southeast Asia (Ng et al., 2009; Manimunda et al., 2011). CHIKV is a member of the Togaviridae family, genus Alphavirus, which are characterized by being enveloped, single-stranded positive polarity RNA viruses (Inoue et al., 2003; Sourisseau et al., 2007; Schwartz and Albert, 2010). In human, CHIKV typically induces symptoms 2–7 d after infection and is characterized by a rapid onset of fever, severe arthralgia, and myalgia, followed by constitutional symptoms (headache, photophobia, nausea, and abdominal pain) and a rash (Bodenmann et al., 2007; Borgherini et al., 2007). In vivo, CHIKV mainly targets the connective tissue (i.e., primarily fibroblast cells), but infection has also been reported in the liver, muscle, and brain in human and mice (Ozden et al., 2007). In vitro, CHIKV is able to infect a wide range of adherent cells, including macrophages, fibroblasts, and HeLa cells (Sourisseau et al., 2007; Schwartz and Albert, 2010). As observed for other alphaviruses (e.g., SINV and Ross River), CHIKV infection has the capacity to induce apoptosis, which has been suggested to be associated with pathogenesis. Notably, apoptotic blebs have been shown to contain CHIKV, which may represent a mechanism of cell-to-cell viral spread (Krejbich-Trotot et al., 2011a).
Apoptosis (or programmed cell death type I) is an energy-dependent process that is regulated by a preformed cascade of proteases called caspases (Danial and Korsmeyer, 2004; Kurokawa and Kornbluth, 2009). Two main pathways are involved in apoptosis: (1) the intrinsic pathway, in which activation of Bak and Bax results in mitochondrial outer membrane permeabilization inducing apoptosome formation and activation of caspase-9 (Kroemer et al., 2007); and (2) the extrinsic pathway that is initiated by death receptor oligomerization and cleavage of caspase-8 (or caspase-10; Wilson et al., 2009; Kurokawa and Kornbluth, 2010). Both pathways result in the activation of executioner caspases (e.g., caspase-3, -6, and -7), leading to the morphological and biochemical modifications recognized as apoptosis (Kurokawa and Kornbluth, 2009). Apoptosis is considered a first line of defense against viral infection, where engagement of intracellular stress pathways may trigger cell death, serving to limit viral replication (Griffin and Hardwick, 1997; Everett and McFadden, 1999; Li and Stollar, 2004). Many viruses have evolved strategies to escape or delay apoptosis, further pointing toward the role of cell death as an antiviral mechanism (Teodoro and Branton, 1997; Kepp et al., 2009).
The autophagic pathway is a bulk degradation system, which controls the clearance and recycling of intracellular constituents for the maintenance of cellular survival (Deretic and Levine, 2009). Autophagy consists of at least three pathways: microautophagy, macroautophagy, and chaperone-mediated autophagy (Crotzer and Blum, 2010; Li et al., 2011). Of those, macroautophagy is best characterized and has been implicated in both innate and adaptive immunity (Deretic and Levine, 2009). For the purpose of this study we will refer to macroautophagy as autophagy. Although the autophagic pathway was originally identified as a process induced by cellular starvation, there has been a strong interest in characterizing the role of autophagy as a mechanism of host defense (Deretic and Levine, 2009; Crotzer and Blum, 2010; Joubert et al., 2011). In vitro, autophagy is able to isolate and degrade Streptococcus pyogenes after it enters in the cytosol (Nakagawa et al., 2004; Joubert et al., 2009). Other in vitro and in vivo examples include the control of Mycobacterium tuberculosis and Listeria monocytogenes (Deretic and Levine, 2009). Some viral proteins are also targeted by autophagy (e.g., Sindbis or Tobacco Mosaic virus; Deretic and Levine, 2009; Orvedahl et al., 2010, 2011). Other roles for autophagy in the host response includes the enhancement of type I IFNs, or the processing and presentation of antigen for MHC I or MHC II presentation and T cell priming (Dengjel et al., 2005; English et al., 2009; Uhl et al., 2009; Crotzer and Blum, 2010). There also exist examples of microbes that are capable of abrogating and/or exploiting autophagic processes to enhance their replication or transmission. Examples include Shigella flexnerii and Listeria monocytogenes, which express virulence factors capable of inhibiting autophagosome formation, herpes simplex 1, which expresses ICP-34.5 for inhibition of initiation step in the autophagic process, and influenza A, which encodes M2 for inhibition of autophagosome/lysosome fusion (Deretic and Levine, 2009). Some viruses manage to be even more subversive, using mechanisms of autophagy for enhanced replication and viral release, including hepatitis C virus, poliovirus, and HIV-1 (Deretic and Levine, 2009; Blanchet et al., 2010).
Recent data suggest cross talk between autophagic and apoptotic pathways; for example, evidence supports interactions between Bcl-2 and Beclin-1 (also known as Atg6), and both may be engaged after induction of similar cell stress pathways (e.g., ER or oxidative stress; Thorburn, 2008; Djavaheri-Mergny et al., 2010; Kang et al., 2011). Herein, we characterized the role of apoptosis and autophagy in the context of CHIKV pathogenesis and report that host mechanisms, including ER and oxidative stress, limit the cytopathic effect (CPE) of CHIKV through the induction of autophagy. Moreover, in the absence of autophagic genes, we observe higher levels of apoptosis, both in vitro and in vivo. With respect to the latter, Atg16LHM mice were more sensitive to CHIKV-mediated lethality. These results define autophagy as a host defense mechanism that limits CHIKV pathogenesis.
RESULTS
CHIKV infection induces autophagosome formation in infected cells
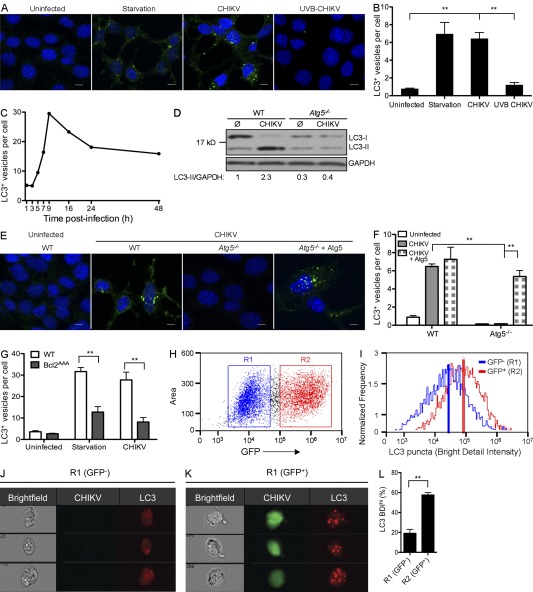
During autophagy, Beclin-1–PI3K-III complex activation results in isolation membrane formation that surrounds its substrates to generate an autophagic vesicle characterized by a double membrane, which is called an autophagosome (Mehrpour et al., 2010). Two ubiquitin-like systems have been shown to be essential for autophagosome formation. In the first, Atg12 (autophagy-related gene 12) is conjugated to Atg5, together forming a complex with Atg16L1, which decorates the outer membrane of the isolation membrane. Microtubule-associated protein 1 LC3 (light chain 3, also known as Atg8) constitutes the second ubiquitin-like system: LC3 conjugates phosphatidylethanolamine (PE) at the outer and inner autophagosomal membrane. Unlike the Atg12–Atg5–Atg16L1 complex that is recycled, the LC3-PE (referred to as LC3-II) remains associated with the inner membrane of autophagosome, making it a useful marker of autophagosomes (Kroemer et al., 2010; Mehrpour et al., 2010). To define the relationship between CHIKV infection and autophagy, we first examined autophagosome formation. Mouse embryonic fibroblasts (MEFs) were infected and LC3 puncta or LC3-II formation was measured by immunofluorescence or Western blot, respectively. Compared with uninfected MEFs, CHIKV-infected cells showed increased numbers of LC3 puncta (Fig. 1, A–C) and stronger staining for LC3-II by Western blotting (Fig. 1 D). Autophagosome formation could be first observed after 5 h and peaked at 9 h after infection (Fig. 1 C), suggesting the requirement for CHIKV replication as the trigger for autophagy induction. Replication-defective CHIKV (achieved by UV-B irradiation) failed to induce autophagosome formation, thus supporting the requirement for viral replication (Fig. 1, A and B). Serum starvation served as a positive control for autophagy induction (Fig. 1, A and B; and not depicted).
Figure 1.
Autophagosome induction within CHIKV-infected cells. (A and B) WT MEFs were incubated for 24 h in control media (uninfected), in nutrient-deprived media (starvation), or in the presence of live or UV-B–treated CHIKV (MOI = 1). (A) Immunofluorescence was performed using anti-LC3 antibody and DAPI. Bars, 10 µm. (B) The number of LC3 puncta (autophagosomes) per cell is depicted. Data shown represent mean ± SEM for triplicate samples of >100 cells per experimental condition. Similar results were observed in three independent experiments. (C) GFP-LC3 transfected cells were infected with CHIKV and the number of GFP dots is represented at different times after infection. Similar results were observed in two independent experiments. (D) WT or Atg5−/− MEFs were infected or not with CHIKV for 24 h and Western blotting was performed using anti-LC3 antibody to distinguish unconjugated LC3-I from PE-ylated LC3-II, and GAPDH to control for protein loading. Band intensity was calculated using ImageJ software and the ratio of LC3-II/GAPDH expression was normalized across the experiment and values are indicated. Similar results were observed in three independent experiments. (E and F) WT or Atg5−/− MEFs were transfected with Atg5 cDNA plasmid. Cells were infected with CHIKV and autophagy was monitored as in A and B. Data shown represent mean ± SEM for triplicate samples of at >100 cells per experimental condition. Similar results were observed in two independent experiments. Bars, 10 µm. (G) WT or Bcl2AAA MEFs were incubated for 24 h in control media (uninfected), in nutrient-deprived media (starvation), or in the presence of CHIKV (MOI = 1). Immunofluorescence was performed using anti-LC3 antibody, and the number of LC3 puncta (autophagosomes) per cell is depicted. Data shown represent mean ± SEM for triplicate samples of >100 cells per experimental condition. Similar results were observed in three independent experiments. (H–L) WT MEFs were infected for 24 h with GFP-expressing recombinant CHIKV (MOI=1) and stained with anti-LC3 antibody. Cells were analyzed using an ImageStreamX multispectral cytometer allowing gating of uninfected (R1) and infected cells (R2) according to GFP intensity (H) while simultaneously assessing the LC3 puncta based on BDI of LC3 staining (I). The mean BDI was marked. Representative uninfected (J) and infected (K) cells are shown. (L) The percentage of BDIhi cells was calculated and plotted for comparison across experiments. More than 10,000 cells per condition were collected. Error bars indicate the SEM. Similar results were observed in three independent experiments. Student’s t test: **, P < 0.05.
To evaluate if the induction of autophagosomes was occurring via a classical macroautophagy pathway, we evaluated CHIKV infection using MEFs deficient in the key autophagy gene Atg5. As predicted, Atg5−/− MEFs showed no evidence of LC3-II conversion upon CHIKV infection (Fig. 1 D). Similarly, immunofluorescence studies demonstrated that LC3 puncta observed during CHIKV infection were dependent on Atg5 expression (Fig. 1, E and F). Restoration of Atg5 expression, achieved by cDNA transfection, rescued the cells’ ability to induce autophagy (Fig. 1, E and F). We observed similar results using cells deficient for Atg7, another key autophagy gene (data not depicted). Furthermore, we investigated the implication of Beclin-1 in CHIKV-induced autophagy using MEFs that express a mutant form of Bcl-2 (MEF-Bcl2AAA). Notably, Bcl-2 directly regulates the activation of Beclin-1 and alanine substitution of the three phosphorylation sites (T69A/S70A/S87A) in Bcl-2 prevents dissociation of the Bcl-2–Beclin-1 complex, selectively inhibiting autophagy induction by Beclin-1 without affecting the antiapoptotic role of Bcl-2 (Wei et al., 2008; He et al., 2012). As expected, MEF-Bcl2AAA cells were not able to induce autophagy under starvation conditions (Fig. 1 G). Similarly, CHIKV-induced autophagy was abrogated in Bcl-2AAA MEFs, indicating that CHIKV-induced autophagy occurs via a Beclin-1–dependent mechanism. To confirm that CHIKV infection induced autophagosome formation in other cell types, we also investigated the appearance of autophagosomes in HeLa cells and human foreskin fibroblasts. Silencing of Atg5 or Atg7 genes using small interference RNA (siRNA) confirmed data shown using MEFs (unpublished data).
