High levels of glutathione peroxidase 3 (GPx3) expression correlate with adverse prognosis in acute myeloid leukemia, and enhance activity of long-term repopulating hematopoietic stem cells in mice.
Abstract
The determinants of normal and leukemic stem cell self-renewal remain poorly characterized. We report that expression of the reactive oxygen species (ROS) scavenger glutathione peroxidase 3 (GPx3) positively correlates with the frequency of leukemia stem cells (LSCs) in Hoxa9+Meis1-induced leukemias. Compared with a leukemia with a low frequency of LSCs, a leukemia with a high frequency of LSCs showed hypomethylation of the Gpx3 promoter region, and expressed high levels of Gpx3 and low levels of ROS. LSCs and normal hematopoietic stem cells (HSCs) engineered to express Gpx3 short hairpin RNA (shRNA) were much less competitive in vivo than control cells. However, progenitor cell proliferation and differentiation was not affected by Gpx3 shRNA. Consistent with this, HSCs overexpressing Gpx3 were significantly more competitive than control cells in long-term repopulation experiments, and overexpression of the self-renewal genes Prdm16 or Hoxb4 boosted Gpx3 expression. In human primary acute myeloid leukemia samples, GPX3 expression level directly correlated with adverse prognostic outcome, revealing a potential novel target for the eradication of LSCs.
The cancer stem cell (CSC) hypothesis postulates that most cancers are heterogeneous in their cellular constituents and that self-renewal potential is restricted to a subset of cells within each tumor. The frequency of CSC appears to vary considerably between tumors and also in time within a given cancer. CSCs are best described in human leukemia in which the leukemia stem cells (LSCs) can be prospectively isolated and transmit the disease when introduced in immuno-compromised mice (Lapidot et al., 1994). A detailed understanding of the molecular bases that regulate LSC self-renewal is lacking. Growing evidence indicates that although certain regulators of self-renewal (e.g., Bmi1) are shared between normal cells and LSCs (Lessard and Sauvageau, 2003), others (e.g., NF-kB and Wnt/β-catenin) are more specific to LSC (Guzman et al., 2002; Wang et al., 2010).
Variation in oxidative stress has been associated with changes in self-renewal potential of normal hematopoietic stem cells (HSCs; Shao et al., 2011). By analogy, oxidative stress may also affect LSC self-renewal but the experimental evidence supporting this possibility remains scarce (Konopleva and Jordan, 2011). Using a series of recently characterized mouse acute myeloid leukemia (AML; Wilhelm et al., 2011) in which the LSC frequency varied from ∼1 in 100–350 to 1 in 1.4 cells, we now show that Gpx3, a reactive oxygen species (ROS) detoxifying enzyme, determines the activity of normal cells and LSCs, extending the function of oxidative stress to regulation of LSC biology in vivo.
RESULTS AND DISCUSSION
High LSC frequency leukemia expresses elevated GPx3
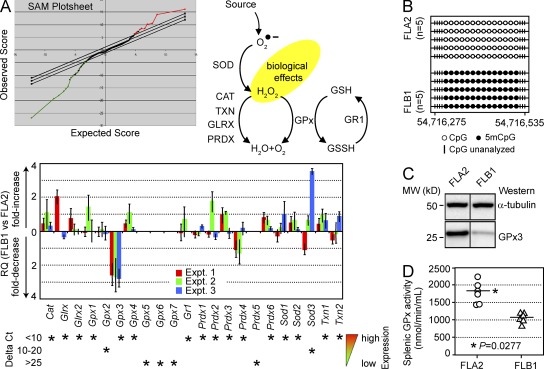
We reasoned that comparative mRNA profiling of FLA2 (LSC at 70% or 1 in 1.4 cells; Wilhelm et al., 2011) to the phenotypically similar FLB1 (0.3% LSC) might identify genes uniquely associated with LSC activity (Fig. 1 A). Microarray-based global gene expression profiling identified very few genes—besides some sex chromosome and immunoglobulin light chain (IgK-C) genes—as having differential expression between these two leukemia specimens. After excluding the sex chromosome genes because of the male and female origin of FLA2 and FLB1, only 16 probe sets exhibited significant changes of signal for a false discovery rate of 2.71% (Table S1). In particular, qRT-PCR confirmed a two- to threefold up-regulation of antioxidant enzyme glutathione peroxidase 3 (Gpx3) in FLA2 compared with FLB1 (Supplemental Text).
Figure 1.
