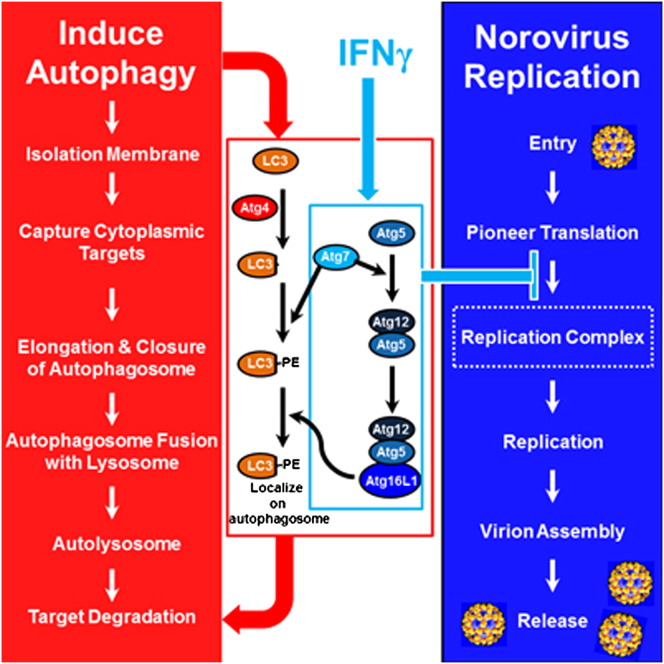
Abstract
Host resistance to viral infection requires type I (α/β) and II (γ) interferon (IFN) production. Another important defense mechanism is the degradative activity of macroautophagy (herein autophagy), mediated by the coordinated action of evolutionarily conserved autophagy proteins (Atg). We show that the Atg5-Atg12/Atg16L1 protein complex, whose prior known function is in autophagosome formation, is required for IFNγ-mediated host defense against murine norovirus (MNV) infection. Importantly, the direct antiviral activity of IFNγ against MNV in macrophages required Atg5-Atg12, Atg7, and Atg16L1, but not induction of autophagy, the degradative activity of lysosomal proteases, fusion of autophagosomes and lysosomes, or the Atg8-processing protein Atg4B. IFNγ, via Atg5-Atg12/Atg16L1, inhibited formation of the membranous cytoplasmic MNV replication complex, where Atg16L1 localized. Thus, the Atg5-Atg12/Atg16L1 complex performs a pivotal, nondegradative role in IFNγ-mediated antiviral defense, establishing that multicellular organisms have evolved to use portions of the autophagy pathway machinery in a cassette-like fashion for host defense.
Graphical Abstract
Highlights
► Atg5-Atg12/Atg16L1 and Atg7 are required for IFNγ to inhibit norovirus replication ► The degradative activity of autophagy is not required for the antiviral activity of IFNγ ► IFNγ, via autophagy proteins, inhibits the formation of the replication complex
Introduction
Previous work using mice lacking the interferon (IFN) α/β receptor, the IFNγ receptor, or both receptors has shown that when compensatory IFNα/β responses are absent, IFNγ is essential for host defense against acute infection with both RNA and DNA viruses (Gil et al., 2001, Karst et al., 2003). Furthermore, IFNγ has direct antiviral activity against many viruses, including single-stranded positive-sense RNA viruses (Kimura et al., 1994, Shrestha et al., 2006). The antiviral role of IFNγ likely provides a host counterpoint to the capacity of many viruses to inhibit the antiviral activities of IFNα/β using highly evolved immune evasion strategies. Autophagy and autophagy proteins play important roles in host defense against infection, development, cellular energy homeostasis, and multiple diseases, including cancer and inflammatory bowel disease (Levine et al., 2011). Since autophagy proteins expressed by macrophages, which are key innate immune cells, have a role in IFNγ-mediated resistance to both mycobacteria and the apicomplexan parasite Toxoplasma gondii (Levine et al., 2011, Zhao et al., 2008), we hypothesized that autophagy proteins also participate in the antiviral activities of IFNγ.
The degradative function of the overall autophagy pathway involves delivery of cytoplasmic cargo contained within double membrane-bound autophagosomes to the lysosome. This process requires the ordered activity of protein complexes that induce autophagosome formation, envelopment of specific cargoes or bulk cytoplasm, elongation and closure of autophagosome membranes, fusion of the outer autophagosomal membrane to the lysosome, and degradation of cargo within the autophagosome by lysosomal enzymes active at low pH (Levine et al., 2011). One protein complex required for autophagy contains Atg16L1 bound to a covalent Atg5-Atg12 conjugate that is generated by the action of Atg7. The known activity of this complex is to promote elongation and closure of the autophagosome via an E3-ligase-like role in the generation of lipidated forms of LC3 (microtubule-associated protein 1 light chain 3, Atg8) family proteins and their localization to the autophagosome membrane (Weidberg et al., 2010, Fujita et al., 2008b). LC3 proteins are processed in preparation for lipidation by Atg4 proteins including, prominently, Atg4B (Mariño et al., 2010).
Noroviruses cause the majority of human nonbacterial epidemic gastroenteritis and are a major cause of food-borne illness and human morbidity (Glass et al., 2009). Relatively little is known about the mechanisms of host resistance for this important class of pathogens. While human noroviruses have not been cultured efficiently and do not infect small animals, murine norovirus (MNV) can be cultured in macrophages and thus provides a model of infection for this important genus of human pathogens (Glass et al., 2009, Wobus et al., 2004, Karst et al., 2003). The replication of MNV in macrophages is, as for other single-stranded positive-sense RNA viruses, associated with extensive membrane rearrangements that generate membranous replication complexes within which viral RNAs and proteins are produced and assembled into infectious virions (Hyde et al., 2009, Wobus et al., 2004). Using the MNV model, we investigated the role of the autophagy pathway and protein components of the autophagy machinery in host antiviral defense. We show here that IFNγ-activated macrophages use portions of the autophagy machinery, in a cassette-like fashion, to block norovirus infection by inhibiting formation of the replication complex.
Results
Atg5 Protects against Lethal MNV Infection in the Absence of IFNα/β Signaling
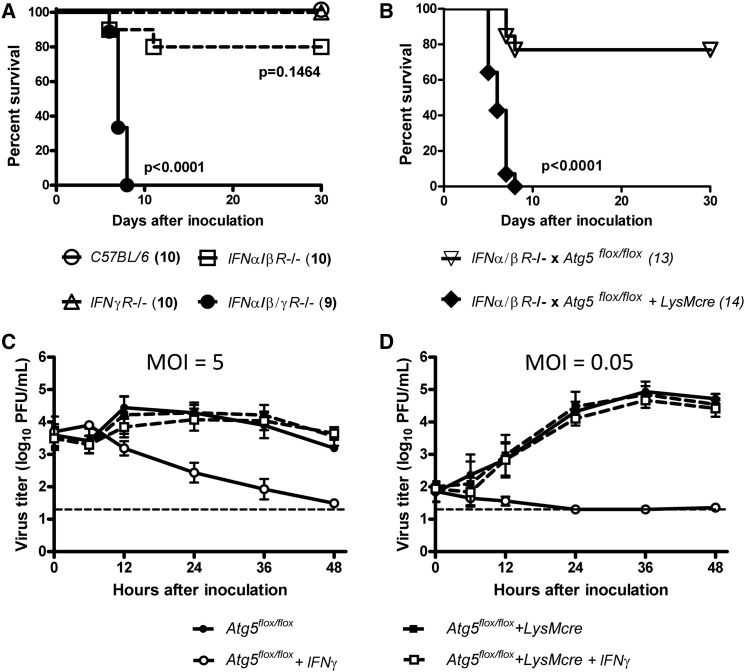
To test the hypothesis that autophagy proteins are involved in IFNγ-mediated antiviral defense, we analyzed the role of Atg5 in control of disease induced by the enteric pathogen MNV. We first confirmed that mice lacking both IFNα/β and IFNγ receptors (IFNα/β/γR−/−) succumb to lethal infection with 3 × 104 plaque forming units (pfu) of MNV (Figure 1A) (Karst et al., 2003, Mumphrey et al., 2007). In contrast, mice lacking either the IFNα/βR or the IFNγR alone are resistant to lethal MNV infection (Figure 1A). These data indicate that IFNα/β and IFNγ share overlapping but nonredundant roles in defense against MNV. As for many viruses, the antiviral role of IFNγ, and therefore proteins required for IFNγ control of MNV infection in vivo, may then be detected when compensatory IFNα/β responses are absent. Atg5 is essential for autophagosome formation and Atg5-deficient mice die soon after birth (Levine et al., 2011). To test the role of Atg5 in IFNγ-dependent resistance to MNV infection, we generated IFNα/βR−/− mice lacking Atg5 in macrophages and neutrophils that express lysozyme M. We crossed Atg5flox/flox mice expressing lysozyme-M promoter driven cre-recombinase (Atg5flox/flox+LysMcre) (Zhao et al., 2008, DeSelm et al., 2011) to IFNα/βR−/− mice, generating IFNα/βR−/− x Atg5flox/flox+LysMcre mice. We then compared MNV infection of IFNα/βR−/− x Atg5flox/flox+LysMcre mice to infection of control IFNα/βR−/− x Atg5flox/flox mice. Mice deficient in Atg5 in macrophages and neutrophils succumbed to lethal MNV infection while control mice did not (Figure 1B). While MNV replication is confined to certain tissues (mesenteric lymph node, intestine, spleen, and liver) in immune-competent mice, MNV replication is widely disseminated in immune-compromised mice (Karst et al., 2003, Mumphrey et al., 2007). Consistent with this, MNV replication was significantly increased in all tissues examined from IFNα/βR−/− x Atg5flox/flox+LysMcre mice compared to tissues of control IFNα/βR−/− x Atg5flox/flox mice (Figures S1A–S1F available online). These data demonstrate that Atg5 expression in macrophages and/or neutrophils was essential for control of MNV infection in mice with compromised IFNα/β signaling.