To analyze whether autophagosome formation was dependent on direct viral infection, we marked active replication using GFP-expressing recombinant CHIKV (Vanlandingham et al., 2005) and analyzed LC3 puncta using ImageStreamX. In brief, multispectral cytometric analysis enables the capture of high-resolution images of cells in flow (up to 500 cells/s) and permits analysis of LC3 puncta (de la Calle et al., 2011). 24 h after infection, GFP-expressing cells were gated (Fig. 1 H, R2), and LC3 bright detail intensity (BDI) was integrated for each cell as a measure of autophagosome formation. For comparison, GFP-negative cells were gated (Fig. 1, H [R1] and I [blue line]), and histogram plots representing LC3 puncta indicate that CHIKV-infected cells (Fig. 1 I, red line) have higher LC3 BDI. Representative ImageStreamX images with median intensity levels of BDI are shown for CHIKV-infected and uninfected cell populations (Fig. 1, J and K), confirming that LC3 puncta (scored based on high BDI) correlated with the presence of both viral-encoded GFP and robust autophagosome accumulation. Using this method, we quantified the percentage of LC3-positive cells (BDIhi) when bulk cultures are segregated for CHIKV infection (Fig. 1 L, R2 vs. R1, P < 0.05). Starved cells were used as positive control for autophagy induction (unpublished data). Based on these data, we conclude that autophagy induction occurs via a Beclin-1–dependent mechanism in a cell-intrinsic manner; in other words, viral replication within the cell, as opposed to secreted factors produced by neighboring infected cell, is the stimulus for autophagy induction.
Autophagosome/lysosome fusion remains intact during CHIKV infection
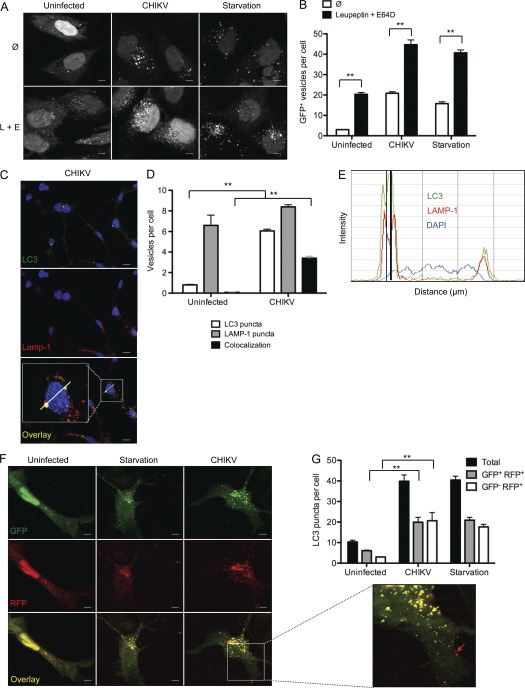
Upon maturation, autophagosomes fuse with late endosomes and lysosomes, which results in the formation of a degradative compartment referred to as autolysosomes (Deretic and Levine, 2009). Some viruses encode inhibitors of this event (e.g., influenza virus), and as a result enhanced numbers of LC3 puncta could be a reflection of basal autophagy accumulation and not de novo autophagosome formation (Gannagé et al., 2009). To discriminate between these two possibilities, we analyzed autophagy in the presence of lysosomal inhibitors leupeptin and E64D in transfected GFP-LC3 HeLa cells (Fig. 2, A and B). As expected, inhibition of autophagosome/lysosome fusion resulted in an increased number of LC3 puncta in control (basal autophagy flux) and starved (induced autophagy) HeLa cells. The presence of leupeptin and E64D also enhanced the number of autophagosomes in infected cells, allowing us to conclude that CHIKV infection induces de novo autophagosome formation (Fig. 2 B). To confirm this finding, we directly assessed colocalization of LC3 and the lysosomal-associated membrane protein LAMP-1 (Fig. 2 C). Optical sectioning of cells was performed and LC3+, LAMP-1+, and double-labeled vesicles were enumerated on a per cell basis. Approximately 45% of the LC3+ puncta colocalized with LAMP-1+ lysosomes in CHIKV-infected cells (Fig. 2 D). Line scan analysis confirmed colocalization and indicated that the lysosomal structures were discrete vesicles within the cell (Fig. 2 E).
Figure 2.
CHIKV induces autophagic flux. (A and B) GFP-LC3–expressing HeLa cells were starved (starvation) or infected with CHIKV (MOI = 1) alone or in the presence of leupeptin and E64D (L+E) for 24 h. The numbers of GFP+ vesicles per cell were enumerated. Data shown represent mean ± SEM for triplicate samples of at least 100 cells per sample. Similar results were observed in four independent experiments. Bars, 5 µm. (C–E) WT MEFs were infected with CHIKV for 24 h. Immunofluorescence was performed using anti-LC3, anti-Lamp-1, and DAPI to assess autophagosome and autolysosome formation. The number of LC3 puncta (green), Lamp-1+ vesicles (red), or costained vesicles (yellow) are depicted (C and D). Data represent mean ± SEM for triplicate samples of at least 100 cells per sample. Similar results were observed in two independent experiments. Bars, 10 µm. (E) Line scale bar analysis of a representative cell indicated in C is depicted. LC3 profiles (green curve), Lamp-1 profiles (red curve), and DAPI staining (blue curve) are shown. The black vertical line indicates colocalization of LC3 and Lamp-1 staining. (F and G) WT MEF cells were transfected with a plasmid encoding a dual-labeled LC3 probe, RFP-GFP-LC3. Cells were again starved (starvation) or infected with CHIKV for 24 h. Representative images are shown and the number of total autophagic vacuoles (GFP+RFP+ + GFP-RFP+), autophagosomes (GFP+RFP+), and autolysosomes (GFP−RFP+) were enumerated. The inset permits visualization of yellow autophagosome (yellow arrow) and red autolysosomes (red arrow). Data represent mean ± SEM for three independents experiments of at least 100 cells per sample. Similar results were observed in three independent experiments. Bars, 5 µm. Student’s test: **, P < 0.05.
As a final measure of autophagic flux, we transfected MEF cells with a construct encoding an RFP-GFP-LC3 polyprotein. In this way, it is possible to distinguish autophagosomes (RFP+ GFP+ puncta) and autolysosomes (RFP+ GFP− vesicles), with the latter being GFP negative as a result of the quenching of signal by the acidic microenvironment of the lysosome. CHIKV infection increased the number of both autophagosomes and autolysosomes compared with uninfected cells (Fig. 2, F and G). Moreover, an equivalent ratio of autophagosomes and autolysosomes was measured in infected cells and starved cells (Fig. 2 G). Together, the data presented in Figs. 1 and 2 provide evidence for CHIKV infection inducing de novo autophagosome formation without inhibition of autophagosome maturation.
ER stress serves as a trigger for autophagy during CHIKV infection
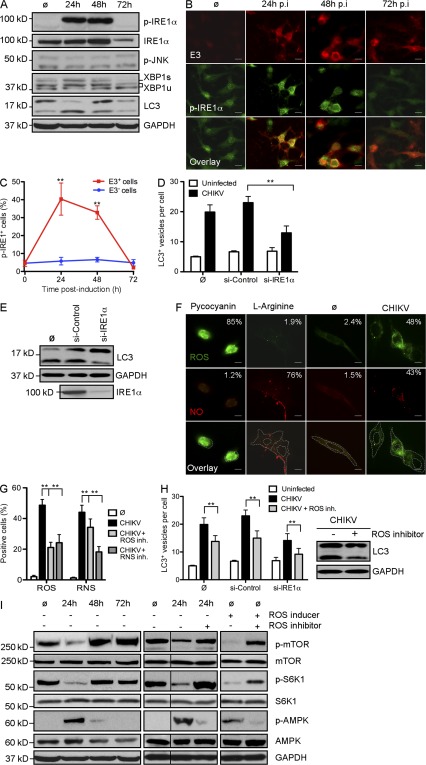
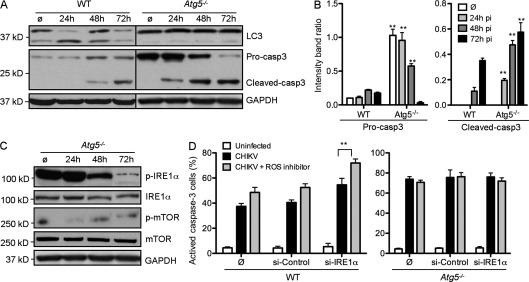
The ER serves as an important sensor of cellular stress. It detects changes in cell homeostasis and responds by triggering pathways referred to as the unfolded protein response (UPR; Lee et al., 2003; McGuckin et al., 2010). Viral infection has been shown to activate UPR as a result of the accumulation of viral proteins. At least three different pathways may be activated during ER stress, which are regulated by the signaling molecules eIF2α, IRE1α, and ATF6, respectively (McGuckin et al., 2010). We screened all three pathways (unpublished data) and identified a critical role for IRE1α. A role for IRE1α activation during CHIKV infection was first demonstrated by analyzing the phosphorylation of IRE1α (p-IRE-1α) at different time points after infection. Western blotting and immunofluorescence indicated that p-IRE1α was observed during the early phase of infection (Fig. 3, A and B) and could be detected only in infected cells (as evaluated based on E3 colocalization; Fig. 3, B and C). These data suggested that CHIKV leads to an intrinsic activation of ER stress. Importantly, p-IRE1α was no longer detected 3 d after infection. This shutdown seems to be the result of a decreased level of IRE1α protein, as indicated by Western blot analysis (Fig. 3 A). Remarkably, the kinetics of IRE1 phosphorylation correlated with the conversion of LC3-I to LC3-II (Fig. 3 A).
Figure 3.
CHIKV-induced autophagy is regulated by ER and oxidative stress. (A–C) WT MEFs were infected with CHIKV at indicated time points and Western blotting was performed to detect phosphorylation of IRE1α (p-IRE1α) and JNK (p-JNK) as well as the formation of spliced form of XBP1 (XBP1s) and the conjugation of LC3 (LC3-II). (A) IRE1α and GAPDH were also followed to control protein expression and loading. Similar results were observed in two independent experiments. (B) Immunofluorescence was performed using anti-pIRE1α and anti-E3 antibody. Bars, 15 µm. (C) The number of cells positive for pIRE1α in the E3+ (infected cells) and E3− (uninfected cells) populations is depicted. Data shown represent mean ± SEM for triplicate samples of >100 cells per experimental condition. Similar results were observed in three independent experiments. (D and E) WT MEFs were pretreated with control siRNA or siRNA against IRE1α for 3 d followed by infection with CHIKV for 24 h at MOI 1. The number of LC3 punctas per cell and the amount of LC3-II are depicted. Data shown represent mean ± SEM for triplicate samples of >100 cells per experimental condition. Similar results were observed in three independent experiments. (F and G) WT MEFs were incubated for 24 h in control media (ø) or in the presence of CHIKV (MOI = 1). (F) Immunofluorescence was performed using an ROS/RNS detection kit that specifically stains oxygen species and free NO. As positive controls for ROS or RNS induction, WT MEFs were incubated for 5 h with pycocyanin and l-arginine, respectively. Bars, 10 µm. (G) Percentage of cells containing ROS or NO among infected with CHIKV and/or pretreated with specific inhibitor of ROS and RNS as indicated is depicted. Data shown represent mean ± SEM for triplicate samples of >100 cells per experimental condition. Similar results were observed in two independent experiments. (H) WT MEFs or cells pretreated with siRNA against IRE1α for 3 d were infected with CHIKV for 24 h in presence of a ROS inhibitor. The number of LC3 punctas per cell and the amount of LC3-II are depicted. Data shown represent mean ± SEM for triplicate samples of >100 cells per experimental condition. Similar results were observed in three independent experiments. (I) WT MEFs were infected with CHIKV at indicated time points and Western blotting was performed to detect phosphorylation of mTOR (p-mTOR), S6KI (p-S6K1), and AMPK (p-AMPK). mTOR, S6K1, AMPK, and GAPDH were also followed to control protein expression and loading. As control to ROS implication, similar experiments were performed in cells pretreated with ROS inducer and/or ROS inhibitor. Black lines indicate that intervening lanes were spliced out. Similar results were observed in three independent experiments. Student’s test: **, P < 0.05.