GPx3 is overexpressed in FLA2 cells. (A) Plot depicting proportions of overexpressed (red points) and underexpressed (green points) genes in FLA2 (high LSC frequency) compared with FLB1 cells (low LSC frequency). RNA isolated from BM cells of leukemic mice (n = 3 per group) was reverse transcribed, and cDNA was hybridized on Affymetrix Mouse Genome 430A microarrays. The stringency of the false discovery rate for the GCRMA-normalized data was adjusted using the SAM software (top left). The antioxidant enzymatic pathway and putative function of each enzyme (top right) is shown. Analyses of antioxidant enzyme expression in FLA2 (high LSC frequency) and FLB1 cells (low LSC frequency) are shown. Results of qRT-PCR assays are presented as FLB1/FLA2 expression ratios (mean ± SD, n = 3). ΔCt values (versus Actb) shown below indicate expression levels of each transcript (bottom). (B) Methylation status of the CpG island in the promoter region of Gpx3 as determined by sequencing HSO3-treated gDNA. Each row represents one leukemic mouse (n = 5 per group). Open circle, nonmethylated CpG; closed circle, methylated CpG. (C) Western blot analysis of GPx3 protein levels in FLA2 and FLB1 cells. α-Tubulin was used as loading control. Black lines indicate that intervening lanes were spliced out. (D) Quantitation of the GPx (glutathione peroxidase) activity in spleens of FLA2 and FLB1 leukemic mice (n = 6, two experiments) reveals significant increase (P = 0.0277) in splenic GPx activity of FLA2 mice. Spleens of FLA2 and FLB1 leukemic mice had similar weights (239 ± 12 and 227 ± 18 mg in FLA2 and FLB1 mice, respectively; mean ± SD, n = 19) and comparable leukemic blast infiltrations (Fig. S1).
Because redox metabolism has been implicated in normal HSC self-renewal (Ito et al., 2004, 2006; Tothova et al., 2007), we compared the mRNA levels of other antioxidant enzymes implicated in ROS detoxification. In contrast to Gpx3, most of these genes are expressed at comparable levels in FLA2 and FLB1 cells (Fig. 1 A), which also express similar levels of an additional 170 potential regulators of stem cell activity analyzed (Supplemental Text). In accordance with the Gpx3 overexpression in FLA2, 14/14 tested CpG sequences in the Gpx3 promoter region were methylated in FLB1 and hypomethylated in FLA2 cells (Fig. 1 B). Higher expression of Gpx3 in FLA2 compared with FLB1 cells resulted in increased protein levels (Fig. 1 C) and elevated glutathione peroxidase activity (1,773 ± 127 nmol/min/ml and 1,077 ± 60 nmol/min/ml, respectively; Fig. 1 D).
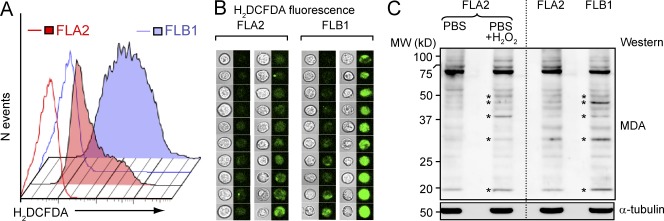
Flow cytometric comparison of the ROS indicator dichlorodihydrofluorescein diacetate (H2DCFDA) fluorescence revealed a relative reduction in ROS levels in FLA2 compared FLB1 leukemia (Fig. 2 A), and confocal analysis confirmed these results (Fig. 2 B). Elevated ROS levels also result in lipid peroxidation and formation of malondialdehyde (MDA) protein adducts, and Western blot analyses demonstrated increase in MDA levels in FLB1 compared with FLA2 cells (Fig. 2 C).
Figure 2.
The ROS level is decreased in FLA2 cells. (A) Intracellular ROS levels in FLA2 and FLB1 evaluated by flow cytometry using H2DCFDA probe immediately after extracting marrow from leukemic mice. Open histograms, DMSO (vehicle control); filled histograms, H2DCFDA. (B) Confocal microscopy of FLA2 and FLB1 cells stained with H2DCFDA. 20 cells of each leukemia were randomly chosen and classified according to H2DCFDA fluorescence. The corresponding phase-contrast pictures are presented. (C) Western blot of MDA-modified protein adducts in FLA2 and FLB1 cells. H2O2-treated FLA2 cells were used as a positive control and α-tubulin as a loading control. Asterisks indicate major MDA protein adduct species. The dotted line shows margins of two blots shown in a merged image.
Members of the FoxO subfamily regulate intracellular ROS through transcriptional up-regulation of antioxidative enzymes (Kops et al., 2002; Tothova et al., 2007). FoxO-deficient HSCs were reported to overexpress several antioxidant enzymes such as Sod1, Sod3, Prdx6, and Gpx4, but not Gpx3 (Tothova et al., 2007). FLA2 and FLB1 leukemias expressed comparable levels of Foxo3a, Foxo4, and Foxo6, whereas Foxo1 was not expressed (Supplemental Text), indicating that FoxO induction is likely not responsible for differential ROS (Supplemental Text) and suggesting that the only differentially expressed antioxidant enzyme GPx3 may be responsible for the decreased ROS observed in FLA2 leukemia.