Figure 1.
Atg5 Was Required for IFNγ-Mediated Suppression of MNV Both In Vivo and In Vitro
(A and B) Survival curves after per oral inoculation with 3 × 104 pfu of MNV. The number of mice used is indicated in parentheses. Experimental groups were compared to C57BL/6 controls (A). Atg5-deficient mice were compared to control mice (B).
(C and D) Growth analysis of MNV in control (circle; Atg5flox/flox) and Atg5-deficient (square; Atg5flox/flox+LysMcre) macrophages after pretreatment with none (filled) or 100 U/ml IFNγ (open) for 12 hr before infection at MOI = 5 (C) and 0.05 (D). Mean virus titers ± SEM from three independent experiments (n = 3) are shown.
See also Figure S1.
The Control of MNV by IFNγ in Macrophages Is Atg5 Dependent
To explain this profound in vivo phenotype, we hypothesized that Atg5 is required for the direct antiviral effects of IFNγ against MNV in macrophages, a cell type in which MNV replicates (Wobus et al., 2004). Bone marrow-derived macrophages were prepared from Atg5flox/flox+LysMcre (henceforth Atg5-deficient) and Atg5flox/flox (henceforth control) mice and inoculated with MNV at high (5) or low (0.05) multiplicity of infection (MOI). No significant differences in MNV replication were observed between control and Atg5-deficient macrophages (Figures 1C and 1D). However, pretreatment with IFNγ inhibited MNV replication by >100–1000 fold in control macrophages but not in Atg5-deficient macrophages (p < 0.05 [Figure 1C] and p < 0.01 [Figure 1D]). In contrast, pretreatment with IFNβ suppressed MNV replication equally in control and Atg5-deficient macrophages (Figure S1G). Thus, Atg5 was not required for the replication of MNV, but was critical for the IFNγ-mediated control of MNV replication in macrophages.
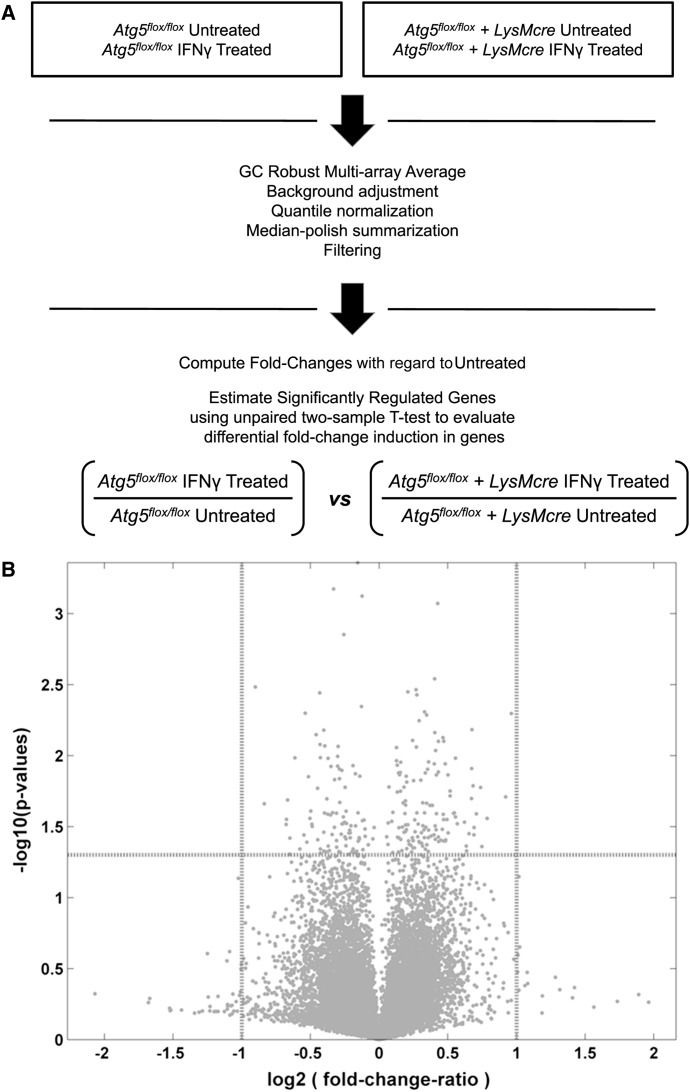
Atg5 Is Not Required for IFNγ-Mediated Cellular Transcriptional Changes
The antiviral effects of IFNγ depend on the expression of IFN-stimulated genes via the Janus-activated kinase (JAK)-signal transducer and activator of transcription (STAT) signaling pathway. It has been reported that autophagy protein-deficient but transformed murine fibroblast cells exhibit abnormal IFNγ-induced JAK-STAT activation (Chang et al., 2010). Therefore, we used microarrays to measure transcript levels in control and Atg5-deficient primary macrophages with or without IFNγ treatment (Figure 2 and Table S1). While IFNγ induced major changes in cellular transcription as expected, no statistically significant differences were observed in the transcriptional profiles induced by IFNγ treatment of control and Atg5-deficient macrophages (Figure 2B). When evaluating the fold change of induced genes between IFNγ treated control and Atg5-deficient macrophages, there were some genes that exceeded either the biological relevance threshold [shown on the x axis as log2 (fold-change-ratio) >1 or <−1; indicated by vertical dashed lines] or the statistical significance threshold [shown on the y axis as significance >–log10 (0.05); indicated by horizontal dashed line]. However, none of these genes exceeded both thresholds (Figure 2B). These data demonstrate that IFNγ-mediated transcriptional modulation in macrophages is not Atg5 dependent. We also considered the possibility that Atg5 deficiency resulted in the general disruption of cell homeostasis or IFNγ-mediated activation of macrophages. However, Atg5-deficient macrophages had normal levels of ATP (Figure S2A), exhibited normal phosphorylation of STAT1 and induction of IRF1 protein (Figure S2B), and were able to inhibit the replication of encephalomyocarditis virus, murine hepatitis virus, and West Nile virus in response to treatment with IFNγ (Figures S2D–S2F). While canonical autophagy can play an important role in cell homeostasis upon various cellular stress or stimulation (Stephenson et al., 2009, Saitoh et al., 2008, Tal et al., 2009, Chang et al., 2010, Levine et al., 2011), these data suggest, consistent with studies in osteoclasts (DeSelm et al., 2011), that there is no general disruption of cellular homeostasis in Atg5-deficient macrophage lineage cells. Importantly, the capacity of Atg5-deficient cells to control the replication of other viruses in response to IFNγ strongly argues against the possibility that the lack of IFNγ effects on MNV replication in these cells is due to a combination of minor disruptions in cell homeostasis. Together, these data indicate that IFNγ-mediated inhibition of MNV replication occurs via a non-transcriptional function of Atg5, and there is no general disruption of cellular responses to IFNγ in the absence of Atg5.
Figure 2.
Transcriptional Analysis of IFNγ-Treated Control and Atg5-Deficient Macrophages
(A) Schematic diagram of transcript analysis in control and Atg5-deficient macrophages with and without IFNγ treatment using microarrays.