To define the molecular events triggered by p-IRE1α, we investigated the activation of XBP1 and c-Jun amino-terminal kinases (JNK; McGuckin et al., 2010). Activation of XBP1 is regulated by a differential splice variant of XBP1 mRNA, which may be evaluated based on the expression of a protein of higher molecular weight and is referred to as XBP1s (for spliced XBP1; Yoshida et al., 2001). This pathway has been shown to favor cell survival (McGuckin et al., 2010). In contrast, IRE1α-induced phosphorylation of JNK is considered a link between cell stress and apoptosis (Urano et al., 2000). During CHIKV infection, we observed an induced expression of XBP1s, but did not detect enhanced phosphorylation of JNK (p-JNK; Fig. 3 A). To examine the functional link between IRE1α in CHIKV-induced autophagy, we silenced expression of IRE1α using siRNA and analyzed CHIKV-induced LC3 puncta as well as LC3-II conversion (Fig. 3, D and E). Reduced IRE1α gene expression was confirmed by Western blot, and shown to result in fewer CHIKV-induced autophagosomes. These data define a role for CHIKV activation of ER stress, which induces autophagy via an IRE1α- and XBP1s-mediated signaling pathway.
CHIKV-induced oxidative stress favors autophagosome production through the inhibition of mTORC1
Oxidative stress, primarily caused by increased levels of reactive oxygen species (ROS) and reactive nitrogen species (RNS), is a feature of the host response to viral infections (Cataldi, 2010). O2− and NO are considered to be the most important mediators among ROS and RNS, respectively. Free oxidative agents are known to induce autophagy and can also lead to cell death during strong and prolonged stimulation (Djavaheri-Mergny et al., 2007; Filomeni et al., 2010; Guo et al., 2010). To assess the impact of ROS/RNS production in CHIKV-induced autophagy, we first investigated whether CHIKV infection induces ROS and/or RNS production. We infected WT MEF for 24 h and monitored the presence of oxygen species and free NO. As positive control, we used pycocyanin and l-arginine, inducers of ROS and NO, respectively (Fig. 3 F). As expected, pycocyanin increased the percentage of cells that produced ROS, in the absence of NO production, whereas l-arginine induced free NO but not ROS. Interestingly, we observed that CHIKV infection led to increased production of both ROS and NO (Fig. 3 F), which could be inhibited using the ROS inhibitor N-acetyl-l-cysteine or the RNS scavenger c-PTIO (Fig. 3 G). Of note, exposure to a RNS scavenger cross-inhibited ROS production, highlighting the interconnectivity between the ROS and RNS pathways. This phenomenon could be explained by NO reacting with O2− to form the oxidant peroxynitrite (ONOO-; Djavaheri-Mergny et al., 2007; Novo and Parola, 2008).
To confirm the implication of ROS/RNS production in CHIKV-induced autophagy, we investigated LC3+ staining and the conversion of LC3-I to LC3-II in infected MEFs pretreated with N-acetyl-l-cysteine (Fig. 3 H). A significant decrease in LC3 puncta was observed, demonstrating the importance of ROS production in CHIKV-induced autophagy. We further evaluated potential overlap with the ER stress pathway by assessing autophagy in cells silenced for IRE1α mRNA and treated with N-acetyl-l-cysteine. Strikingly, we observed an additive inhibitory effect that reduced the number of autophagosomes per cell to near baseline levels (Fig. 3 H). This data suggests that ER stress and oxidative stress act via independent mechanisms to induce autophagy during CHIKV infection.
Although oxidative stress is known to induce autophagy upon microbial infection, the precise mechanism remains poorly documented. Based on recently established links between ROS and mTORC1 inhibition, which appears to be dependent on TSC2 (tuberous sclerosis complex 2), itself regulated by the AMP-activated protein kinase (AMPK; Alexander et al., 2010a,b), we investigate the regulation of this complex during CHIKV infection. Importantly, phosphorylated mTOR can be integrated into two different complexes, called mTORC1 and mTORC2, depending on its interactions with Raptor and Rictor, respectively (Zoncu et al., 2011). Although both complexes are implicated in protein synthesis, only mTORC1 is linked to autophagy (Thomson et al., 2009). To discriminate the activation of mTORC1 from mTORC2, we analyzed both mTOR phosphorylation and the induction of p-S6K1, a specific substrate of mTORC1. As shown, we observed a diminished level of both p-mTOR and p-S6K1 24 h after infection (Fig. 3 I). This inhibition was transient, and 2–3 d after infection, a strong induction of the mTOR–S6K1 pathway could be detected. The kinetics of mTOR–S6K1 inhibition correlated with conversion of LC3-I to LC3-II, suggesting a role for mTORC1 as a mediator of CHIKV-induced autophagy. To define how CHIKV-induced ROS is capable of inhibiting mTORC1, we next evaluated the activation of AMPK. Strikingly, the active form of AMPK was detected 24 h after infection, coincident with the inhibition of mTORC1 (Fig. 3 I). Moreover, the implication of the AMPK pathway in ROS-mediated inhibition of mTORC1 could be confirmed using N-acetyl-l-cysteine (Fig. 3 I). Together, these data provide a mechanistic understanding of CHIKV-induced autophagy.
Autophagy is a prosurvival mechanism that limits CHIKV-induced cell death
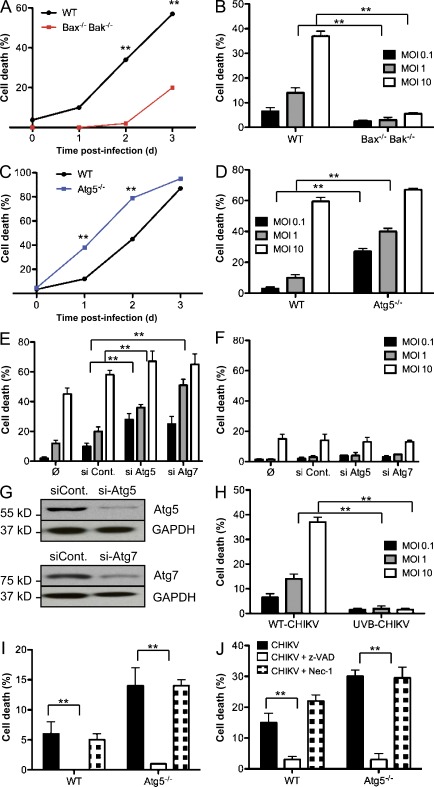
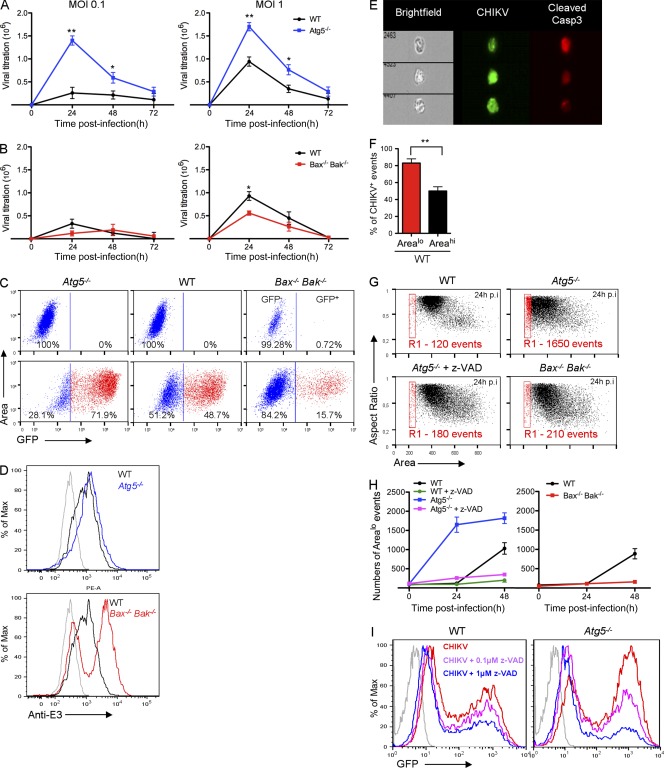
In addition to autophagy, other forms of cell stress may be triggered as a result of viral infection, including activation of cell death pathways. Increasing evidence suggests that cell stress pathways intersect and in some instances cross-inhibit each other (Thorburn, 2008). As CHIKV infection triggers a pronounced CPE (Sourisseau et al., 2007), it was important to investigate the function of autophagy on CHIKV-induced cell death. Atg5−/− MEFs and cells unable to engage the intrinsic apoptosis pathway (Bax−/− Bak−/− MEFs) were infected with CHIKV and loss of membrane integrity was analyzed. Whereas CHIKV infection triggered cell death in WT cells in a time- and dose-dependant manner, Bax−/− Bak−/− MEFs remained refractory, showing only minimal evidence of CPE at day 3 (Fig. 4, A and B). In contrast, Atg5−/− MEFs showed a dramatic increase in cell death compared with its WT control (Fig. 4, C and D). Notably, the enhanced CPE was most prominent at low multiplicity of infection (MOI), suggesting a link between cell death and viral propagation throughout the cultured MEFs. Similar results were obtained by silencing expression of autophagy genes in WT MEFs (Fig. 4, E and G). To explore alternative cell death pathways, autophagy genes were silenced in the Bax−/− Bak−/− MEFs. Importantly, the absence of both autophagy and apoptosis pathways did not further sensitize the cells to alternative forms of cell death (Fig. 4 F). Finally, we tested the importance of viral replication for cell death induction. WT MEFs were infected with live or UVB-inactivated CHIKV, and cell death was evaluated. As shown, replication defective CHIKV fails to induce cell death (Fig. 4 H).
Figure 4.
Enhanced CHIKV-induced cell death in the absence of autophagy. (A–D) WT, Bax−/− Bak−/−, or Atg5−/− MEFs were infected with CHIKV at indicated doses and time points. Percentage of cell death was measured using a membrane-permeable fluorescent probe and assessed by cytometry. Death curves over the 3 d after infection are shown for a representative experiment (A and C). The induction of cell death was also evaluated as a function of the MOI (B and D). In all conditions ≥10,000 cells were acquired. Similar results were observed in five independent experiments. (E and F) WT or Bax−/− Bak−/− MEFs were pretreated with the indicated siRNA for 3 d, followed by infection with CHIKV for 24 h at the indicated doses. Cells death is analyzed as described for A–D. Similar results were observed in three independent experiments. (G) WT MEFs were treated with si-Atg5 or si-Atg7, and Western blotting was performed using anti-Atg5 or anti-Atg7 antibodies. Similar results were observed in two independent experiments. (H) WT MEFs were infected with CHIKV or UVB-treated CHIKV at indicated doses. Percentage of cell death was measured using a membrane permeable fluorescent probe and assessed by cytometry. Similar results were observed in two independent experiments. (I and J) WT or Atg5−/− MEFs were pretreated with z-VAD or necrostatin-1 (Nec-1) before infection by CHIKV. Percentage of cell death is depicted. Error bars indicate mean values ± SD from three independent experiments. Student’s test: **, P < 0.05.