GPx3 promotes LSC in vivo competitiveness
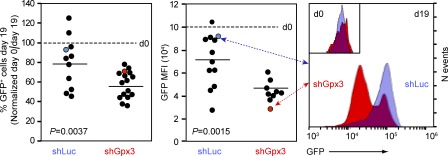
Correlation between GPx3 levels and LSC frequency could reflect functional dependence of LSC on this enzyme. We exploited retroviruses encoding short hairpin RNAs (shRNAs) and IRES-GFP reporter to explore the in vivo activity of FLA2 cells after GPx3 knockdown (Fig. 3 A). Gene transfer in these experiments ranged from 42 to 66%, and because FLA2 cells poorly survive cell sorting, untransduced leukemia-initiating cells were co-transplanted together with transduced cells. This resulted in competitive repopulation experiments in which both the GFP+ shRNA (either Luciferase or Gpx3)-transduced and GFP− untransduced FLA2 cells contributed to leukemia development in vivo. To assess the effect of shRNA treatment on relative contribution of each cell population to clinical leukemia 19 d later, we compared changes in proportions (Fig. 3 B, left) and mean fluorescence intensity (MFI; Fig. 3 B, middle and right) of GFP+ FLA2 cells at two different time points (t = 0 and 19 d). The decrease in proportion of GFP+ FLA2 cells contributing to fulminant leukemia (∼19 d) below the values determined for the initial transplant was at least twofold higher after Gpx3 knockdown compared with shLuc control (Fig. 3 B, left). Moreover, the shGpx3-infected FLA2 cells remaining at day 19 displayed a significant reduction in MFI of GFP compared with controls (Fig. 3 B, middle and right). These results indicate that GFPhigh cells were selectively depleted (Fig. 3 B, right, inset), and suggest that Gpx3 is critical for the competitiveness of LSCs.
Figure 3.
Gpx3 depletion decreased LSC competitiveness. Left: effect of the RNAi-mediated depletion of Gpx3 on the competitiveness of FLA2 leukemic cells. 2 × 105 fresh FLA2 BM cells were co-cultured with shLuc or shGpx3 retrovirus-producing GP+E-86 cells. After infection (d0), cells were partitioned between flow cytometric analysis of GFP+ frequency and transplantation of unsorted cells into sublethally irradiated recipients (shLuc, n = 12; shGpx3, n = 16). Contribution of GFP+ (shRNA-transduced) cells to leukemia was determined on day 19. Results are shown as normalized values: the GFP+ frequency in inoculum was considered 100% (dotted line, d0), and the day 19 GFP+ frequency was calculated as a percentage of that value. Horizontal bars represent mean values determined for each experimental group. Middle: day 19 MFIs of GFP+ cells in recipients of shLuc- and shGpx3-infected FLA2 leukemic cells. The dotted line represents MFI of GFP+ cells in shLuc and shGpx3 inocula. Horizontal bars represent mean values determined for each experimental group. Right: GFP fluorescence intensity in representative recipients of shLuc- or shGpx3-infected FLA2 cells. The inset demonstrates comparable MFI GFP fluorescence in shLuc and shGpx3 inocula. *, P = 0.0037 and P = 0.0015, respectively (Wilcoxon’s test).
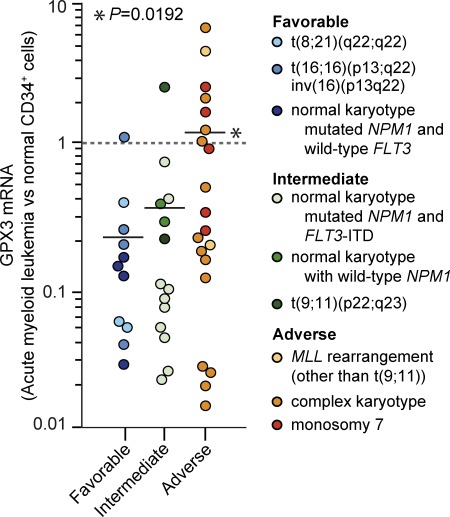
We quantified GPX3 expression in 48 acute myeloid leukemia patients categorized into cytogenetically and molecularly defined risk subgroups designated as favorable, intermediate, and adverse (Mrózek et al., 2007). Within the AML samples, expression of GPX3 increased with the severity of disease, and the highest expression levels were associated with adverse risk group of patients (P = 0.019; Fig. 4). The worst clinical outcomes were thus observed in AML enriched for phenotypically defined LSC (van Rhenen et al., 2005) and high GPX3 levels, indicating that GPX3 may be relevant for human leukemia.
Figure 4.
GPX3 expression in marrow samples from AML patients classified into cytogenetic and molecular risk subgroups. Expression of GPX3 was quantified by qRT-PCR (ΔCt method versus GAPDH and HPRT) in marrow leukemic cells (leukoblasts > 95%) from AML patients and FACS-sorted marrow CD34+ cells from healthy volunteers. Results are presented as relative expression (RQ = 2−ΔΔCt) of GPX3 versus normal marrow CD34+ cells (dotted line, ΔCt = 10.10 ± 0.58, n = 5 for normal marrow CD34+ cells). Horizontal bars represent mean GPX3 levels determined for individual risk subgroup. *, P = 0.0192 (adverse versus favorable risk subgroups; Wilcoxon’s test).