(B) Transcriptional profiles induced by IFNγ treatment of control and Atg5-deficient macrophages. x axis: the ratio (in log2 scale) of fold changes induced in IFNγ-treated Atg5-deficient cells to fold changes induced in IFNγ-treated control macrophages. y axis: the significance (p values in –log10 scale) of the difference in means of fold change induction.
The Catabolic Activity of the Autophagy Pathway Is Not Required for Control of MNV by IFNγ
Since Atg5 is essential for autophagy, we examined whether IFNγ inhibits the replication of MNV in macrophages through the modulation of the canonical autophagy pathway. To monitor the flux of the autophagy pathway in response to IFNγ, we analyzed the conversion (lipidation) of LC3-I (unconjugated) to LC3-II (conjugated to phosphatidylethanolamine) in the presence and absence of chloroquine. Chloroquine blocks the degradation of LC3-II that occurs after fusion of lysosomes and autophagosomes (Mizushima et al., 2010). Treatment of macrophages with chloroquine led to the accumulation of LC3-II (Figure S3A). As a control we showed that the autophagy-inducing drug rapamycin increased the conversion of LC3-I to LC3-II, and accumulation of LC3-II in combination with chloroquine, in treated cells compared to untreated cells (Mizushima et al., 2010). In contrast, neither high (100 U/ml) nor low (1 U/ml) doses of IFNγ affected the conversion of LC3-I to LC3-II, suggesting that effects of IFNγ on MNV replication are not associated with significant changes in bulk autophagy. It has been shown that IFNγ induces autophagy in cell lines and that autophagy facilitates IFNγ-induced Jak2-STAT1 signaling in specific transformed cell lines by negatively regulating reactive oxygen species (ROS) (Levine et al., 2011, Delgado et al., 2008, Chang et al., 2010). However, in primary mouse macrophages, we did not detect a significant difference in IFNγ signaling in the absence of Atg5 (Figures 2 and S2B) and IFNγ did not significantly induce autophagy (Figure S3A). Furthermore, downregulation of ROS using antioxidant treatment (Tal et al., 2009) did not effect IFNγ-mediated control of MNV replication (Figure S2C). This discrepancy between our findings and published data may be due to differences in experimental design or to differences between primary cells and transformed cell lines.
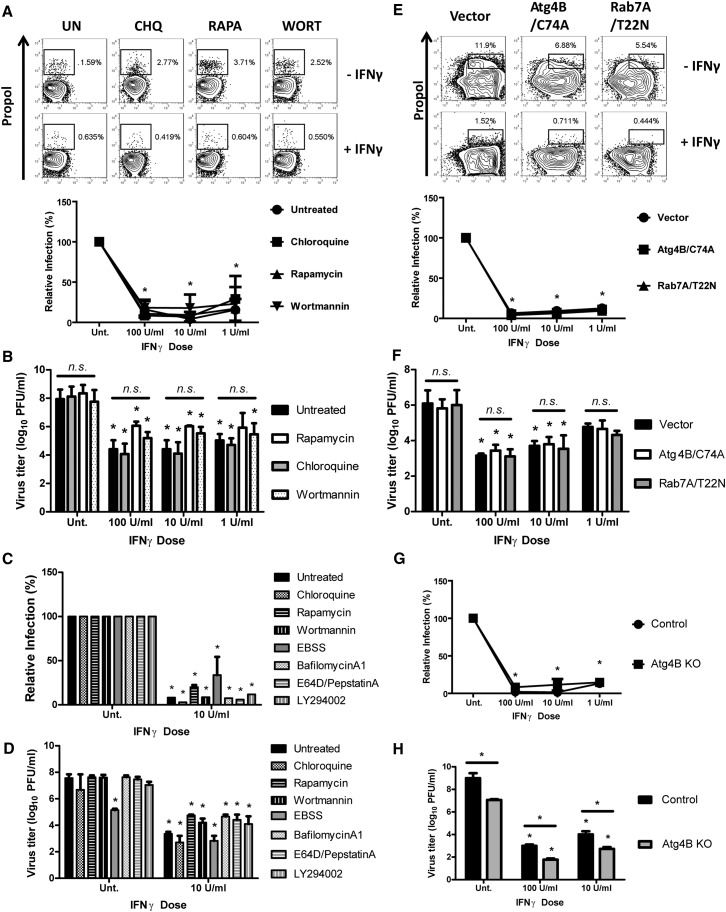
Next, we directly examined whether inducing or inhibiting autophagy would alter MNV replication or its control by IFNγ. To analyze the replication of MNV in macrophages within a single replication cycle, we developed a flow cytometry-based assay in which cells were inoculated with MNV (MOI = 5) and stained with an antibody to the MNV polymerase (Figure S3B). Treatment of macrophages with rapamycin, chloroquine (blocks lysosomal degradation), or wortmannin (inhibits the initiation of autophagy in nutrient-rich conditions (Wu et al., 2010) either before or after viral infection had the expected effects on autophagy (Figure S3G) but did not significantly affect MNV replication or its control by IFNγ (Figures 3A and 3B). To examine the role of later steps in canonical autophagy in antiviral activity mediated by IFNγ, we used lentiviral transduction to overexpress a dominant negative form of ras-related protein 7A (Rab7A/T22N), which blocks fusion of autophagosomes and lysosomes, as well as bacterial autophagy (Yamaguchi et al., 2009). As expected, Rab7/T22N expression inhibited degradative autophagy leading to the accumulation of p62, which is a well-known substrate degraded through autophagy (Figure S3C). However, this inhibitor of degradative autophagy did not prevent IFNγ-mediated control of MNV replication (Figures 3E and 3F). Similarly, inhibitors of lysosomal proteases or lysosomal acidification (E64D/PepstatinA and BafilomycinA1) blocked the degradative activity of the autophagy pathway (Figure S3G) but did not block the antiviral effects of IFNγ against MNV (Figures 3C and 3D). Further, induction or inhibition of autophagy initiation (starvation, wortmannin, LY294002) also did not significantly affect the IFNγ-mediated control of MNV replication (Figures 3C and 3D). Lastly, protein kinase R (PKR) is involved in autophagic control of herpes simplex virus replication (Levine et al., 2011), but PKR was not required for IFNγ-mediated blockade of MNV replication (Figure S3D). Taken together, these data demonstrated that, while Atg5 is required for IFNγ-mediated inhibition of MNV replication, the degradative autophagy pathways, as well as other pathways known to be involved in the antiviral effects of autophagy, were not involved in the control of MNV replication by IFNγ.
Figure 3.
Catabolic Autophagy Was Not Required for the IFNγ-Mediated Suppression of MNV Replication
(A) Flow cytometry analysis of MNV replication (n = 3) at 12 hpi (MOI = 5) upon treatment with autophagy modulatory drugs. Chloroquine (CHQ, 20 μM), rapamycin (Rapa, 10 μM), wortmannin (WORT, 50 nM), or none (UN). Top: Representative flow data of MNV replication after no treatment (–IFNγ) or 100 U/ml of IFNγ pretreatment (+IFNγ) for 12 hr. Bottom: Quantitation of relative MNV replication after normalization to the level in untreated BMDMs. ∗ indicates statistically significant difference (p < 0.05) compared to the untreated (100%). This convention is carried through all figures. Mean virus titers ± SEM from three independent experiments (n = 3) are shown.
(B) Growth analysis of MNV (n = 3) in drug-treated BMDMs at 24 hpi (MOI = 0.05). n.s. indicates no statistically significant difference among samples (p > 0.05).
(C) Same analysis (n = 2) shown in (A) with more autophagy-modulating drugs: EBSS (Earle's balanced salt solution), BafilomycinA1 (100 nM), E64D/PepstatinA (10 μg/ml), LY294002 (10 μM).
(D) Same analysis (n = 2) shown in (B) with the drugs shown.
(E) Flow cytometry analysis of MNV replication (n = 3) upon the expression of dominant negative forms of autophagy proteins.
(F) Growth analysis of MNV (n = 3) in the transduced BMDMs.
(G) Flow cytometry analysis of MNV replication (n = 2) in control and Atg4BKO BMDMs.
(H) Growth analysis of MNV (n = 2) in control and Atg4BKO BMDMs.
In all graphs, data represent mean ± SEM. See also Figure S3.