To define the form of programmed cell death responsible for the CPE in WT and Atg5−/− MEFs, we used pharmacological inhibitors of apoptosis (z-VAD, a broad spectrum caspase inhibitor) or necroptosis (necrostatin-1, an inhibitor of RIPK1). Infected cells treated with necrostatin-1 exhibited a similar level of cell death as compared with infected control cells, whereas z-VAD rescued both the WT and Atg5−/− MEFs from CHIKV-induced CPE. (Fig. 4, I and J). These data, as well as the absence of CPE in Bax−/− Bak−/− MEFs, demonstrate that the principle form of cell death induced by CHIKV is caspase-mediated apoptosis.
Autophagy delays both intrinsic and extrinsic apoptosis pathways
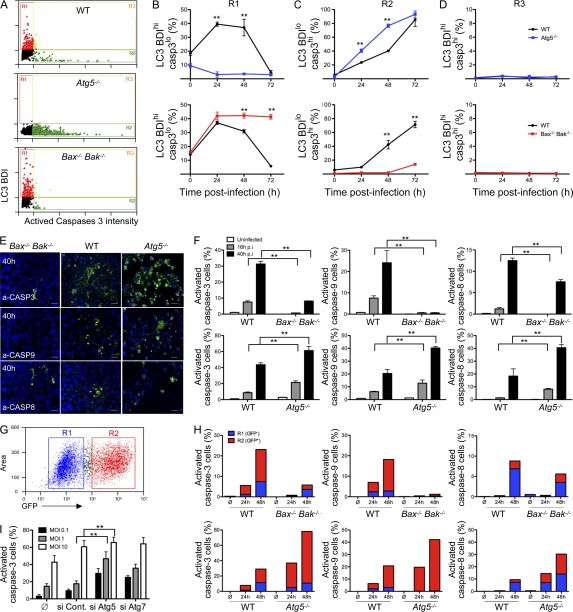
To define the interaction between autophagy and apoptosis at the single cell level, WT and Atg5−/− or Bax−/− Bak−/− cells were infected with CHIKV. As previously, LC3 BDI was used as a measure for autophagy induction, and apoptosis activity was characterized by labeling with antibodies specific for the active, cleaved form of caspase-3 (de la Calle et al., 2011). Data are represented in Fig. 5 A with each dot indicating a single cell. Regions were established as detailed previously (de la Calle et al., 2011), and the percentage of autophagic cells (LC3 BDIhi, cleaved caspase-3lo; defined by R1), apoptotic cells (LC3 BDIlo, cleaved caspase-3hi; defined by R2), or cells with evidence for both processes (LC3 BDIhi, cleaved caspase-3hi; defined by R3) were enumerated and represented graphically (Fig. 5, B–D). Supporting immunofluorescence results (Fig. 1), CHIKV infection triggered an increase in LC3 puncta in WT and Bax−/− Bak−/− MEFs, but not in Atg5−/− MEFs (Fig. 5 B). Strikingly, the number of cells exhibiting active caspase-3 was increased upon CHIKV infection as compared with uninfected cells, shown at 0 h (Fig. 5 C). The number of LC3+ cells was dramatically reduced in WT cells 3 d after infection. This observation correlated with high level of apoptosis and suggests that autophagy and apoptosis are mutually exclusive processes (Fig. 5, compare B with C). This observation was further supported by the absence of double-positive cells (region R3; Fig. 5 D).
Figure 5.
Autophagy limits CHIKV-induced apoptosis. (A–D) WT, Bax−/− Bak−/−, or Atg5−/− MEFs were infected with CHIKV (MOI = 1) for 24 h and were stained for LC3 and activated caspase-3. (A) Representative ImageStreamX dot plots from WT, Bax−/− Bak−/−, or Atg5−/− infected MEFs are depicted and the gating strategy is indicated. (B–D) The relative percentage of autophagic cells (LC3 BDIhi, cleaved caspase-3lo; defined by R1), apoptotic cells (LC3 BDIlo, cleaved caspase-3hi; defined by R2), or cells with evidence for both processes (LC3 BDIhi, cleaved caspase-3hi; defined by R3) is shown. Error bars indicate mean ± SD of three independent experiments. (E and F) WT, Bax−/− Bak−/−, or Atg5−/− MEFs were infected with CHIKV (MOI = 1) at the indicated time points, and activated caspase-3 (a-CASP3), activated caspase-9 (a-CASP9), or activated caspase-8 (a-CASP8) were stained from parallel cultures. Representative microscopic images are shown (E), and the percentage of positive cells were determined for >100 cells per condition (F). Error bars show mean ± SD of three independent experiments. Bars, 25 µm. (G and H) WT, Bax−/− Bak−/−, or Atg5−/− MEFs were infected with GFP-expressing recombinant CHIKV (MOI = 1) and stained with anti–active capase-3, -8, and -9 antibody. Cells were gated as uninfected (R1) and infected (R2) cells according to GFP intensity (G) while simultaneously assessing the active capase-3, -8, and -9 staining (H). Similar results were observed in three independent experiments. (I) HFFs were infected with CHIKV (MOI = 1) for 24 h, and the percentage of activated caspase-3 cells was determined for >100 cells per condition. Error bars show mean ± SD of three independent experiments.
Two important observations emerged from the study of the knockout MEFs. The first intriguing finding concerned a marked increase in active caspase-3+ cells when Atg5 is absent (Fig. 5 C). These data suggested an important role for autophagy in the regulation of CHIKV-induced apoptosis. The second discovery concerned the possibility of the Bax−/− Bak−/− cells to activate caspase-3; the timing of apoptosis onset showed interexperimental variation with cells showing detectable levels of cleaved caspase-3 between 48 and 72 h (Fig. 5, C, E, and F). Evidence for caspase-3 activation in Bax−/− Bak−/− cells indicated activation of the extrinsic apoptosis pathway, which may be induced independently of mitochondrial outer membrane permeabilization.
We then investigated whether autophagy is able to regulate apoptotic cell death in human fibroblasts cells (HFF), which are known cell targets of CHIKV infection (Sourisseau et al., 2007). We down-regulated the expression of Atg5 and Atg7 genes in HFF by an siRNA strategy and analyzed the activation of caspase-3 after 24 h of infection with different viral inputs (Fig. 5 I). Inhibition of both Atg5 and Atg7 dramatically increased the percentage of cleaved caspase-3–positive cells according to viral doses, demonstrating that the antiapoptotic function of autophagy is also observed in human cells.
To distinguish the role of autophagy in limiting the distinct apoptosis pathways, we evaluated cleaved caspase-9 (a marker of the intrinsic pathway) and cleaved caspase-8 (indicative of activation of the extrinsic pathway) during CHIKV infection (Fig. 5, E and F). Early during the kinetics of viral infection (16 h), caspase-9 activation was evident in WT cells in the absence of detectable levels of cleaved caspase-8. In comparison, Atg5−/− MEFs displayed a twofold increase in the percentage of active caspase-9–positive cells, as well as early evidence for cleaved caspase-8. By 40 h after infection, both the intrinsic and extrinsic pathways were engaged in the WT cells and, again, Atg5−/− cells demonstrated higher levels of activation for both cell death pathways (Fig. 5 F).
Using GFP-expressing CHIKV, we next evaluated the relationship between infection and activation of caspase-3, -9, and -8 (Fig. 5, G and H). Whereas the intrinsic apoptotic pathway was engaged primarily in CHIKV+ cells, the extrinsic pathway was detectable in both infected and bystander uninfected cells. These data suggest that CHIKV infection induces apoptosis through both intra- and extracellular factors (Fig. 5 H) and that autophagy, acting in a cell-intrinsic manner, preferentially protects infected cells from apoptosis.
CHIKV-induced apoptosis is independent of ER and oxidative stress
As introduced in the previous sections, both ER and oxidative stress may trigger proapoptotic pathways (McGuckin et al., 2010). To determine the relative contribution of stress pathway induction on autophagy versus apoptosis, we again used Atg5−/− MEFs. We first confirmed the antiapoptotic effect of autophagy by investigating the cleavage of pro–caspase-3 in WT and Atg5−/− MEFs (Fig. 6, A and B). Whereas pro–capase-3 was cleaved only after 48 h of infection in WT cells, the active form of caspase-3 was detected 24 h after infection in Atg5−/− MEFs. Moreover, the ratio of active caspase-3/GAPDH was increased in Atg5−/− cell during all time points investigated (Fig. 6 B).
Figure 6.
ER and oxidative stress don’t enhance in CHIKV-induced apoptosis. (A and B) WT or Atg5−/− MEFs were infected with CHIKV at the indicated time points. (A) Western blotting was performed using anti-LC3 and anti–caspase-3 (Casp3) to discriminate pro–caspase-3 and cleaved caspase-3, and GAPDH to control for protein loading. Black lines indicate that intervening lanes were spliced out. (B) Band intensity was calculated using ImageJ software and the ratio of pro–caspase-3/GAPDH and cleaved-caspase-3/GAPDH was represented by graphs. Similar results were observed in three independent experiments. (C) Atg5−/− MEFs were infected with CHIKV at indicated time points and Western blotting was performed to detected phosphorylation of IRE1α (p-IRE1α) and mTOR (p-mTOR). IRE1α, mTOR, and GAPDH were followed to control protein expression and loading. Similar results were observed in two independent experiments. (D) WT and Atg5−/− MEFs were pretreated or not with siRNA against IRE1α for 3 d and were then infected by CHIKV for 48 h in control media or in the presence of ROS inhibitor. Activated caspase-3 was stained and the percentage of positive cells was determined. Error bars show mean ± SD of three independent experiments. Student’s test: **, P < 0.05.
We next investigated the activation of IRE1α and inhibition of mTOR in Atg5−/− MEFs. Interestingly, both pathways were similarly regulated by CHIKV infection as compared with WT MEFs (Fig. 6 C compared with Fig. 3, A and I). To ascertain the impact of these pathways on apoptosis, we evaluated by immunofluorescence the percentage of active caspase-3–positive cells in WT or Atg5−/− MEFs in which ER and oxidative stress had been inhibited (Fig. 6 D). IRE1α and/or ROS inhibition increased the percentage of WT cells with active caspase-3. Importantly, no change in active caspase-3 expression was observed in Atg5−/− MEFs. These data suggest that ER and oxidative stress occur before the role of Atg5 in facilitating autophagy and are not critical for the induction of CHIKV-induced apoptosis.
Together, the data presented in Figs. 1–6 demonstrate that during the early phase of CHIKV infection, autophagy is induced via the activation of ER stress and the inhibition of mTOR by ROS production. By triggering autophagy, CHIKV-induced cell stress indirectly limits apoptotic cell death. Nevertheless, after 48 h of infection, ER stress is blunted, in part secondary to IRE1α degradation, and mTOR becomes hyperphosphorylated. These events result in decreased autophagic tone and, via a still undefined mechanism, apoptosis pathways dominate, in turn resulting in pronounced CHIKV-induced CPE.
Regulation of apoptosis and in vitro CHIKV propagation
Both autophagy and apoptosis have been related to the regulation of viral replication and/or propagation. Although autophagy can result in the degradation of viral proteins (e.g., Sindbis) without significantly affecting viral infection, it has also been reported to enhance viral replication (e.g., HCV; Dreux and Chisari, 2009). Similarly, apoptosis has been implicated in both pro- and antiviral responses (Li and Stollar, 2004). As both pathways are engaged by CHIKV infection, it was important to evaluate the effect of autophagy and apoptosis on viral propagation. To achieve this, we measured viral load in the supernatant of WT, Atg5−/−, or Bax−/− Bak−/− MEFs. CHIKV titers seemed enhanced in supernatant of Atg5−/− cells, suggesting that autophagy could restrict viral release (Fig. 7 A). However, modest viral titers were lower in Bax−/− Bak−/− MEFs as compared with WT controls at day 1 after infection, thereby suggesting a role for apoptotic cell death in viral release during early phase infection (Fig. 7 B). Similar results were obtained in HeLa cells using siRNA specific for Atg5 or Atg7 (unpublished data).