Gpx3 influences normal HSC activity
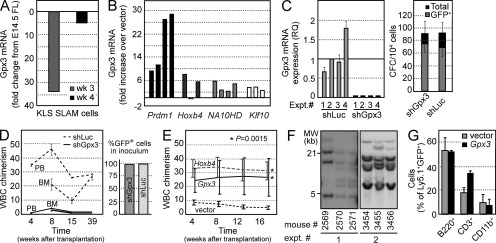
Adult BM-derived HSCs are less proliferative than their fetal liver (FL)–derived counterparts (Rebel et al., 1996). Quantitative RT-PCR (qRT-PCR) assays revealed that HSC isolated from BM of 3- and 4-wk-old mice express 39- to 6-fold, respectively, lower Gpx3 levels than HSC sorted from embryonic day 14.5 (E14.5) FL (Fig. 5.A), suggesting a correlation between Gpx3 expression and HSC activity. Interestingly, Gpx3 levels were also elevated in BM HSC/progenitor cells engineered to overexpress genes associated with self-renewal such as Hoxb4, NA10HD, Klf10, and Prdm16 (Sauvageau et al., 1995; Ohta et al., 2007; Deneault et al., 2009). In these transduced cells, levels of Gpx3 expression increased 3.2- to 19.2-fold compared with controls (Fig. 5 B).
Figure 5.
Gpx3 regulates the self-renewal capacity of normal HSCs. (A) Gpx3 expression in HSC isolated from 3- or 4-wk-old mice (KLS SLAM cells) compared with HSC from E14.5 FL (SLAM Sca+Mac1+) cells. Mean ± SD, n = 2. (B) Empty vector (n = 3), Prdm16 (n = 4), Hoxb4 (n = 3), NA10HD (n = 4), and Klf10 (n = 3) were overexpressed in Lin−CD150+CD48− sorted BM cells as previously described (Deneault et al., 2009). Gpx3 mRNA was evaluated by qRT-PCR. (C) 5-FU–treated BM cells were transduced with shLuc or shGpx3. Gpx3 mRNA was evaluated by qRT-PCR in four experiments (left). CFC output was assessed in two experiments (right; mean ± SD; n = 8). GFP+ cells were shRNA-transduced cells. (D) Evaluation of short-term and LTR ability of KLS CD150+CD48−Ly5.1+ BM cells expressing shLuc or shGpx3 in competitive transplantations into congenic Ly5.2 mice. Results are presented from both peripheral blood (PB) and BM, and represent mean ± SEM at the indicated time points for 8 mice per group. The level of gene transfer was near 100% (right) and Gpx3 silencing was obtained in all experiments (n = 4). Two independent shGpx3 constructs (#5 and #6) were used. (E) Gpx3, Hoxb4, or empty vector was overexpressed in BM KLS CD150+CD48− sorted cells (as described in Deneault et al., 2009), cells were expanded for 7 d ex vivo, and competitive transplantations in Ly5.2 congenic mice were performed. Results are shown as mean ± SEM of peripheral blood reconstitution at the indicated time points for three mice per group in five experiments. *, P = 0.0015 (Wilcoxon’s test). (F) Southern blot analyses of Gpx3 proviral integrations in DNA isolated from BM from mice reconstituted 20 wk earlier with Gpx3-transduced cells. n = 2 experiments, three mice per experiment. (G) Analysis of differentiation potential of vector or Gpx3-transduced cells in vivo. Blood cells were collected from recipients transplanted 20 wk earlier and stained with antibodies specific for B, T, and myeloid cells (B220, CD3, and CD11b, respectively). Values are mean ± SD (vector: n = 5; Gpx3, n = 2).
We next infected BM HSC/progenitor cells with retroviruses carrying two different Gpx3 hairpins with the ability to achieve ∼95% Gpx3 knockdown (Fig. 5 C, left). Clonogenic progenitor cell activity was not affected by this decrease in Gpx3 expression (Fig. 5 C, right), and there was no increase in apoptosis in the shGpx3 cell population compared with shLuc controls (not depicted). shRNA-mediated Gpx3 silencing, however, dramatically impeded the ability of transplanted HSC to repopulate the hematopoietic system of recipient mice at early (8 wk) and late (39 wk) time points after transplantation (Fig. 5 D). We also transplanted the shGpx3- and shLuc-transduced 5-FU–derived BM cells and analyzed their ability to home in the BM 48 h after injection. These experiments showed no difference between homing abilities of shGpx3- and shLuc-transduced cells. Together, these results identify Gpx3 as a critical regulator of short- and long-term repopulating (LTR) HSC.
In reciprocal gain-of-function experiments, BM HSC/progenitor cells engineered to overexpress Gpx3 showed a marked competitive advantage over controls when transplanted after a 7-d ex vivo culture step (Fig. 5 E). Proviral integration analyses of six recipients confirmed polyclonal hematopoiesis (Fig. 5 F), thus ruling out the possibility of insertional mutagenesis. Moreover, some mice (#2570 and #2571; Fig. 5 F) were reconstituted by the same clones, indicating that self-renewal occurred in vitro before transplantation. GFP+ Gpx3-transduced cells contributed to production of lymphoid and myeloid peripheral blood leukocytes at >30 wk after transplantation, confirming the multipotential nature of the reconstituting clones (Fig. 5 G). These mice were hematologically normal and showed no evidence of myeloproliferation or leukemia. Together, these results indicate that Gpx3 enhances HSC expansion ex vivo possibly through modulation of self-renewal activity. Unfortunately, such gain-of-function experiments were not possible with our FLB1 leukemia model as we have not yet identified the in vitro conditions that would allow their effective transduction.