Atg12 Conjugation to Atg5 Is Required for IFNγ-Mediated Control of MNV
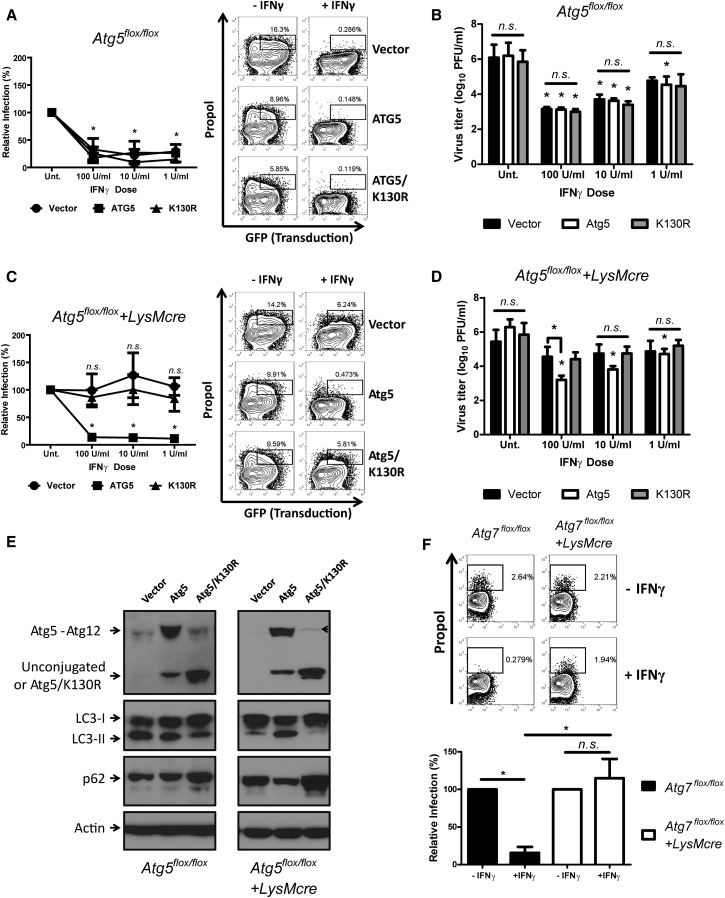
To define the molecular basis for the required role for Atg5 in IFNγ-mediated inhibition of MNV replication, we reconstituted Atg5-deficient macrophages by expressing wild-type and mutant forms of Atg5 (Figure 4E). Atg5 functions in autophagy as a covalent conjugate with Atg12. To examine the role of Atg5-Atg12 conjugation, we expressed Atg5/K130R, which cannot be conjugated to Atg12 (Levine et al., 2011). Expression of Atg5 in Atg5-deficient macrophages significantly restored expression of the Atg5-Atg12 conjugate, the conversion of LC3-I to LC3-II, and reduction of p62 level (Figure 4E). In contrast, expression of Atg5/K130R generated an unconjugated form of Atg5, did not restore the conversion of LC3-I to LC3-II and increased, rather than decreased, the level of p62. IFNγ suppressed the replication of MNV in all lentivirus-transduced control macrophages (Figures 4A and 4B). Importantly, while Atg5 reconstituted IFNγ control of MNV replication in Atg5-deficient macrophages, Atg5/K130R did not (Figures 4C and 4D), indicating a role for the Atg5-Atg12 conjugate in IFNγ-mediated inhibition of MNV replication.
Figure 4.
Specific Requirement of Atg5 and Atg12 Conjugation in the IFNγ-Mediated Suppression of MNV Replication
(A and C) Flow cytometry analysis of MNV replication and its control by IFNγ (n = 5) in control (A) and Atg5-deficient (C) macrophages at 12 hpi (MOI = 5) upon the expression of Atg5 or Atg5/K130R. Right: A representative flow data. Left: Quantitation of relative MNV replication.
(B&D) Growth analysis of MNV (n = 5) in transduced control (B) and Atg5-deficient (D) macrophages at 24 hpi (MOI = 0.05).
(E) A representative western blot analysis of autophagy status in macrophages (n = 3) after lentiviral transduction with control, Atg5, or Atg5/K130R. The arrowhead indicates the uncleaved form of Atg5/K130R-T2A-copGFP.
(F) Flow cytometry analysis (n = 3) of MNV replication and its control by IFNγ (10 U/ml) in control (Atg7flox/flox) and Atg7-deficient (Atg7flox/flox+LysMcre) macrophages at 12 hpi (MOI = 5). Top: Representative flow data. Bottom: Quantitation of relative MNV replication.
In all graphs, data represent mean ± SEM. See also Figure S4.
Atg16L1 Is Required for IFNγ-Mediated Suppression of MNV
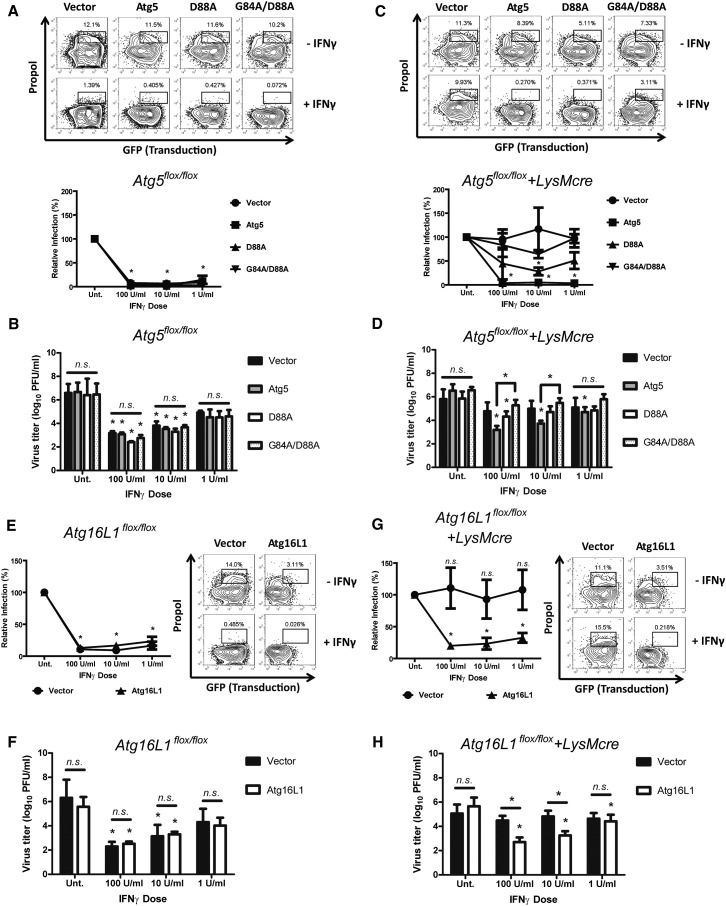
The Atg5-Atg12 conjugate plays its role in autophagy as part of a complex that includes Atg16L1 (Levine et al., 2011). We therefore determined whether the binding of Atg5-Atg12 to Atg16L1 was required for IFNγ-mediated control of MNV using two approaches. First, based on the structure of the yeast Atg5 and Atg16L1 complex (Matsushita et al., 2007), we generated Atg16L1 binding-defective mutants of Atg5. Several single amino acid substitutions in Atg5 (R41A, G84A, D88A, G242A) reduced the binding of Atg5 to Atg16L1, but none of them completely blocked the interaction (Figure S4B). Therefore, we made an Atg5/G84A/D88A mutant combining two single amino acid substitutions, and we observed that this mutant exhibited a substantial deficit in Atg16L1 binding (Figure S4B). We then tested the effects of two Atg5 mutants, with differing ability to bind Atg16L1, in control and Atg5-deficient macrophages. In control macrophages, neither mutant significantly affected the replication of MNV or its control by IFNγ (Figures 5A and 5B). However, in Atg5-deficient macrophages Atg5/D88A partially restored the control of MNV replication by IFNγ but Atg5/G84A/D88A did not (Figures 5C and 5D). Although we cannot rule out the possibility that this partial restoration by Atg5/G84A but not by Atg5/G84A/D88A might be caused by differences in expression (Figure S4C), given their differing capacity to bind Atg16L1 (Figure S4B), these data suggest that Atg5 binding to Atg16L1 was also required for IFNγ to control MNV replication.
Figure 5.
Atg16L1 Was Required for the IFNγ-Mediated Suppression of MNV Replication
(A and C) Flow cytometry analysis (n = 3) of the effect of Atg16L1 binding-defective Atg5 mutants (D88A and G84A/D88A) on MNV replication and its control by IFNγ in transduced control (A) and Atg5-deficient (C) macrophages at 12 hpi (MOI = 5). Top: Representative data. Bottom: Quantitation of relative MNV replication.
(B and D) Growth analysis of MNV (n = 3) in the transduced control (B) and Atg5-deficient (D) macrophages at 24 hpi (MOI = 0.05).