Figure 7.
Regulation of apoptotic cell death limits CHIKV propagation in vitro. (A and B) WT, Bax−/− Bak−/−, or Atg5−/− MEFs were infected with CHIKV at indicated MOI and extracellular viral titers were determined during the 3 d after infection. Results were expressed as TCID50/ml. Error bars indicate mean viral titer ± SD of four independent experiments. (C) WT, Bax−/− Bak−/−, or Atg5−/− MEFs were infected with GFP-expressing recombinant CHIKV (MOI = 0.1). ImageStreamX dot plots based on GFP intensity are shown. Similar results were observed in three independent experiments. (D) WT, Atg5−/−, or Bax−/− Bak−/− MEFs were infected at MOI = 1 and viral proteins were stained using anti-E3 Ab and analyzed by cytometry. Similar results were observed in three independent experiments. (E and F) WT MEFs were infected with GFP-CHIKV (MOI = 0.1) for 48 h and arealo events were gated (R1 in G). (E) GFP-CHIKV or active caspase-3 was stained in WT MEFs and three representative events of R1 are depicted. (F) The percentages of GFP+ event in R1 (arealo) or in live cell (areahi; all events except R1) are shown. Error bars indicate SEM. Similar results were observed in three independent experiments. (G and H) WT, Bax−/− Bak−/−, or Atg5−/− MEFs were infected with CHIKV (MOI = 0.1) alone or in the presence of z-VAD for 48 h and arealo events were numerated by ImageStreamX cytometry (G). The numbers of events in R1 are shown (H). Error bars show mean ± SD of four independent experiments. (I) WT or Atg5−/− MEFs were pretreated with z-VAD before infection with GFP-expressing recombinant CHIKV (MOI = 0.1) and GFP intensity was analyzed by cytometry. Similar results were observed in two independent experiments. Student’s test: **, P < 0.05; *, P < 0.08.
To further analyze the function of autophagy and apoptosis in viral infection, we assessed the percentage of infected cells after 24 h of infection by using GFP-expressing recombinant CHIKV as a marker of active infection (Fig. 7 C). At low viral dose (MOI = 0.1), Atg5−/− MEFs showed greater infection than WT controls. This difference is a result of enhanced infection and not differential viral entry, as suggested by a kinetic study of infected cells (data not depicted). Results for Bax−/− Bak−/− MEFs were even more striking as only a minority of cells were GFP+ cells, supporting that apoptotic cell death influences CHIKV propagation (Fig. 7 C). Cytometric assessment of the Bax−/− Bak−/− MEFs indicated that although fewer cells were infected, those cells that were infected showed higher expression of CHIKV E3 proteins as compared with its WT control (Fig. 7 D). This suggested that a delay in cell death allowed for greater per cell viral replication, but failure to undergo rapid cell death resulted in fewer infected cells at the population level. In contrast, autophagy-deficient cells expressed similar expression intensity to CHIKV E3 proteins as compared with its WT control, indicating that autophagy had minimal impact on virally encoded GFP expression within the cell (Fig. 7 D). These data were further confirmed in a secondary culture–based assay; cell supernatants from the respective cell lines were exposed to uninfected WT MEFs and GFP expression was scored after an additional 24-h incubation, indicating higher levels of infectious virus in the absence of autophagic flux and lower levels in apoptosis-deficient cells (unpublished data). Together, these data suggested that autophagy could regulate viral propagation by limiting the release of virus induced by apoptotic cell death.
To support this observation, we established an image analysis script using the ImageStreamX, which integrated cell area (unpublished data), thereby permitting quantification of cells in the final stages of apoptosis based on their being small and pyknotic. Importantly, analysis of the pyknotic cells indicated that ∼80% expressed viral capsid protein as compared with larger, less dead cells, of which ∼45% stained positive for E3 (Fig. 7, E and F), suggesting that ingestion of apoptotic bodies could contribute to infection of phagocytic neighboring cells (Krejbich-Trotot et al., 2011a). Apoptotic body accumulation upon CHIKV infection was next determined in WT or Atg5−/− MEFs (Fig. 7 G, R1). We also confirmed that accumulation of small area events was dependent on an apoptotic process, as use of z-VAD eliminated this population of cells (Fig. 7, G and H). Following from previous results, cells deficient for autophagy genes accumulated more apoptotic bodies as compared with WT MEFs (Fig. 7, G and H), whereas the Bax−/− Bak−/− cells showed fewer numbers of events in the R1 gate (Fig. 7 H).
To establish the importance of apoptosis induction in viral propagation, WT and Atg5−/− MEFs were treated with an apoptosis inhibitor and the percentage of infected cells was assessed (Fig. 7 I). Interestingly, apoptosis inhibition dramatically decreased the percentage of infected cells, demonstrating that apoptotic cell death, and consequently apoptotic body formation, is an important mechanism to enhance CHIKV propagation in cell culture. Based on these data we conclude that mechanistically, propagation of CHIKV can be controlled by autophagy, which acts via the limitation of infection-induced apoptosis. Although these data are in apparent contradiction with the recent study of Krejbich-Trotot et al. (2011b), they do not follow infectious particles, instead drawing their conclusions based on E1 expression and CHIKV mRNA transcription.
Although cell stress, cell death and viral propagation appear linked, one potential caveat is that autophagy induction enhances type I IFN production, thus providing an alternative explanation for decreased viral infection. Indeed, prior data suggests an intersection between Atg5 and RIG-I although, arguably, autophagy enhanced vesicular stomatitis virus (VSV) replication by limiting the innate immune response (Jounai et al., 2007). To assess whether IFN-β expression is altered by autophagy induction, Atg5−/− MEFs were infected by CHIKV and the culture supernatant was assayed at 24, 48, and 72 h after infection (unpublished data). Despite differences in viral propagation, there was no difference in IFN-β production in Atg5−/− MEFs as compared with the WT control cells.
In vivo CHIKV infection of Atg16LHM mice results in increased apoptosis and greater lethality
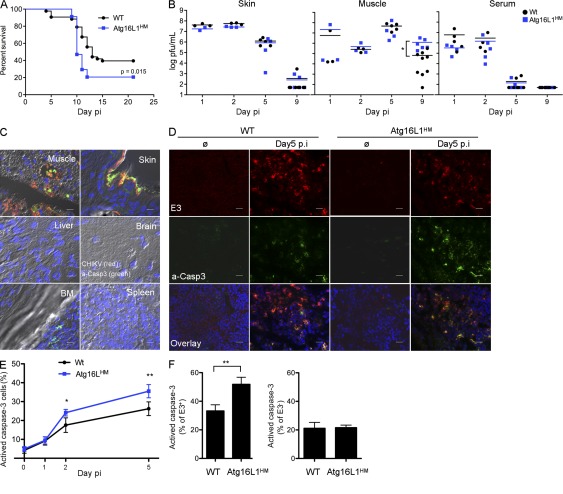
Given the importance of autophagy in controlling CHIKV-induced apoptosis in vitro, we next investigated the role of autophagy in vivo using a neonatal model of infection. The disruption of autophagy genes results in embryonic lethality (Cadwell et al., 2008). Therefore, to analyze the potential role of autophagy in CHIKV pathogenesis we used Atg16L hypomorphic mice (Atg16LHM) in which Atg16L1 gene has been modified by gene trap mutagenesis. These mice display hypomorphic expression of Atg16L protein and reduced autophagy (not depicted; Cadwell et al., 2008). 9-d-old WT and Atg16LHM mice were infected with CHIKV and followed for lethality (Fig. 8 A). Although only 60% of WT mice succumbed to CHIKV, we observed a increase in lethality in Atg16LHM mice. These data indicate that autophagy has a prosurvival function during CHIKV infection and limits disease pathogenesis. To understand how autophagy enhances survival, we first analyzed viral load in infected tissues in WT and Atg16LHM mice (Fig. 8 B). Viral load in skin muscle and serum, important targets of CHIKV infection, were similar in WT and Atg16LHM mice, suggesting that autophagy did not significantly affect in vivo viral infection. Similar results were obtained by analyzing the viral load in lung, liver, brain, and spleen (unpublished data). Interestingly, the one observed difference concerned a delayed clearance of CHIKV in the muscle at day 9 after infection. This is consistent with failure to thrive as a cause of death in neonatal animals (Couderc et al., 2008). In the context of our in vitro data, and the fact that the Atg16LHM have only a partial block in autophagy (Cadwell et al., 2008), we suggest that the muscle tissue may be highly sensitive to the autophagy-mediated protective effects of CHIKV infection.
Figure 8.
Autophagy limits apoptotic induction in CHIKV infected tissues and delays lethality of mice. (A–F) WT (n = 43) and Atg16LHM mice (n = 34) were infected at 9 d of age with 4 × 105 PFU CHIKV subcutaneously. (A) Mice were monitored for lethality for 21 d with data displayed as Kaplan-Meier curves. (B) Skin, muscle, and serum were collected after days 1 (n = 3), 2 (n = 3), 5 (n = 4), and 9 (n = 5) of infection, homogenized, and viral titers were determined by standard plaque assay. Median values for WT (black bars) or Atg16LHM (blue bars) mice are depicted. (C) Muscle, skin, liver, brain, BM, and spleen were collected from WT infected mice at day 5. Tissues were fixed in PFA, frozen in OCT blocks, and stained for cleaved capsase-3 (green) and E3 (red). Similar results were observed in two independent experiments. Bars: (muscle) 50 µm; (other tissues) 20 µm. (D–F) Muscle from infected WT or Atg16LHM mice was collected at days 1, 2, and 5 after infection and stained for cleaved caspase-3, E3, or DAPI for nucleus staining (D). The percentage of cleaved capsase-3–positive cells was numerated in all population (E) or in E3+ and E3− cells (F). Error bars mean ± SD of three independent experiments. Bars, 60 µm. Student’s test: **, P < 0.05.
To understand how autophagy could protect mice against CHIKV-induced CPE, we further investigated the impact of autophagy on apoptosis in infected tissues. We first analyzed whether apoptosis could be detected during CHIKV pathogenesis. 9-d-old WT mice were infected and, after 5 d, muscles, skin, liver, brain, BM, and spleen were collected and stained for the expression of E3 (Fig. 8 C, red) and active caspase-3 (a-CASP3; Fig. 8 C, green). Both E3 and a-casp3 could be readily detected in muscle and skin but not in other tissues. Of note, a-casp3 was detected in the BM; however, this could be attributed to higher basal apoptosis as uninfected animals had similar numbers of a-casp3–positive cells. Interestingly, apoptosis induction in infected tissues was observed in infected cells (determined by colocalization of E3 and a-casp3) as well as in bystander cells (no E3 staining), suggesting, as seen in our in vitro studies, that both intrinsic and extrinsic apoptosis could be induced. These studies were extended to Atg16LHM mice, and although similar levels of E3-expressing cells were found in the muscle and skin, we observed higher levels of a-casp3–positive cells (Fig. 8, D and E). Remarkably, the enhanced levels of apoptosis were observed only in infected cells and did not alter the level of bystander cell death, again supporting the relevance of our in vitro findings (Fig. 8 F and Fig. 5). Similar observations were shown in skin (unpublished data). These results demonstrate the relevance of our findings and support a role for autophagy as a regulator of apoptosis, thus characterizing a novel mechanism of host response during CHIKV infection.
DISCUSSION
The recent CHIKV outbreak has exposed how little we understand about the pathogenesis of this virus or the ability to harness host responses to enhance the control of arboviral infection. Immunological studies have suggested that early events of viral–host cell interactions determine whether CHIKV achieves disseminated infection, or if replication is limited and controlled by innate defense mechanisms. In our prior studies, we evaluated the role of pattern recognition receptors and type I IFNs as mediators of viral control (Schilte et al., 2010). Here, we examined an alternative stress response pathway that is activated upon infection, making the exciting discovery that engagement of the autophagy pathway limits viral-induced cell death as a mechanism for slowing viral propagation. In vivo data in Atg16LHM mice support an antiapoptotic effect for autophagy and indicate that in the absence of autophagic genes, there is an increased susceptibility to severe forms of Chikungunya disease.