In summary, identification of an antioxidant enzyme such as GPx3 acting as a stem cell activity factor in leukemia raises the possibility that other ROS scavenging systems have regulatory importance in other CSS. A low ROS level in breast CSCs has recently been associated with increased expression of Foxo1 and the glutathione biosynthesis genes Gss and Gclm (Diehn et al., 2009). In normal hematopoiesis, ROSlow HSCs have a higher self-renewal potential than ROShigh HSCs (Jang and Sharkis, 2007), whereas loss of ATM (ataxia telangiectasia mutated) or a FoxO transcription factor correlates with elevated ROS levels and limits self-renewal capacity of HSC during serial transplantations (Ito et al., 2004, 2006; Tothova et al., 2007). ROS can function as signaling molecules in several cell types (Finkel and Holbrook, 2000; Owusu-Ansah et al., 2008; Takeda et al., 2008) and can modulate the activation of signal transduction pathways involved in cellular proliferation and differentiation (Sundaresan et al., 1995; Sattler et al., 1999; Owusu-Ansah and Banerjee, 2009).
Although the current study highlights the importance of GPx3 in normal and leukemia hematopoiesis, the implication of ROS as the key modulator of GPx3 activity has not been established. This is particularly critical in the context that ROS scavenger (N-acetyl-l-cysteine treatment) and p38 inhibition failed to consistently increase LSC frequency of FLB1 leukemia in vivo (unpublished data). Nonetheless, our results indicate that in some conditions (Hoxb4 or Prdm16 gain of function) Gpx3 expression may represent a converging pathway that determines the activity of normal and leukemic stem cells.
MATERIALS AND METHODS
Mice.
Donor C57BL/6:Pep3b (Ly5.1) and recipient C57BL/6 (Ly5.2) mice were bred and maintained as previously described (Kroon et al., 1998). Experimental procedures were revised and approved by the University of Montreal animal ethics committee (Comité de Déontologie de l’Expérimentation sur les Animaux de l’Université de Montréal).
Human cells.
AML tumor samples and normal marrow samples were obtained from the Quebec Leukemia Cell Bank (Montreal, Quebec) and the Orthopedic Surgery Department of the University Hospital of Tours (Tours, France), respectively. All these cells were obtained according to approved protocols of the institutional review boards after informed consent. Patients were classified in three risk groups for cytogenetic and molecular abnormalities (Mrózek et al., 2007).
Isolation of mouse hematopoietic cell subpopulations.
Cells were first purified for Sca1 using magnetic activated cell sorting as per the manufacturer’s instructions (Miltenyi Biotec). Further antibody staining and sorting to purify the c-Kit+, lineage− (CD45R−, CD3−, Gr-1−, Ter119−) population were performed as described previously (Okada et al., 1992). The LTR-HSC from BM (c-Kit+Lin−Sca1+CD150+CD48−, KLS SLAM) or E14.5 d.p.c. FL (CD150+CD48-Sca1+Mac1+ Lin−, SLAM Sca+Mac+), and HSC/progenitor cells (CD150+CD48-Lin−) from BM were sorted using FACSAria cytometer (BD).
Retroviral infection and transplantation.
Infection and transplantation procedures were performed as previously described (Thorsteinsdottir et al., 1999). Infection medium consisted of DMEM, 15% FCS, 100 ng/ml IL-11, 50 ng/ml Flt-3 ligand, 100 ng/ml SF, 10 µg/ml ciprofloxacin (Serologicals), and 6 µg/ml polybrene (hexadimethrine bromide; Sigma-Aldrich).
Methylcellulose cultures.
Progenitor assays were plated and scored as previously described (Kroon et al., 1998).
Competitive repopulation unit enumeration assays.
Stem cell quantification assays were performed essentially as previously described (Antonchuk et al., 2001).
Fluorescence studies.
The studies were performed with monoclonal antibodies specific for the following: Sca-1 (D7), Mac1 (M1/70), and Gr-1 (RB6-8C5; BD); kit (2B8), CD48 (HM48-1), and CD45R (RA3-6B2; eBioscience); and CD150 (TC15-12F12.2), B220 (30-F11), CD3 (17A2), CD11b (M1/70), and TER-119 (BioLegend). Staining with H2DCFDA was performed according to the manufacturer’s instructions (Invitrogen) using fresh, not purified leukemic cells immediately after BM harvest. The peak excitation wavelength for oxidized H2DCFDA was 488 nm, and emission was 525 nm. Flow cytometry and confocal analyses were performed using an LSRII cytometer (BD) and an LSM 510 laser-scanning confocal microscope (Carl Zeiss), respectively.