(E and G) Flow cytometry analysis (n = 3) of the replication of MNV and its control by IFNγ in control (E; Atg16flox/flox) and Atg16L1-deficient (G; Atg16flox/flox+LysMcre) macrophages at 12 hpi of MOI = 5. Right: Representative data. Left: Quantitation of relative MNV replication.
(F and H) Growth analysis of MNV (n = 3) in transduced control (F) and Atg16L1-deficient (H) macrophages at 24 hpi (MOI = 0.05).
In all graphs, data represent mean ± SEM. See also Figure S4.
To directly investigate the necessity of Atg16L1 for the antiviral function of IFNγ, we generated a conditional knockout (KO) allele of mouse Atg16L1 and deleted Atg16L1 specifically in macrophages using LysMcre (Atg16flox/flox+LysMcre). Autophagy was defective in Atg16L1-deficient macrophages (e.g., accumulation of p62), but expression of wild-type Atg16L1 in the Atg16L1-deficient macrophages restored autophagy (Figure S4D). Similar to Atg5-deficient macrophages, MNV replication was controlled by IFNγ in control macrophages (Figures 5E and 5F) but not in Atg16L1-deficient macrophages (Figures 5G and 5H). However, upon reintroduction of Atg16L1 in Atg16L1-deficient macrophages, MNV replication was controlled by IFNγ treatment. These data demonstrate that Atg16L1 was required for IFNγ to control MNV replication. Taken together, the data shown in Figures 4 and 5 indicate that the Atg5-Atg12/Atg16L1 protein complex is required for IFNγ to control MNV replication.
Atg7 Is Required for IFNγ-Mediated Suppression of MNV through Generation of the Atg5-Atg12 Conjugate
Since Atg7 is the enzyme responsible for Atg5-Atg12 conjugation (Levine et al., 2011), we determined the requirement for Atg7 in IFNγ-mediated inhibition of MNV replication using Atg7-deficient macrophages derived by crossing Atg7flox/flox mice to LysMcre mice (DeSelm et al., 2011) (Figure S4A). There was no significant difference in MNV replication between control and Atg7-deficient macrophages (Figure 4F). However, IFNγ-mediated inhibition of MNV replication required Atg7 (Figure 4F). In addition to generating the Atg5-Atg12 conjugate, Atg7 participates in canonical autophagy through its required role in the lipidation of LC3 family members. To determine the role of this function in control of MNV replication, we studied macrophages lacking LC3β (Cann et al., 2008), but we detected no effect of LC3β deficiency on either growth of MNV or the effect of IFNγ on MNV replication (Figure S3E). The lack of a role for LC3β might be explained by the extensive redundancy of LC3 family members (Reggiori et al., 2010, Weidberg et al., 2010). Therefore, we examined the role of Atg4B, which is the predominant Atg4 family member involved in canonical autophagy via its role in preparing LC3 proteins for conjugation to phosphatidylethanolamine through proteolytic processing. Accordingly, we overexpressed the dominant negative form of Atg4B (AtgB/C74A) (Fujita et al., 2008a), which blocks the proper processing and lipidation of LC3 proteins. As expected, this protein inhibited autophagy (Figure S3C). However, expression of this dominant negative form of Atg4B had no significant effect on MNV replication or its control by IFNγ (Figures 3E and 3F). Furthermore, Atg4B deficient macrophages isolated from mutant mice (Mariño et al., 2010) exhibited a striking deficiency in autophagy (Figure S3F), but exhibited no significant change in MNV replication or the control of MNV replication by IFNγ (Figures 3G and 3H). These data indicate that a highly selective portion of the autophagy machinery, including only one of the two known activities of Atg7, is required for the antiviral effects of IFNγ against MNV replication.
Atg16L1 Is Specifically Localized in the MNV Replication Complex
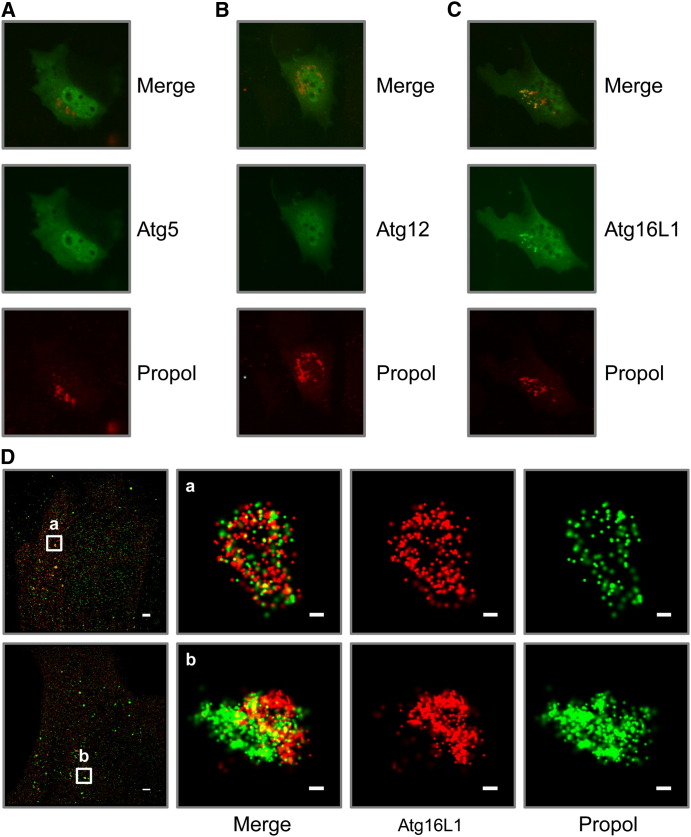
To further understand the role of Atg5-Atg12/Atg16L1 and Atg7 in IFNγ-mediated inhibition of MNV replication, we investigated the localization of epitope tagged Atg5, Atg12, and Atg16L1 in murine embryonic fibroblast (MEF) cells transfected with a plasmid that expresses the nonstructural proteins of MNV. We utilized this system because localizing endogenous autophagy proteins in infected macrophages has been difficult using the antibodies available. We confirmed that IFNγ inhibits the replication of MNV in MEFs in an Atg5-dependent manner (Figure S5A). In wild-type MEFs, Atg5 and Atg12 were localized throughout the cell (Figures 6A and 6B), while Atg16L1 had a punctate localization, which colocalized with the MNV polymerase (Figure 6C). Interestingly, this colocalization of Atg16L1 with the MNV polymerase was also detected in Atg5 KO and Atg7 KO MEFs (Figure S5E). Although it is not clear whether Atg5 and Atg12 specifically colocalize with the MNV polymerase due to their diffuse cytoplasmic localization, the punctate localization of Atg16L1 with the MNV polymerase strongly suggests that Atg16L1 localizes on the MNV replication complex. To examine this colocalization at high resolution, we used stochastic optical reconstruction microscopy (STORM), a nanometer-resolution fluorescence microscopy method that is based on high-accuracy localization of photoswitchable fluorophores (Rust et al., 2006). Using STORM, we confirmed that Atg16L1 colocalized and was tightly coassociated with the MNV polymerase (Figure 6D). Interestingly, we observed a spectrum of colocalization between Atg16L1 and the MNV polymerase with the two proteins in some cases closely adjacent but separated while in other cases they were comingled (Figure 6D and Movie S1). These localization data place a protein, Atg16L1, which is required for the blockade of MNV replication by IFNγ, at the MNV replication complex, a membranous structure required for viral replication.
Figure 6.
The Localization of Atg5, Atg12, and Atg16L1 in Relation to MNV Replication Complex
(A–C) Conventional immunofluorescence microscopy image of FLAG-tagged Atg5 (A), Atg12 (B), and Atg16L1 (C) in relation to MNV replication complex (as propol+) in cotransfected MEFs.
(D) High-resolution STORM images of FLAG-Atg16L1 and MNV replication complex in cotransfected MEFs. The scale bars inside images represent 100 nm, except the leftmost images (2 μm).