The role for autophagy in host defense has been documented for bacteria and several viruses (Deretic and Levine, 2009); however, the results from our current study identify a previously uncharacterized role for autophagy as a mechanism of limiting disease pathogenesis. Importantly, we demonstrate that autophagy is triggered in a cell-intrinsic manner by direct CHIKV infection, which leads to the induction of both ER and oxidative stress (Figs. 1 and 3). Specifically, among the ER stress pathways that are induced during viral infection, only IRE1α/XBP1s is involved in autophagic regulation (Fig. 3, A–D; and not depicted). Oxygen species produced during CHIKV infection are able to independently enhance autophagy via the inhibition of mTORC1 (Fig. 3, E–I). The autophagy pathway induced is dependent on Beclin-1 and proceeds unimpeded, with ultimate fusion between autophagosomes and degradative lysosomes (Figs. 1 G and 2). Using novel, single cell assays that permitted simultaneous assessment of autophagy and apoptosis, we were able to demonstrate that at all time points studied, the two biological processes are mutually exclusive (Fig. 5, A–D). That said, kinetic studies, as well as assessment of viral protein expression in dying cells, suggest that as viral infection progresses, the autophagy pathway is overwhelmed and cells began to undergo apoptosis (Fig. 5).
Role of apoptosis in acute viral infection
Apoptosis after viral infection is considered to be an important mechanism of host defense, as autodestruction may limit viral replication (Everett and McFadden, 1999). RNA viruses responsible for acute infection may have a distinct agenda, and growing evidence suggests that a subset of agents may have evolved to actively trigger programmed cell death (Yatim and Albert, 2011). Members of the alphavirus family have been shown to actively induce programmed cell death. For example, Sindbis virus (SinV) may trigger both intrinsic and extrinsic apoptosis (Li and Stollar, 2004). Although transfection of SinV nsp2 or the E1, pE2, and E3 structural proteins may trigger cell death, the precise mechanism remains unknown (Zhang et al., 1997; Li and Stollar, 2004). Mathematical models argued that a virus will evolve toward increasing cytopathicity when the mean lifetime of a cell is high and the rate of viral budding is low (Krakauer and Payne, 1997). Indeed, CHIKV infects primarily fibroblasts and other long-lived stromal cells (Powers and Logue, 2007). In accordance with this prediction, it was recently demonstrated by Krejbich-Trotot et al. (2011a) that apoptotic cell death is required for efficient CHIKV propagation. Our findings support this observation, as multispectral analysis of CHIKV-infected cells indicated the early engagement of the intrinsic cell death pathway (Fig. 5 F). Similar to SinV, CHIKV infection also resulted in caspase-8 activation (Fig. 5 F), suggesting a possible role for TNFSF receptor engagement (e.g., TRAIL-R). Interestingly, in contrast to caspase-9 activation, there was evidence for uninfected cells with active caspase-8 (Fig. 5 H).
Using z-VAD or gene silencing of caspase-3, we further showed that inhibition of apoptosis significantly inhibited CHIKV propagation (Fig. 7, B, C, and I; and not depicted). Similar results were observed in cells deficient for Bax−/− Bak−/−, albeit with a less dramatic effect as a result of the fact that the extrinsic pathway remains intact in these cells (Fig. 7). Thus, we suggest that both the intrinsic and extrinsic pathways participate in enhancing CHIKV propagation.
Autophagy as an antiviral mechanism
The autophagy pathway and/or autophagy proteins have been recently shown to participate in the host response to intracellular pathogens (Deretic and Levine, 2009). Physical degradation of the microbe or microbial components may be achieved through their isolation in autophagic vesicles that in turn fuse with lysosomes, which is referred to as xenophagy (Orvedahl and Levine, 2008). A selective mechanism that leads to their capture is regulated, in part, by the poly-ubiquitin binding adaptor proteins p62 and NDP52. Components of the SinV are also sequestered by autophagy in a p62-dependant manner (Orvedahl et al., 2010). In vivo, genetic inactivation of Atg5 in virally infected mouse neurons resulted in delayed clearance of SinV capsid, the accumulation of p62 aggregates, increased neuronal apoptosis, and rapid mortality, all in the apparent absence differing viral load.
A second mechanism by which autophagy may participate in the host response to infection relates to the cross talk between autophagy genes and pattern recognition receptors. Lee et al. (2007) demonstrated in a seminal study that autophagy sequesters cytosolic viral genome and/or host RNA during VSV infection. This allowed ligand to access the luminal domain of TLR7 in pDCs and enhanced production of IFN-α/β and other proinflammatory cytokines. Although this pathway is unlikely to contribute to the control of CHIKV during infection of fibroblast or epithelial cells, as a result of their lack of TLR7 expression, it is possible that autophagy my impact RIG-I signaling, although as discussed previously, the intersection between Atg5 and RIG-I results in a blunting of the antiviral response (Jounai et al., 2007). However, we did not observe altered IFN-β production in cells lacking Atg5 or altered IFN-α induction in mice hypomorphic for Atg16L (unpublished data), suggesting that this mechanism of regulation did not account for autophagy-mediated suppression of CHIKV replication in fibroblasts and epithelial cells.
Our study provides mechanistic information for another strategy by which autophagy participates in the host response to limit cell mortality after viral infection, thus controlling the release of infectious viral particles secondary to apoptotic cell death. We demonstrate that autophagy promotes cell survival by limiting apoptotic cell death. Importantly, autophagy induction is dependent on CHIKV replication; consequently, the delay in apoptosis occurs in a cell-intrinsic manner. In other words, neighboring uninfected cells remain sensitive to putative extracellular inducers of the extrinsic pathway (Fig. 5 H). The mechanism accounting for autophagy induction is linked to the activation of host stress responses during acute infection, arguing that autophagy is induced by cells to regulate these stress responses. Regarding the oxidative stress, autophagy has been described to restrict ROS production, for example by eliminating damaged mitochondria. Indeed, this is occurring in the context of CHIKV infection, as indicated by their being higher levels of ROS production in infected Atg5−/− cells (unpublished data). Nonetheless, autophagy did not succeed to completely control the high levels of oxidative stress induced by viral infection and replication. For both ER and oxidative stress, it has been suggested that their induction may result in autophagy and apoptosis, depending on the signal intensity or duration of the stress inducer. In the case of IRE1α, we demonstrate that during the early phase of CHIKV infection, IRE1α favors XBP1 mRNA splicing and autophagy, yet does not lead to proapoptotic JNK phosphorylation. Future studies will be required to ascertain if this signaling event is dictated by virally encoded proteins or a reflection of the concentration of unfolded proteins during CHIKV replication. It will also be valuable to establish a better understanding of how autophagy is terminated, with an absolute switch to an apoptotic phenotype as suggested by our single cell analysis (Fig. 5 A).
Cross talk between autophagy and apoptosis
Our study provides an interesting link between autophagy, apoptosis, and viral propagation, also highlighting the active investigation into cross talk between cell stress and cell death. Several mechanisms may account for autophagy limiting apoptotic cell death. For example, degradation of protein aggregates and damaged mitochondria that result from viral infection may delay induction of apoptosis (Kroemer et al., 2010). Additionally, it has been suggested that release of bcl-2 and FLIP from Atg3 protein complexes may actively inhibit the intrinsic and extrinsic pathways of apoptosis (Thorburn, 2008; Kroemer et al., 2010). Conversely, apoptosis may inhibit autophagy as calpain has been shown to cleave Atg5 and caspase-3 cleaves Atg4D, an enzyme implicated in the delipidation of Atg protein (Kang et al., 2011). Many molecular details must still be defined, yet the available results have led to the concept that autophagy and apoptosis are antagonistic events that are cross inhibitory. That said, prior studies evaluating these antagonistic mechanisms used bulk assays such as Western blotting and thus could not discern activation of both pathways in a single cell.
One of the unique aspects presented here is the use of multispectral images cytometer, which permitted unbiased medium throughput and simultaneous assessment of viral protein expression, LC3 puncta, and caspase activation at the single cell level. Consistent with the prevailing dogma, but different from other viral infections such as influenza A (de la Calle et al., 2011), we demonstrate the absence of cells showing features of both autophagy and apoptosis. Even in situations where both pathways are active in the bulk population of cells, the two pathways were mutually exclusive. This result suggests that viral infection may trigger either autophagy or apoptosis and that the molecular decision is a proximal event. One obvious candidate is Beclin-1 (Atg6), which interacts with bcl-2, regulating both pathways. In resting cells, Beclin-1–bcl-2 complexes limit autophagy without altering the antiapoptotic effects of bcl-2. Nutrient starvation, ER stress, or oxidative damage may disrupt these interactions, favoring the association of Beclin-1 with PI3K-III and the induction of autophagy (Kang et al., 2011). If, however, apoptosis precedes autophagy, caspase cleavage of Beclin-1 will prevent its induction (Kang et al., 2011). Interestingly, Beclin-1 has been shown to limit SinV propagation (Liang et al., 1998), suggesting a more general role for autophagy as a mechanism for delaying apoptosis and protecting the host from acute alphaviral infections. Integration of these data in the context of ER and oxidative stress may reveal novel mechanisms by which viruses modulate host pathways. Our data characterizing late phases of CHIKV infection (e.g., 48–72 h) identified mTOR overexpression and IRE1α degradation as possible clues for how these stress pathways may be perturbed.
In sum, we have provided evidence for a novel mechanism by which autophagy participates in host defense. In the context of acute infection by CHIKV, apoptotic cell death is important for viral propagation, and autophagy, in a cell-intrinsic manner, protects cells from apoptosis and may limit viral release. These data suggest that inducers of autophagy may be useful therapeutics for limiting the pathogenesis of acute Chikungunya disease or protecting neonates from mother-to-child transmission.
MATERIALS AND METHODS
Cells and mice.
WT and Bax−/− Bak−/− MEF cells from the Korsmeyer laboratory (Dana-Farber Cancer Institute, Boston, MA) were derived from C57BL/6 mice (H-2b). Atg5−/− cells from the Kroemer Laboratory (NSERM U848, Institut Gustave Roussy, Villejuif, France) were derived from C57BL/6 mice. GFP-LC3-HeLa cells were obtained after transfection with peGFP-LC3 plasmid (peGFP-LC3, Addgene; from the Yoshimori laboratory, Okazaki, Japan) using Lipofectamine 2000 reagent (Invitrogen). A stable GFP-LC3-HeLa clone was established by limited dilution in 0.5 mM geneticin-selective medium. All cell lines were maintained at 37°C in humidified atmosphere containing 5% CO2 in medium supplemented with 10% heat inactivated fetal calf serum, 100 µg/ml penicillin (Invitrogen), 100 U/ml streptomycin (Invitrogen), and 1× MEM nonessential amino acid (Invitrogen). HFFs were obtained from American Type Culture Collection. WT mice were obtained from Charles River or The Jackson Laboratory. Atg16LHM mice on the C57BL/6 background, fully backcrossed by speed congenics, were generated by H. Virgin (Washington University School of Medicine, St. Louis, MO) and have been previously described (Cadwell et al., 2008). These mice were received into an enhanced barrier facility with specialized features as previously described (Cadwell et al., 2010). Mouse studies at the Institut Pasteur and at Washington University were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and according to the International Guiding Principles for Biomedical Research Involving Animals. The protocols were approved by the Animal Studies Committee at Washington University (#20090287) and the Institutional Committees on Animal Welfare of the Institut Pasteur (OLAW assurance # A5476-01). All efforts were made to minimize suffering.
siRNA treatment.