DNA and RNA preparation.
Genomic DNA was isolated using DNAzol and total cellular RNA with TRIzol, according to the manufacturer’s instructions (Invitrogen).
Southern blot analysis.
All blots were prepared and analyzed as previously described (Kroon et al., 1998).
Bisulfite conversion and sequencing.
Genomic DNA was converted using the methylSEQr Bisulfite Conversion kit (Applied Biosystems) according to the manufacturer’s instructions. The Gpx3 region was amplified by PCR and sequenced using an M13 tail (BigDye Terminator v3.1; Applied Biosystems). Purified sequencing products were analyzed on a 3730 Genetic Analyzer (Applied Biosystems).
Hybridization.
RNA samples were sent to the microarray facility at the Ottawa Hospital Research Institute (Ottawa, ON), where the RNA was reverse transcribed, labeled, and hybridized to Affymetrix 430A arrays according to manufacturer’s protocols.
Affymetrix gene expression profiling.
Affymetrix CEL files from biological triplicates for FLA2 and FLB1 were normalized using either RMA or GCRMA, using the LIMMA (Linear Modes for MicroArray data) package for R/Bioconductor. Normalized data were then used to compute a linear model to derive differentially expressed genes, or exported to perform SAM (significance analysis of microarrays) analysis. SAM analysis on both normalized datasets was performed using version 3.01 of the SAM software, with 100 permutations and a twofold minimum change in both cases. Using delta calibration values of 1.236 (GCRMA) and 0.567 (RMA), lists of 33 or 31 genes which showed differential expression (22 genes in common) were generated with false discovery rates of 2.71 and 3.31%, respectively. The ArrayExpress accession no. is E-MEXP-1597.
qRT-PCR.
qRT-PCR was performed using two strategies: the SYBR green dye and Universal ProbeLibrary assays designed with the ProbeFinder software (Roche) as described using Hprt, Actb, GAPDH, or HPRT as endogenous controls. Ct values were determined using the advanced relative quantification algorithm for each target (Cttarget) and endogenous control reference genes (Ctreference). The expression level of the target gene was expressed as ΔCt value (ΔCt = Cttarget − Ctreference). RQ (relative quantities) were calculated as RQ = (ΔCttest population/ΔCtcalibrating population). Calibrators used for calculation of various RQ values are indicated in the corresponding figure legends. To ensure that our qRT-PCR analyses accurately reflected expression of genes encoding antioxidant enzymes in the LTR-HSC compartment, only sorted HSC populations comprised of ∼50% of LTR-HSC as determined by competitive CRU were processed for cDNA synthesis and gene expression studies presented in Fig. 5 A.
Protein extraction and immunoblots.
Cells were lysed in 10 mmol/liter Hepes, pH 7.9, 10 mmol/liter KCl, 0.1 mmol/liter EDTA, 0.1 mmol/liter EGTA, and 10% (vol/vol) NP-40 containing protease inhibitors. Western blot analyses were done as previously described (Krosl and Sauvageau, 2000) using antibodies against GPx3 (R&D Systems) and MDA (Abcam).
Glutathione peroxidase assay.
The enzymatic activity of glutathione peroxidase in the spleen of mice was quantified using the Glutathione Peroxidase Assay kit (Cayman Chemical Company) according to the manufacturer’s recommendations.
Construction of shRNA retroviral vectors.
Two shRNAs were selected as single-stranded oligonucleotides also incorporating mir-30 flanking arms using the RNAi Central shRNA psm2 design tool (G. Hannon, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY). Amplicons were digested with XhoI–EcoRI (Invitrogen) and subcloned into the LMP vector according to the manufacturer’s guidelines (Invitrogen). Confirmatory sequencing was performed on all cloned shRNAs: shGpx3 #5, 5′-TGCTGTTGACAGTGAGCGCTTCTACACTTTCCTGAAGAACTAGTGAAGCCACAGATGTAGTTCTTCAGGAAAGTGTAGAATTGCCTACTGCCTCGGA-3′; shGpx3 #6, 5′-TGCTGTTGACAGTGAGCGCGCAGTATGCAGGCAAATATATTAGTGAAGCCACAGATGTAATATATTTGCCTGCATACTGCTTGCCTACTGCCTCGGA-3′; and shLuc, 5′-TGCTGTTGACAGTGAGCGCCGATATGGGCTGAATACAAATTAGTGAAGCCACAGATGTAATTTGTATTCAGCCCATATCGTTGCCTACTGCCTCGGA-3′.
Apoptosis assay.
Phosphatidylserine externalization was analyzed 4 d after retroviral infection by labeling GFP+ cells with annexin V–Alexa Fluor 350 and propidium iodide. Flow cytometry analyses were performed using an LSRII cytometer.
Homing assay.