Atg5 Is Required for IFNγ to Prevent MNV from Forming Replication Complex
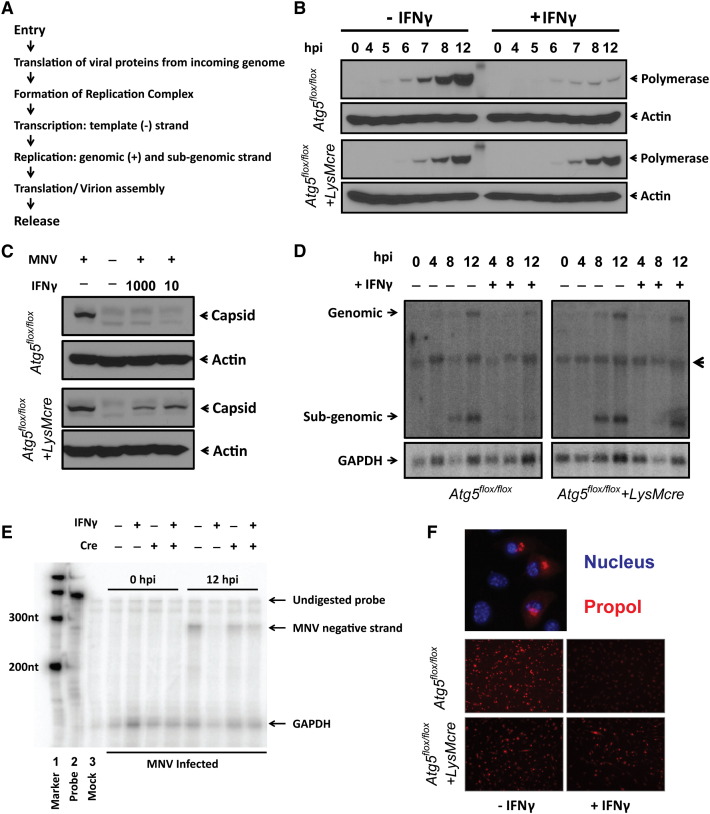
To understand this mechanism of Atg5-Atg12/Atg16L1-dependent inhibition of MNV replication, we first defined the stage of the MNV life cycle at which IFNγ exerts Atg5-dependent antiviral effects, focusing on steps in the life cycle upstream and downstream of the formation of the replication complex (flow chart in Figure 7A). As observed in a previous study, expression of the nonstructural MNV polymerase in untreated control macrophages was detected by western blot by 5 to 6 hours postinfection (hpi) (Changotra et al., 2009). Treatment with IFNγ did not alter this initial burst of viral protein synthesis in either control or Atg5-deficient cells (Figure 7B). Of note, early synthesis of viral proteins was detected even after pretreatment with 1000 U/ml of IFNγ treatment (Figure S5B) or in the presence of 200 U/ml of IFNγ throughout the course of infection (data not shown), demonstrating that detection of normal levels of polymerase expression at early time points was not due to insufficient amounts of IFNγ. These data further indicated that the initial steps in MNV infection are not altered by IFNγ although it is blocked by IFNβ (Changotra et al., 2009). It has been reported that autophagy increases the degradation of herpesvirus proteins in interferon treated cells (Levine et al., 2011). Thus, we examined the stability of the MNV polymerase in control cells treated with IFNγ. We allowed infection to proceed to a point at which polymerase was expressed and then added cycloheximide at a dose that prevents protein synthesis (Figure S5C). When polymerase levels were followed through the rest of the MNV replication cycle, there was no significant degradation of MNV polymerase in IFNγ-treated control cells (Figure S5D). In contrast to the lack of effect on early steps in the MNV life cycle or polymerase stability, the increase in MNV polymerase expression observed 7–12 hr after inoculation was inhibited by IFNγ and this inhibition required Atg5 (Figure 7B).
Figure 7.
The Life Cycle of MNV that Is Suppressed by IFNγ Dependently on Atg5
(A) Simple flow chart of the MNV life cycle.
(B) Western blot analysis of MNV polymerase expression in infected cells. A representative blot (n = 2) is shown.
(C) Western blot analysis of MNV capsid expression in infected cells. Samples used in (B) were reprobed with anti-VP1 (viral capsid) antibody.
(D) Northern blot analysis of genome and subgenome of MNV using RNA probe against viral capsid gene. GAPDH probe was used as loading control. A representative blot (n = 3) is shown. The arrowhead indicates nonspecific signal.
(E) RNase protection assay of negative strand transcription of MNV. Lane 2 contains undigested MNV and GAPDH probe. RNA from mock-inoculated macrophages served as a control (lane 3). A representative blot (n = 4) is shown.
(F) Immunofluorescence analysis of MNV (n = 3) using anti-propol antibody. Top: Representative MNV replication complexes (40×) in relation to nucleus. Bottom: Representative MNV replication complexes (20×) at 12 hpi (MOI = 5) with (+IFNγ) or without (–IFNγ) pretreatment of 100 U/ml of IFNγ. All images were taken using the same settings.
See also Figure S5.
We next examined the replication of viral RNAs. Both positive-sense genomic and subgenomic RNAs were detected at 8 and 12 hpi (Figure 7D). IFNγ inhibited expression of both positive-sense genomes (encoding the polymerase protein and other nonstructural proteins) and subgenomic RNAs (encoding the capsid proteins) in an Atg5-dependent manner (Figure 7D). Furthermore, IFNγ inhibited expression of the negative-sense viral RNA used as a template for positive-sense genomic and subgenomic RNA transcription (Rohayem et al., 2006) in an Atg5-dependent manner (Figure 7E). Consistently, the expression of MNV capsid protein, which is expressed after viral replication, was inhibited by IFNγ in an Atg5-dependent fashion (Figure 7C). Viruses with single-strand positive-sense RNA genomes, including MNV, form cytoplasmic membranous replication complexes for efficient generation of negative-sense and positive-sense viral RNAs (Hyde et al., 2009, Wobus et al., 2004). The MNV replication complex is a membranous structure consisting of all MNV nonstructural proteins, including the viral polymerase, as well as components from the ER, Golgi, and endosomes, but not lysosomes (Hyde et al., 2009). Since IFNγ blocked synthesis of viral RNAs in an Atg5-dependent fashion without effects on early steps in the viral life cycle, we examined the effects of IFNγ on the replication complex by analyzing the subcellular distribution of the MNV polymerase. By 12 hr after inoculation, we observed the MNV replication complex around the nuclei of macrophages (Figure 7F). However, the formation of detectable MNV replication complexes was inhibited by treatment with IFNγ in an Atg5-dependent manner (Figure 7F), likely explaining the effects of IFNγ on subsequent steps in the viral life cycle including production of infectious virus.
Discussion
IFNγ plays a major role in immune responses to infection but, in contrast to its antibacterial and antiparasitic effects, the mechanisms responsible for its direct antiviral effects against many viruses are not understood. Here we report that a subset or “cassette” of the machinery involved in canonical autophagy is essential for the antiviral effects of IFNγ against MNV, a positive-sense single-stranded RNA virus within a genus including the human pathogens responsible for the majority of epidemic viral gastroenteritis world-wide. We found that expression of Atg5 in macrophages and perhaps neutrophils is required for resistance to MNV in the absence of the IFNαβ receptor, a situation in which the host relies heavily on IFNγ to prevent death and disease. This could be explained by the failure of IFNγ to control MNV replication in the macrophage in vivo, as shown in our in vitro experiments. Alternatively, an independent autophagy gene-dependent function of macrophages or neutrophils, such as secretion of cytokines (Saitoh et al., 2008, Dupont et al., 2011), might play a determining role in MNV resistance.
In IFNγ-activated macrophages, the Atg5-Atg12/Atg16L1 complex generated by Atg7 is responsible for blocking MNV replication complex formation, explaining Atg5-dependent effects on multiple subsequent steps in the viral life cycle including synthesis of negative-sense RNAs, synthesis of positive-sense RNAs and viral structural proteins, and viral replication. Previous work from our laboratory (Wobus et al., 2004) has shown that the membranous MNV replication complex is morphologically quite distinct from autophagosomes. Unlike autophagosomes, the replication complex contains complex arrangements of convoluted and sometimes closely apposed membranes without a concentric double membraned structure or obvious intra-vesicular cytoplasmic contents or organelles.
Remarkably, extensive experiments detected no role for the portion of the autophagy pathway downstream of Atg7 involving Atg4B, lysosome-autophagosome fusion, or the degradative function of the autophagy pathway, indicating that the host has evolved to use specific portions or “cassettes” of the autophagy machinery in host defense against specific pathogens. Analysis of macrophage homeostasis, signaling, and gene expression indicates that the role of the Atg5-Atg12/Atg16L1 proteins in IFNγ-mediated inhibition of MNV replication is posttranscriptional. Previously, we and others have shown that Atg5 is required not for the expression of, but for the recruitment of, the IFNγ-inducible p47 GTPase (IIGP1 or Irga6) to the parasitophorous vacuole membrane in order to clear Toxoplasma gondii infection, without the apparent direct involvement of autophagosomes (Zhao et al., 2008, Zhao et al., 2009). Here we report a distinct role for autophagy proteins comprising the Atg5-12/Atg16L1 complex in IFNγ-mediated alterations in intracellular membranes—in this case preventing the formation of the membranous norovirus replication complex. Collectively, these data support the concept that the Atg5-12/Atg16L1 complex may function as a versatile antipathogen module for IFNγ-dependent blockade of membrane rearrangements required for the replication of certain pathogens.