SMARTpool siRNA targeting Atg5, Atg7, or caspase-3 and control siRNA were obtained from Thermo Fisher Scientific. 0.106 HeLa or MEF cells were cultured in 6-well plates for 1 d in OptiMEM (Invitrogen) containing 10% FCS and transfected with 30 nM of indicated siRNA using Lipofectamine RNAiMAX (Invitrogen). In some experiments, Atg5 or Atg7 cDNA was transfected after 2 d of siRNA treatment. For all experiments, CHIKV infection was performed after 3 d of siRNA incubation.
CHIKV infection and UV inactivation.
The preparation of CHIKV from clinical samples has been previously described (Schuffenecker et al., 2006). CHIKV-21 strain was propagated in C6/36 cells and supernatants were harvested and frozen at −80°C before titration and further use. MEF or HeLa cells (plate at ∼50% confluence in 24- or 48-wells plates) were exposed to the indicated viruses for 1–2 h at 37°C, extensively washed with PBS, and cultivated for various periods of time before further analysis. The MOI was defined as the amount of CHIKV infectious units (calculated on BHK cells as PFU) per target cell. 3′CHIKV-GFP was generated using a full-length infection cDNA clone provided by S. Higgs (Vanlandingham et al., 2005). Infectious virus was obtained by transfection of BHK21 cells with RNA-derived cDNA, as described. UV inactivation of the virus was performed as described previously (Jan and Griffin, 1999; Krejbich-Trotot et al., 2011a) by exposure of the viral suspension to UV light (2 J over 10 min at room temperature) from UV transilluminator (Spectra; The Daavlin Company).
Immunofluorescent study.
MEFs were fixed with 4% paraformaldehyde (PFA) for 20 min and stained for LC3 (anti-LC3 Ab, clone 4E12; MBL), activated caspase-3 (anticleaved caspase-3; Cell Signaling Technology), activated caspase-8 (anticleaved caspase-8; Cell Signaling Technology), activated caspase-9 (anticleaved caspase-9; Cell Signaling Technology), p-IRE1 (rabbit polyclonal; Abcam), or Lamp-1 (anti–Lamp-1; Cell Signaling Technology) according to the experiments. The nucleus was stained with 300 nM DAPI (Invitrogen) for 5 min at 37°C in culture media. For some experiments, cells were treated with 1 µg/ml leupeptin hemisulfate (Invitrogen) and 1 µg/ml E64D (Invitrogen) 15 min before CHIKV infection or starvation. Cells were observed using an AxioCam MRm version 3 (Carl Zeiss) with a magnification of 63 (oil immersion) and analyzed using AxioVision software (Carl Zeiss). The number of vesicles per cell was analyzed from one single optical section per cell using the ApoTome (Carl Zeiss) and numerated using ImageJ software (National Institutes of Health). The RFP-GFP-LC3 construct (ptfLC3, Addgene, from the Yoshimori laboratory) was transiently expressed in HeLa cells 48 h before starvation or CHIKV infection. For all experiments, the numbers of vesicles were determined form at least 100 cells per samples.
Western blot analysis.
Lysates were prepared in 1× Dulbecco’s Phosphate Buffer Saline (DPBS; Invitrogen) containing 1% Nonidet P 40 substitute (NP40; Sigma-Aldrich) and protease inhibitor cocktail (Roche). Total protein was determined by Lowry’s method and 25 mg was loaded on a 4–12% gradient SDS–PAGE (Invitrogen). Proteins were transferred to PVDF membrane and blotted with anti-LC3 (mouse monoclonal; Cell Signaling Technology), anti-Atg5 (mouse monoclonal; Cell Signaling Technology), anti-ATG7 (mouse monoclonal; Cell Signaling Technology), anti-IRE1 (rabbit polyclonal; Abcam), anti-pIRE1 (rabbit polyclonal; Abcam), anti-pJNK (rabbit polyclonal; Abcam), anti-XBP1 (rabbit polyclonal; Abcam), anti-pmTOR (rabbit polyclonal; Abcam), anti-mTOR (rabbit polyclonal; Abcam), anti-pS6K1 (rabbit polyclonal; Abcam), anti-pAMPK (rabbit polyclonal; Abcam), or anti-GAPDH (rabbit polyclonal; Cell Signaling Technology). Secondary HRP-coupled Abs were detected using ECL Plus (GE Healthcare).
Cell viability assays and flow cytometry.
MEFs or HeLA cells were infected with CHIKV at the indicated time and fixed with 4% PFA for 20 min. After fixation, cells were stained with live/dead fixable violet cell stain kit (405 nm excitation; Invitrogen) and the percentage of cell death was measured by flow cytometry using FACSCanto (BD) and FlowJo software (Tree Star). For detection of intracellular CHIKV proteins, cells were permeabilized with Cytofix/Cytoperm (BD) before labeling with anti-E3 (from the Lecuit laboratory, Microorganismes et barrières de l’hôte, Institue Pasteur, France) or anti-capsid (from the Schwartz laboratory, viruses et immunité, URA 3015 Centre National de la Recherche Scientifique, Institue Pasteur, France).
CHIKV titers in cell lines.
MEFs or HeLa cells were infected with CHIKV-21 and supernatants were recovered at indicated time points. Viral samples were titrated as TCID50 endpoint on Vero cells using a standard procedure. Serial 10-fold dilutions (100 µg/liter) of supernatants were added in six replicates in 96-well plates seeded with 104 Vero cells. The CPE was scored 5 d after infection and the titers were calculated by determining the last dilution, giving 50% of wells with cells displaying a CPE. Results were expressed as TCID50/ml.
ImageStreamX procedure.
Please refer to de la Calle et al. (2011) for details on the methods of sample preparation, ImageStreamX acquisition, and analysis. For detection of intracellular CHIKV proteins, cells were permeabilized with Cytofix/Cytoperm before labeling with anti-capsid (from the Schwartz laboratory). To detect infected cells, MEFs were infected with CHIKV-GFP and GFP intensity was analyzed according to ImageStreamX procedure.
In vivo infection, titration, and staining.
WT littermate controls were used for all experiments (C57BL/6). 9-d-old C57BL/6 WT (n = 43) and Atg16LHM mice (n = 34) were infected with 4 × 105 PFU CHIKV s.c. in the upper chest and were followed for lethality for 25 d after infection. Litters were weight matched at the start of the experiments. For viral titration (n = 3), tissues were collected, homogenized, and viral samples were titrated by standard plaque assay on BHK cells. For histology blocks, skin (n = 3) and muscle (n = 3) from the site of infection was fixed in PFA and frozen in OCT blocks. Tissues were sectioned and stained for activated caspase-3 and CHIKV E3 protein and image analysis was performed as previously described.
Acknowledgments
The authors would like to acknowledge Herbert Virgin III for providing Atg16LHM mice, and the Centre d’Immunologie Humaine at Institut Pasteur for support of experimental study. We also thank F. Rey, M. Lecuit, and members of the Immunobiologie des Cellules Dendritiques laboratory for their helpful advice and review of the paper.
This work was supported by the Institut Pasteur, Institut National de la Santé et de la Recherche Médicale, Agence Nationale de Recherches sur le Sida et les hépatites virales, the European Research Council, and the French National Research Agency (ANR).
None of the authors have competing financial interests.
Footnotes
Abbreviations used:
- BDI
- bright detail intensity
- CHIKV
- Chikungunya virus
- CPE
- cytopathic effect
- MOI
- multiplicity of infection
- PE
- phosphatidylethanolamine
- RNS
- reactive nitrogen species
- ROS
- reactive oxygen species
References
- Alexander A., Cai S.L., Kim J., Nanez A., Sahin M., MacLean K.H., Inoki K., Guan K.L., Shen J., Person M.D., et al. 2010a. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. USA. 107:4153–4158 10.1073/pnas.0913860107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander A., Kim J., Walker C.L. 2010b. ATM engages the TSC2/mTORC1 signaling node to regulate autophagy. Autophagy. 6:672–673 10.4161/auto.6.5.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet F.P., Moris A., Nikolic D.S., Lehmann M., Cardinaud S., Stalder R., Garcia E., Dinkins C., Leuba F., Wu L., et al. 2010. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 32:654–669 10.1016/j.immuni.2010.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmann P., Althaus F., Burnand B., Vaucher P., Pécoud A., Genton B. 2007. Medical care of asylum seekers: a descriptive study of the appropriateness of nurse practitioners’ care compared to traditional physician-based care in a gatekeeping system. BMC Public Health. 7:310 10.1186/1471-2458-7-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgherini G., Poubeau P., Staikowsky F., Lory M., Le Moullec N., Becquart J.P., Wengling C., Michault A., Paganin F. 2007. Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin. Infect. Dis. 44:1401–1407 10.1086/517537 [DOI] [PubMed] [Google Scholar]
- Cadwell K., Liu J.Y., Brown S.L., Miyoshi H., Loh J., Lennerz J.K., Kishi C., Kc W., Carrero J.A., Hunt S., et al. 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 456:259–263 10.1038/nature07416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K., Patel K.K., Maloney N.S., Liu T.C., Ng A.C., Storer C.E., Head R.D., Xavier R., Stappenbeck T.S., Virgin H.W. 2010. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 141:1135–1145 10.1016/j.cell.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi A. 2010. Cell responses to oxidative stressors. Curr. Pharm. Des. 16:1387–1395 10.2174/138161210791033969 [DOI] [PubMed] [Google Scholar]
- Couderc T., Chrétien F., Schilte C., Disson O., Brigitte M., Guivel-Benhassine F., Touret Y., Barau G., Cayet N., Schuffenecker I., et al. 2008. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog. 4:e29 10.1371/journal.ppat.0040029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotzer V.L., Blum J.S. 2010. Autophagy and adaptive immunity. Immunology. 131:9–17 10.1111/j.1365-2567.2010.03321.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial N.N., Korsmeyer S.J. 2004. Cell death: critical control points. Cell. 116:205–219 10.1016/S0092-8674(04)00046-7 [DOI] [PubMed] [Google Scholar]
- de la Calle C., Joubert P.E., Law H.K., Hasan M., Albert M.L. 2011. Simultaneous assessment of autophagy and apoptosis using multispectral imaging cytometry. Autophagy. 7:1045–1051 10.4161/auto.7.9.16252 [DOI] [PubMed] [Google Scholar]
- Dengjel J., Schoor O., Fischer R., Reich M., Kraus M., Müller M., Kreymborg K., Altenberend F., Brandenburg J., Kalbacher H., et al. 2005. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA. 102:7922–7927 10.1073/pnas.0501190102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Levine B. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 5:527–549 10.1016/j.chom.2009.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavaheri-Mergny M., Amelotti M., Mathieu J., Besançon F., Bauvy C., Codogno P. 2007. Regulation of autophagy by NFkappaB transcription factor and reactives oxygen species. Autophagy. 3:390–392 [DOI] [PubMed] [Google Scholar]
- Djavaheri-Mergny M., Maiuri M.C., Kroemer G. 2010. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 29:1717–1719 10.1038/onc.2009.519 [DOI] [PubMed] [Google Scholar]
- Dreux M., Chisari F.V. 2009. Autophagy proteins promote hepatitis C virus replication. Autophagy. 5:1224–1225 10.4161/auto.5.8.10219 [DOI] [PubMed] [Google Scholar]
- English L., Chemali M., Duron J., Rondeau C., Laplante A., Gingras D., Alexander D., Leib D., Norbury C., Lippé R., Desjardins M. 2009. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 10:480–487 10.1038/ni.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink M. 2006. Infectious diseases. Massive outbreak draws fresh attention to little-known virus. Science. 311:1085 10.1126/science.311.5764.1085a [DOI] [PubMed] [Google Scholar]
- Everett H., McFadden G. 1999. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 7:160–165 10.1016/S0966-842X(99)01487-0 [DOI] [PubMed] [Google Scholar]