After retroviral infection, Ly5.1+ cells were transplanted via tail vein injection into irradiated (1,600 cGy, 137 Cs gamma radiation) C57BL/6J (Ly5.2) mice. A small aliquot of cells was plated at this time in methylcellulose to assess the percentage of transduced CFCs in the inoculum (see Methylcellulose cultures). 4 d after transplant, recipient BM was harvested and the percentage of GFP+Ly5.1+ cells was assessed by flow cytometry with the acquisition of a minimum of 106 events (LSRII cytometer). BM was also plated in methylcellulose for later enumeration and scoring of GFP+ and GFP− CFCs (see Methylcellulose cultures).
Statistical analyses.
Statistical analyses were performed using Wilcoxon’s test. The level of significance was set at 0.05.
Online supplemental material.
Fig. S1 shows representative images of spleens from recipients of FLA2 and FLB1 cells at 19 d after transplantation. Table S1 shows FLA2 gene expression profiling. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20102386/DC1.
Acknowledgments
The authors are grateful to Dr. Trang Hoang and Jana Krosl for critical reading of the manuscript. Thanks to Raphaelle Lambert (Genomic Core Facility, Institute for Research in Immunology and Cancer [IRIC]) for assistance in qRT-PCR assays, Danièle Gagné (Flow Cytometry Core Facility, IRIC) for cell sorting, and Christine Vignon and Philippe Rosset (UPRES EA3855) and Yves Levern (INRA PAIB core facility, Nouzilly, France) for assistance in analysis of normal human marrow samples.
This work was supported by grants from the National Cancer Institute of Canada with funds from the Canadian Cancer Society, Stem Cell Network and the Fonds de la Recherche en Santé du Québec to G. Sauvageau and from the Cancer Research Network of the Fonds de la Recherche en Santé du Québec to J. Hébert. G. Sauvageau holds a Canada Research Chair in the Molecular Genetics of Stem Cells, and J. Hébert is a Research Chair in Leukemia supported by Industrielle-Alliance (Université de Montréal). M.A. Andrade-Navarro is a Canada Research Chair in Bioinformatics, and J. Hébert a Research Chair in Leukemia supported by Industrial Alliance. O. Herault was supported by the CANCEN association, the Ligue Nationale Contre le Cancer (Comité d’Indre-et-Loire, France), and the International Rotary Club of Blois.
The authors declare no competing financial interest.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- CSC
- cancer stem cell
- FL
- fetal liver
- H2DCFDA
- dichlorodihydrofluorescein diacetate
- HSC
- hematopoietic stem cell
- LSC
- leukemia stem cell
- LTR
- long-term repopulating
- MDA
- malondialdehyde
- MFI
- mean fluorescence intensity
- ROS
- reactive oxygen species
- shRNA
- short hairpin RNA
References
- Antonchuk J., Sauvageau G., Humphries R.K. 2001. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Exp. Hematol. 29:1125–1134 10.1016/S0301-472X(01)00681-6 [DOI] [PubMed] [Google Scholar]
- Deneault E., Cellot S., Faubert A., Laverdure J.P., Fréchette M., Chagraoui J., Mayotte N., Sauvageau M., Ting S.B., Sauvageau G. 2009. A functional screen to identify novel effectors of hematopoietic stem cell activity. Cell. 137:369–379 10.1016/j.cell.2009.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehn M., Cho R.W., Lobo N.A., Kalisky T., Dorie M.J., Kulp A.N., Qian D., Lam J.S., Ailles L.E., Wong M., et al. 2009. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 458:780–783 10.1038/nature07733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T., Holbrook N.J. 2000. Oxidants, oxidative stress and the biology of ageing. Nature. 408:239–247 10.1038/35041687 [DOI] [PubMed] [Google Scholar]
- Guzman M.L., Swiderski C.F., Howard D.S., Grimes B.A., Rossi R.M., Szilvassy S.J., Jordan C.T. 2002. Preferential induction of apoptosis for primary human leukemic stem cells. Proc. Natl. Acad. Sci. USA. 99:16220–16225 10.1073/pnas.252462599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Hirao A., Arai F., Matsuoka S., Takubo K., Hamaguchi I., Nomiyama K., Hosokawa K., Sakurada K., Nakagata N., et al. 2004. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 431:997–1002 10.1038/nature02989 [DOI] [PubMed] [Google Scholar]
- Ito K., Hirao A., Arai F., Takubo K., Matsuoka S., Miyamoto K., Ohmura M., Naka K., Hosokawa K., Ikeda Y., Suda T. 2006. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 12:446–451 10.1038/nm1388 [DOI] [PubMed] [Google Scholar]
- Jang Y.Y., Sharkis S.J. 2007. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 110:3056–3063 10.1182/blood-2007-05-087759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva M.Y., Jordan C.T. 2011. Leukemia stem cells and microenvironment: biology and therapeutic targeting. J. Clin. Oncol. 29:591–599 10.1200/JCO.2010.31.0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J., Dansen T.B., Polderman P.E., Saarloos I., Wirtz K.W., Coffer P.J., Huang T.T., Bos J.L., Medema R.H., Burgering B.M. 2002. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 419:316–321 10.1038/nature01036 [DOI] [PubMed] [Google Scholar]