While our studies did not detect a role for canonical autophagy in IFNγ-mediated control of MNV, the degradative effects of autophagy play a key role in control of herpes simplex virus, Sindbis virus infection, and human immunodeficiency virus (Kyei et al., 2009, Orvedahl et al., 2007, Orvedahl et al., 2010, Dreux and Chisari, 2010). In fact, the degradative activity of the canonical autophagy pathway likely contributes to many of the antipathogen roles of autophagy proteins (Levine et al., 2011, Levine and Deretic, 2007, Dreux and Chisari, 2010). However, as shown here for IFNγ-mediated control of MNV replication, other studies also support the concept that individual autophagy proteins or cassettes of autophagy proteins may function in infection or host defense in other ways. For example, autophagy proteins, but not canonical autophagy, function in phagocytosis and presentation of viral antigens to the immune system (Sanjuan et al., 2007, Lee et al., 2010, Huang et al., 2009). Atg5 and Atg7, but not canonical autophagy, are required for the secretory function of osteoclasts, a macrophage-lineage cell type related to the macrophages analyzed here (DeSelm et al., 2011). Further, it has been reported that Atg5 can interact with IFNα/β-inducing molecules RIG-I and IPS-1 to negatively regulate the IFNα/β pathway and enhance viral infection (Levine et al., 2011). The autophagy proteins Beclin-1, Atg4B, Atg5, and Atg12 have a role in initiating infection with Hepatitis C virus (Dreux and Chisari, 2010). The formation of replicative compartments utilized by Brucella abortus for generation of bacteria and cell-to-cell spread is dependent on Atg14L1, Beclin-1, and ULK1 but independent of Atg5, Atg7, and Atg16L1 (Starr et al., 2012). Similarly LC3α and β, a subset of the autophagy pathway proteins, but not Atg5 and Atg7, are required for coronavirus replication (Reggiori et al., 2010, Zhao et al., 2007). Thus, proteins required for the highly conserved and essential autophagy pathway likely have, in addition to their essential role in degradative autophagy, key independent roles in multiple aspects of infection and host defense.
Since degradative autophagy is conserved in single-celled eukaryotic organisms, it is interesting to speculate that the multiple roles of autophagy and autophagy proteins in host defense reflects diversification of the physiologic roles of autophagy and autophagy proteins as multicellular organisms developed (Levine et al., 2011). In this way specific cassettes of autophagy proteins could have developed additional functions important for host defense. As a countermeasure, certain microorganisms have evolved to co-opt cassettes of the autophagy machinery to foster replication or spread within the host (Starr et al., 2012). Targeting these individual cassettes of the autophagy machinery to control specific infections without manipulating a central process such as autophagy that plays such a key role in cell and tissue homeostasis, may allow targeting of treatments for while minimizing potentially deleterious side effects of antimicrobial therapy.
Experimental Procedures
Mice and Cells
Atg5flox/flox and Atg7flox/flox mice were previously described (Zhao et al., 2008, DeSelm et al., 2011). Atg16L1flox/flox mouse was generated using embryonic stem cell line purchased from the European Conditional Mouse Mutagenesis Program (EUCOMM) (unpublished data). All mice were housed and bred at Washington University (St. Louis, MO) under specific-pathogen-free conditions in accordance with federal and university guidelines as previously described (Cadwell et al., 2010). 293T, RAW 264.7, and MEF cells were used for production of lentiviruses, for plaque assay, and for immunofluorescence study, respectively. See the Supplemental Experimental Procedures for the details and the generation of bone marrow-derived macrophages (BMDMs).
Viruses and Infection
The following viruses were used for this study: MNV-1.CW3 (Thackray et al., 2007), EMCV K strain, MHV A59 strain, and WNV North American isolate, lineage 1 WNV strain 3000.0259. See the Supplemental Experimental Procedures for the details of in vitro viral infection. For in vivo experiments, 6- to 8-week-old mice were orally inoculated with 3 × 104 pfu MNV and survival was monitored for 30 days for three independent experiments. Statistical significance was determined with a log-rank test. For quantification of tissue viral burden, mice were sacrificed 5 days after inoculation. Organs were harvested and homogenized as previously described (Chachu et al., 2008) and virus was titrated by plaque assay.
Microarray
For identification of genes that are regulated by IFNγ in an Atg5-dependent manner, RNA collected from IFNγ treated and untreated control and Atg5-deficient macrophages was profiled with Affymetrix Mouse M430 2.0 (Figure 2A). The Gene Expression Omnibus accession number is GSE34863. See the Supplemental Experimental Procedures for analysis details.
Protein Analysis by Western Blot and Immunoprecipitation
Total cellular proteins were harvested with sample buffer (0.1 M Tris [pH 6.8], 4% SDS, 4 mM EDTA, 286 mM 2-mercaptoethanol, 3.2 M glycerol, 0.05% bromophenol blue), and proteins were analyzed as previously described (Hwang et al., 2009). For immunoprecipitation (IP), transfected 293T cells were lysed in IP buffer (50 mM Tris [pH 7.5], 1% NP-40, 150 mM sodium chloride + protease inhibitors) and subjected to IP with anti-FLAG antibody as previously described (Hwang et al., 2009). See the Supplemental Experimental Procedures for details.
Lentiviral Transduction
Modified lentiviral pCDH-MCS-T2A-copGFP-MSCV (System Biosciences, Mountain View, CA; CD523A-1) vector was used as backbone to express genes of interest. The T2A sequence in the vector is an 18 amino acid sequence that enables self-cleavage during translation and allows simultaneous production of copGFP. Lentivirus was generated in 293T by transfecting the lentiviral vector plasmids with packaging vector (psPAX2) and pseudotyping vector (pMD2.G) using the calcium phosphate precipitation method. Produced lentivirus was filtrated through 0.45 μm syringe filter (Millipore, MA) and added onto BMDMs twice on day 3 and day 4 until day 7.
Flow Cytometry and Immunofluorescence Analysis
Infected BMDMs were fixed at 12 hpi using 4% formaldehyde (Ted Pella, CA; 18505) for 10 min at room temperature (RT) and permeabilized with PBS/0.2% TritionX-100 (PBSTX) for overnight at 4°C. For flow cytometry, cells were stained with rabbit anti-propol antibody and then DyLight 649 Donkey anti-rabbit IgG (Biolegend, CA; 406406) in flow blocking buffer (PBS/0.2% TritionX-100/1% normal goat serum/1% normal mouse serum). For immunofluorescence, cells were stained with rabbit anti-propol antibody and then Alexa Fluor 555 goat anti-rabbit antibody in blocking buffer (PBS/0.2% TritionX-100/1% normal goat serum/10% normal mouse serum). See the Supplemental Experimental Procedures for the details.
STORM Setup and Imaging
The STORM setup and imaging was similar to that described before (Dani et al., 2010). See the Supplemental Experimental Procedures for details.
RNA Analysis by Northern Blot and RNase Protection Assay
BMDMs were incubated with IFNγ (1 U/ml) or media alone for 12 hr then infected with MNV at MOI = 5. Total RNAs were harvested at indicated times with Trizol reagent (Invitrogen) according to manufacturer's instructions. Northern blotting was performed using NorthernMax Kit (Applied Biosytems; AM1940) according to manufacturer's instruction. The synthesis and/or accumulation of negative strand RNA was analyzed using two-cycle RNase protection assay. See the Supplemental Experimental Procedures for details.
Statistical Analysis
All data were analyzed with Prism software (GraphPad, San Diego, CA) with an unpaired/paired t test or one-way analysis of variation (Tukey post test). All differences not specifically stated to be significant were not significant (p > 0.05).
Acknowledgments
This work was supported by the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases (U54 AI065982), National Institutes of Health grants AI054483, CA096511, and AI084887, and the Rheumatic Diseases Core Center (NIH P30 AR48335). We thank the Genome Technology Access Center at Washington University School of Medicine for help with genomic analysis (supported by NCI Cancer Center Support Grant number P30 CA91842 and ICTS/CTSA grant number UL1RR024992). Washington University and H.W.V. receive income based on licenses for MNV technology. C.L.O. was supported by grants from Ministry of Science and Innovation-Spain, FP7 (Microenvimet), and Fundacion M. Botin. We would like to thank Masaaki Komatsu (Tokyo Metropolitan Institute of Medical Science, Japan) for Atg7F/F mouse, Noboru Mizushima (Tokyo Medical and Dental University, Japan) for Atg5F/F mouse, Tamotsu Yoshimori (Osaka University, Japan) for Atg4B/C74A and Rab7A/T22N constructs. We thank Virgin lab members for their comments on the manuscript and D. Kreamalmeyer and M. White for managing mouse colonies.