- Filomeni G., Desideri E., Cardaci S., Rotilio G., Ciriolo M.R. 2010. Under the ROS…thiol network is the principal suspect for autophagy commitment. Autophagy. 6:999–1005 10.4161/auto.6.7.12754 [DOI] [PubMed] [Google Scholar]
- Gannagé M., Dormann D., Albrecht R., Dengjel J., Torossi T., Rämer P.C., Lee M., Strowig T., Arrey F., Conenello G., et al. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 6:367–380 10.1016/j.chom.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérardin P., Guernier V., Perrau J., Fianu A., Le Roux K., Grivard P., Michault A., de Lamballerie X., Flahault A., Favier F. 2008. Estimating Chikungunya prevalence in La Réunion Island outbreak by serosurveys: two methods for two critical times of the epidemic. BMC Infect. Dis. 8:99 10.1186/1471-2334-8-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.E., Hardwick J.M. 1997. Regulators of apoptosis on the road to persistent alphavirus infection. Annu. Rev. Microbiol. 51:565–592 10.1146/annurev.micro.51.1.565 [DOI] [PubMed] [Google Scholar]
- Guo W.J., Ye S.S., Cao N., Huang J., Gao J., Chen Q.Y. 2010. ROS-mediated autophagy was involved in cancer cell death induced by novel copper(II) complex. Exp. Toxicol. Pathol. 62:577–582 10.1016/j.etp.2009.08.001 [DOI] [PubMed] [Google Scholar]
- He C., Bassik M.C., Moresi V., Sun K., Wei Y., Zou Z., An Z., Loh J., Fisher J., Sun Q., et al. 2012. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 481:511–515 10.1038/nature10758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Morita K., Matias R.R., Tuplano J.V., Resuello R.R., Candelario J.R., Cruz D.J., Mapua C.A., Hasebe F., Igarashi A., Natividad F.F. 2003. Distribution of three arbovirus antibodies among monkeys (Macaca fascicularis) in the Philippines. J. Med. Primatol. 32:89–94 10.1034/j.1600-0684.2003.00015.x [DOI] [PubMed] [Google Scholar]
- Jan J.T., Griffin D.E. 1999. Induction of apoptosis by Sindbis virus occurs at cell entry and does not require virus replication. J. Virol. 73:10296–10302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert P.E., Meiffren G., Grégoire I.P., Pontini G., Richetta C., Flacher M., Azocar O., Vidalain P.O., Vidal M., Lotteau V., et al. 2009. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 6:354–366 10.1016/j.chom.2009.09.006 [DOI] [PubMed] [Google Scholar]
- Joubert P.E., Pombo Grégoire I., Meiffren G., Rabourdin-Combe C., Faure M. 2011. [Autophagy and pathogens: “Bon appétit Messieurs!”]. Med. Sci. (Paris). 27:41–47 10.1051/medsci/201127141 [DOI] [PubMed] [Google Scholar]
- Jounai N., Takeshita F., Kobiyama K., Sawano A., Miyawaki A., Xin K.Q., Ishii K.J., Kawai T., Akira S., Suzuki K., Okuda K. 2007. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA. 104:14050–14055 10.1073/pnas.0704014104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R., Zeh H.J., Lotze M.T., Tang D. 2011. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 18:571–580 10.1038/cdd.2010.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepp O., Senovilla L., Galluzzi L., Panaretakis T., Tesniere A., Schlemmer F., Madeo F., Zitvogel L., Kroemer G. 2009. Viral subversion of immunogenic cell death. Cell Cycle. 8:860–869 10.4161/cc.8.6.7939 [DOI] [PubMed] [Google Scholar]
- Krakauer D.C., Payne R.J. 1997. The evolution of virus-induced apoptosis. Proc. Biol. Sci. 264:1757–1762 10.1098/rspb.1997.0243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejbich-Trotot P., Denizot M., Hoarau J.J., Jaffar-Bandjee M.C., Das T., Gasque P. 2011a. Chikungunya virus mobilizes the apoptotic machinery to invade host cell defenses. FASEB J. 25:314–325 10.1096/fj.10-164178 [DOI] [PubMed] [Google Scholar]
- Krejbich-Trotot P., Gay B., Li-Pat-Yuen G., Hoarau J.J., Jaffar-Bandjee M.C., Briant L., Gasque P., Denizot M. 2011b. Chikungunya triggers an autophagic process which promotes viral replication. Virol. J. 8:432 10.1186/1743-422X-8-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Galluzzi L., Brenner C. 2007. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87:99–163 10.1152/physrev.00013.2006 [DOI] [PubMed] [Google Scholar]
- Kroemer G., Mariño G., Levine B. 2010. Autophagy and the integrated stress response. Mol. Cell. 40:280–293 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M., Kornbluth S. 2009. Caspases and kinases in a death grip. Cell. 138:838–854 10.1016/j.cell.2009.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M., Kornbluth S. 2010. Stalling in mitosis and releasing the apoptotic brake. EMBO J. 29:2255–2257 10.1038/emboj.2010.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.H., Iwakoshi N.N., Glimcher L.H. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23:7448–7459 10.1128/MCB.23.21.7448-7459.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Lund J.M., Ramanathan B., Mizushima N., Iwasaki A. 2007. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 315:1398–1401 10.1126/science.1136880 [DOI] [PubMed] [Google Scholar]
- Li M.L., Stollar V. 2004. Alphaviruses and apoptosis. Int. Rev. Immunol. 23:7–24 10.1080/08830180490265529 [DOI] [PubMed] [Google Scholar]
- Li W., Yang Q., Mao Z. 2011. Chaperone-mediated autophagy: machinery, regulation and biological consequences. Cell. Mol. Life Sci. 68:749–763 10.1007/s00018-010-0565-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Kleeman L.K., Jiang H.H., Gordon G., Goldman J.E., Berry G., Herman B., Levine B. 1998. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 72:8586–8596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manimunda S.P., Mavalankar D., Bandyopadhyay T., Sugunan A.P. 2011. Chikungunya epidemic-related mortality. Epidemiol. Infect. 139:1410–1412 10.1017/S0950268810002542 [DOI] [PubMed] [Google Scholar]
- Mason P.J., Haddow A.J. 1957. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952-53; an additional note on Chikungunya virus isolations and serum antibodies. Trans. R. Soc. Trop. Med. Hyg. 51:238–240 10.1016/0035-9203(57)90022-6 [DOI] [PubMed] [Google Scholar]
- McGuckin M.A., Eri R.D., Das I., Lourie R., Florin T.H. 2010. ER stress and the unfolded protein response in intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 298:G820–G832 10.1152/ajpgi.00063.2010 [DOI] [PubMed] [Google Scholar]
- Mehrpour M., Esclatine A., Beau I., Codogno P. 2010. Overview of macroautophagy regulation in mammalian cells. Cell Res. 20:748–762 10.1038/cr.2010.82 [DOI] [PubMed] [Google Scholar]
- Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., et al. 2004. Autophagy defends cells against invading group A Streptococcus. Science. 306:1037–1040 10.1126/science.1103966 [DOI] [PubMed] [Google Scholar]
- Ng L.F., Chow A., Sun Y.J., Kwek D.J., Lim P.L., Dimatatac F., Ng L.C., Ooi E.E., Choo K.H., Her Z., et al. 2009. IL-1beta, IL-6, and RANTES as biomarkers of Chikungunya severity. PLoS ONE. 4:e4261 10.1371/journal.pone.0004261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo E., Parola M. 2008. Redox mechanisms in hepatic chronic wound healing and fibrogenesis. Fibrogenesis Tissue Repair. 1:5 10.1186/1755-1536-1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Levine B. 2008. Viral evasion of autophagy. Autophagy. 4:280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., MacPherson S., Sumpter R., Jr, Tallóczy Z., Zou Z., Levine B. 2010. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 7:115–127 10.1016/j.chom.2010.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Sumpter R., Jr, Xiao G., Ng A., Zou Z., Tang Y., Narimatsu M., Gilpin C., Sun Q., Roth M., et al. 2011. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 480:113–117 10.1038/nature10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozden S., Huerre M., Riviere J.P., Coffey L.L., Afonso P.V., Mouly V., de Monredon J., Roger J.C., El Amrani M., Yvin J.L., et al. 2007. Human muscle satellite cells as targets of Chikungunya virus infection. PLoS ONE. 2:e527 10.1371/journal.pone.0000527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A.M., Logue C.H. 2007. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J. Gen. Virol. 88:2363–2377 10.1099/vir.0.82858-0 [DOI] [PubMed] [Google Scholar]
- Schilte C., Couderc T., Chretien F., Sourisseau M., Gangneux N., Guivel-Benhassine F., Kraxner A., Tschopp J., Higgs S., Michault A., et al. 2010. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J. Exp. Med. 207:429–442 10.1084/jem.20090851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuffenecker I., Iteman I., Michault A., Murri S., Frangeul L., Vaney M.C., Lavenir R., Pardigon N., Reynes J.M., Pettinelli F., et al. 2006. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 3:e263 10.1371/journal.pmed.0030263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz O., Albert M.L. 2010. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 8:491–500 10.1038/nrmicro2368 [DOI] [PubMed] [Google Scholar]
- Simon F., Tolou H., Jeandel P. 2006. [The unexpected Chikungunya outbreak]. Rev. Med. Interne. 27:437–441 (The unexpected Chikungunya outbreak) 10.1016/j.revmed.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Sourisseau M., Schilte C., Casartelli N., Trouillet C., Guivel-Benhassine F., Rudnicka D., Sol-Foulon N., Le Roux K., Prevost M.C., Fsihi H., et al. 2007. Characterization of reemerging chikungunya virus. PLoS Pathog. 3:e89 10.1371/journal.ppat.0030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teodoro J.G., Branton P.E. 1997. Regulation of apoptosis by viral gene products. J. Virol. 71:1739–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson A.W., Turnquist H.R., Raimondi G. 2009. Immunoregulatory functions of mTOR inhibition. Nat. Rev. Immunol. 9:324–337 10.1038/nri2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn A. 2008. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis. 13:1–9 10.1007/s10495-007-0154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl M., Kepp O., Jusforgues-Saklani H., Vicencio J.M., Kroemer G., Albert M.L. 2009. Autophagy within the antigen donor cell facilitates efficient antigen cross-priming of virus-specific CD8+ T cells. Cell Death Differ. 16:991–1005 10.1038/cdd.2009.8 [DOI] [PubMed] [Google Scholar]
- Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H.P., Ron D. 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 287:664–666 10.1126/science.287.5453.664 [DOI] [PubMed] [Google Scholar]
- Vanlandingham D.L., Tsetsarkin K., Hong C., Klingler K., McElroy K.L., Lehane M.J., Higgs S. 2005. Development and characterization of a double subgenomic chikungunya virus infectious clone to express heterologous genes in Aedes aegypti mosquitoes. Insect Biochem. Mol. Biol. 35:1162–1170 10.1016/j.ibmb.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Watson R. 2007. Europe witnesses first local transmission of chikungunya fever in Italy. BMJ. 335:532–533 10.1136/bmj.39332.708738.DB [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Pattingre S., Sinha S., Bassik M., Levine B. 2008. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell. 30:678–688 10.1016/j.molcel.2008.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson N.S., Dixit V., Ashkenazi A. 2009. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat. Immunol. 10:348–355 10.1038/ni.1714 [DOI] [PubMed] [Google Scholar]
- Yatim N., Albert M.L. 2011. Dying to replicate: the orchestration of the viral life cycle, cell death pathways, and immunity. Immunity. 35:478–490 10.1016/j.immuni.2011.10.010 [DOI] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 107:881–891 10.1016/S0092-8674(01)00611-0 [DOI] [PubMed] [Google Scholar]
- Zhang X., Qu J., Ding M., Ren X., Qiu Y. 1997. [The mutiplication of Sindbis virus and host BHK-21 cell apoptosis]. Wei Sheng Wu Xue Bao. 37:165–170 [PubMed] [Google Scholar]
- Zoncu R., Efeyan A., Sabatini D.M. 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12:21–35 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]