- Kroon E., Krosl J., Thorsteinsdottir U., Baban S., Buchberg A.M., Sauvageau G. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17:3714–3725 10.1093/emboj/17.13.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krosl J., Sauvageau G. 2000. AP-1 complex is effector of Hox-induced cellular proliferation and transformation. Oncogene. 19:5134–5141 10.1038/sj.onc.1203897 [DOI] [PubMed] [Google Scholar]
- Lapidot T., Sirard C., Vormoor J., Murdoch B., Hoang T., Caceres-Cortes J., Minden M., Paterson B., Caligiuri M.A., Dick J.E. 1994. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 367:645–648 10.1038/367645a0 [DOI] [PubMed] [Google Scholar]
- Lessard J., Sauvageau G. 2003. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 423:255–260 10.1038/nature01572 [DOI] [PubMed] [Google Scholar]
- Mrózek K., Marcucci G., Paschka P., Whitman S.P., Bloomfield C.D. 2007. Clinical relevance of mutations and gene-expression changes in adult acute myeloid leukemia with normal cytogenetics: are we ready for a prognostically prioritized molecular classification? Blood. 109:431–448 10.1182/blood-2006-06-001149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H., Sekulovic S., Bakovic S., Eaves C.J., Pineault N., Gasparetto M., Smith C., Sauvageau G., Humphries R.K. 2007. Near-maximal expansions of hematopoietic stem cells in culture using NUP98-HOX fusions. Exp. Hematol. 35:817–830 10.1016/j.exphem.2007.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Nakauchi H., Nagayoshi K., Nishikawa S., Miura Y., Suda T. 1992. In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood. 80:3044–3050 [PubMed] [Google Scholar]
- Owusu-Ansah E., Banerjee U. 2009. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 461:537–541 10.1038/nature08313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Yavari A., Mandal S., Banerjee U. 2008. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat. Genet. 40:356–361 10.1038/ng.2007.50 [DOI] [PubMed] [Google Scholar]
- Rebel V.I., Miller C.L., Eaves C.J., Lansdorp P.M. 1996. The repopulation potential of fetal liver hematopoietic stem cells in mice exceeds that of their liver adult bone marrow counterparts. Blood. 87:3500–3507 [PubMed] [Google Scholar]
- Sattler M., Winkler T., Verma S., Byrne C.H., Shrikhande G., Salgia R., Griffin J.D. 1999. Hematopoietic growth factors signal through the formation of reactive oxygen species. Blood. 93:2928–2935 [PubMed] [Google Scholar]
- Sauvageau G., Thorsteinsdottir U., Eaves C.J., Lawrence H.J., Largman C., Lansdorp P.M., Humphries R.K. 1995. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 9:1753–1765 10.1101/gad.9.14.1753 [DOI] [PubMed] [Google Scholar]
- Shao L., Li H., Pazhanisamy S.K., Meng A., Wang Y., Zhou D. 2011. Reactive oxygen species and hematopoietic stem cell senescence. Int. J. Hematol. 94:24–32 10.1007/s12185-011-0872-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan M., Yu Z.X., Ferrans V.J., Irani K., Finkel T. 1995. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 270:296–299 10.1126/science.270.5234.296 [DOI] [PubMed] [Google Scholar]
- Takeda S., Gapper C., Kaya H., Bell E., Kuchitsu K., Dolan L. 2008. Local positive feedback regulation determines cell shape in root hair cells. Science. 319:1241–1244 10.1126/science.1152505 [DOI] [PubMed] [Google Scholar]
- Thorsteinsdottir U., Sauvageau G., Humphries R.K. 1999. Enhanced in vivo regenerative potential of HOXB4-transduced hematopoietic stem cells with regulation of their pool size. Blood. 94:2605–2612 [PubMed] [Google Scholar]
- Tothova Z., Kollipara R., Huntly B.J., Lee B.H., Castrillon D.H., Cullen D.E., McDowell E.P., Lazo-Kallanian S., Williams I.R., Sears C., et al. 2007. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 128:325–339 10.1016/j.cell.2007.01.003 [DOI] [PubMed] [Google Scholar]
- van Rhenen A., Feller N., Kelder A., Westra A.H., Rombouts E., Zweegman S., van der Pol M.A., Waisfisz Q., Ossenkoppele G.J., Schuurhuis G.J. 2005. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin. Cancer Res. 11:6520–6527 10.1158/1078-0432.CCR-05-0468 [DOI] [PubMed] [Google Scholar]
- Wang Y., Krivtsov A.V., Sinha A.U., North T.E., Goessling W., Feng Z., Zon L.I., Armstrong S.A. 2010. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 327:1650–1653 10.1126/science.1186624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm B.T., Briau M., Austin P., Faubert A., Boucher G., Chagnon P., Hope K., Girard S., Mayotte N., Landry J.R., et al. 2011. RNA-seq analysis of 2 closely related leukemia clones that differ in their self-renewal capacity. Blood. 117:e27–e38 10.1182/blood-2010-07-293332 [DOI] [PubMed] [Google Scholar]