Published: April 18, 2012
Footnotes
Supplemental Information includes one table, one movie, Supplemental Experimental Procedures, and five figures and can be found with this article online at doi:10.1016/j.chom.2012.03.002.
Accession Numbers
The Gene Expression Omnibus accession number for the microarray data reported in this paper is GSE34863.
Supplemental Information
List of genes differentially Regulated by IFNγ in the Absence or Presence of Atg5
References
- Cadwell K., Patel K.K., Maloney N.S., Liu T.C., Ng A.C., Storer C.E., Head R.D., Xavier R., Stappenbeck T.S., Virgin H.W. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann G.M., Guignabert C., Ying L., Deshpande N., Bekker J.M., Wang L., Zhou B., Rabinovitch M. Developmental expression of LC3alpha and beta: absence of fibronectin or autophagy phenotype in LC3beta knockout mice. Dev. Dyn. 2008;237:187–195. doi: 10.1002/dvdy.21392. [DOI] [PubMed] [Google Scholar]
- Chachu K.A., LoBue A.D., Strong D.W., Baric R.S., Virgin H.W. Immune mechanisms responsible for vaccination against and clearance of mucosal and lymphatic norovirus infection. PLoS Pathog. 2008;4:e1000236. doi: 10.1371/journal.ppat.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.P., Tsai C.C., Huang W.C., Wang C.Y., Chen C.L., Lin Y.S., Kai J.I., Hsieh C.Y., Cheng Y.L., Choi P.C., et al. Autophagy facilitates IFN-gamma-induced Jak2-STAT1 activation and cellular inflammation. J. Biol. Chem. 2010;285:28715–28722. doi: 10.1074/jbc.M110.133355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changotra H., Jia Y., Moore T.N., Liu G., Kahan S.M., Sosnovtsev S.V., Karst S.M. Type I and type II interferons inhibit the translation of murine norovirus proteins. J. Virol. 2009;83:5683–5692. doi: 10.1128/JVI.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani A., Huang B., Bergan J., Dulac C., Zhuang X. Superresolution imaging of chemical synapses in the brain. Neuron. 2010;68:843–856. doi: 10.1016/j.neuron.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado M.A., Elmaoued R.A., Davis A.S., Kyei G., Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSelm C.J., Miller B.C., Zou W., Beatty W.L., van Meel E., Takahata Y., Klumperman J., Tooze S.A., Teitelbaum S.L., Virgin H.W. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell. 2011;21:966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M., Chisari F.V. Viruses and the autophagy machinery. Cell Cycle. 2010;9:1295–1307. doi: 10.4161/cc.9.7.11109. [DOI] [PubMed] [Google Scholar]
- Dupont N., Jiang S., Pilli M., Ornatowski W., Bhattacharya D., Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell. 2008;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil M.P., Bohn E., O'Guin A.K., Ramana C.V., Levine B., Stark G.R., Virgin H.W., Schreiber R.D. Biologic consequences of Stat1-independent IFN signaling. Proc. Natl. Acad. Sci. USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass R.I., Parashar U.D., Estes M.K. Norovirus gastroenteritis. N. Engl. J. Med. 2009;361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Canadien V., Lam G.Y., Steinberg B.E., Dinauer M.C., Magalhaes M.A., Glogauer M., Grinstein S., Brumell J.H. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl. Acad. Sci. USA. 2009;106:6226–6231. doi: 10.1073/pnas.0811045106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S., Kim K.S., Flano E., Wu T.T., Tong L.M., Park A.N., Song M.J., Sanchez D.J., O'Connell R.M., Cheng G., Sun R. Conserved herpesviral kinase promotes viral persistence by inhibiting the IRF-3-mediated type I interferon response. Cell Host Microbe. 2009;5:166–178. doi: 10.1016/j.chom.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J.L., Sosnovtsev S.V., Green K.Y., Wobus C., Virgin H.W., Mackenzie J.M. Mouse norovirus replication is associated with virus-induced vesicle clusters originating from membranes derived from the secretory pathway. J. Virol. 2009;83:9709–9719. doi: 10.1128/JVI.00600-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S.M., Wobus C.E., Lay M., Davidson J., Virgin H.W., 4th STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- Kimura T., Nakayama K., Penninger J., Kitagawa M., Harada H., Matsuyama T., Tanaka N., Kamijo R., Vilcek J., Mak T.W., Taniguchi T. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- Kyei G.B., Dinkins C., Davis A.S., Roberts E., Singh S.B., Dong C., Wu L., Kominami E., Ueno T., Yamamoto A., et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.K., Mattei L.M., Steinberg B.E., Alberts P., Lee Y.H., Chervonsky A., Mizushima N., Grinstein S., Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H.W. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño G., Fernández A.F., Cabrera S., Lundberg Y.W., Cabanillas R., Rodríguez F., Salvador-Montoliu N., Vega J.A., Germanà A., Fueyo A., et al. Autophagy is essential for mouse sense of balance. J. Clin. Invest. 2010;120:2331–2344. doi: 10.1172/JCI42601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita M., Suzuki N.N., Obara K., Fujioka Y., Ohsumi Y., Inagaki F. Structure of Atg5.Atg16, a complex essential for autophagy. J. Biol. Chem. 2007;282:6763–6772. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumphrey S.M., Changotra H., Moore T.N., Heimann-Nichols E.R., Wobus C.E., Reilly M.J., Moghadamfalahi M., Shukla D., Karst S.M. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J. Virol. 2007;81:3251–3263. doi: 10.1128/JVI.02096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., Burns D., Leib D.A., Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Orvedahl A., MacPherson S., Sumpter R., Jr., Tallóczy Z., Zou Z., Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Verheije M.H., Calì T., Ulasli M., Bianchi S., Bernasconi R., de Haan C.A., Molinari M. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohayem J., Robel I., Jäger K., Scheffler U., Rudolph W. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J. Virol. 2006;80:7060–7069. doi: 10.1128/JVI.02195-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Fujita N., Jang M.H., Uematsu S., Yang B.G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Sanjuan M.A., Dillon C.P., Tait S.W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S., Green D.R. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- Shrestha B., Wang T., Samuel M.A., Whitby K., Craft J., Fikrig E., Diamond M.S. Gamma interferon plays a crucial early antiviral role in protection against West Nile virus infection. J. Virol. 2006;80:5338–5348. doi: 10.1128/JVI.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T., Child R., Wehrly T.D., Hansen B., Hwang S., López-Otin C., Virgin H.W., Celli J. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11:33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson L.M., Miller B.C., Ng A., Eisenberg J., Zhao Z., Cadwell K., Graham D.B., Mizushima N.N., Xavier R., Virgin H.W., Swat W. Identification of Atg5-dependent transcriptional changes and increases in mitochondrial mass in Atg5-deficient T lymphocytes. Autophagy. 2009;5:625–635. doi: 10.4161/auto.5.5.8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal M.C., Sasai M., Lee H.K., Yordy B., Shadel G.S., Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray L.B., Wobus C.E., Chachu K.A., Liu B., Alegre E.R., Henderson K.S., Kelley S.T., Virgin H.W., 4th Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 2007;81:10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus C.E., Karst S.M., Thackray L.B., Chang K.O., Sosnovtsev S.V., Belliot G., Krug A., Mackenzie J.M., Green K.Y., Virgin H.W. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.T., Tan H.L., Shui G., Bauvy C., Huang Q., Wenk M.R., Ong C.N., Codogno P., Shen H.M. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J. Biol. Chem. 2010;285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Nakagawa I., Yamamoto A., Amano A., Noda T., Yoshimori T. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathog. 2009;5:e1000670. doi: 10.1371/journal.ppat.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Thackray L.B., Miller B.C., Lynn T.M., Becker M.M., Ward E., Mizushima N.N., Denison M.R., Virgin H.W., 4th Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Fux B., Goodwin M., Dunay I.R., Strong D., Miller B.C., Cadwell K., Delgado M.A., Ponpuak M., Green K.G., et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.O., Khaminets A., Hunn J.P., Howard J.C. Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNgamma-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog. 2009;5:e1000288. doi: 10.1371/journal.ppat.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes differentially Regulated by IFNγ in the Absence or Presence of Atg5