Abstract
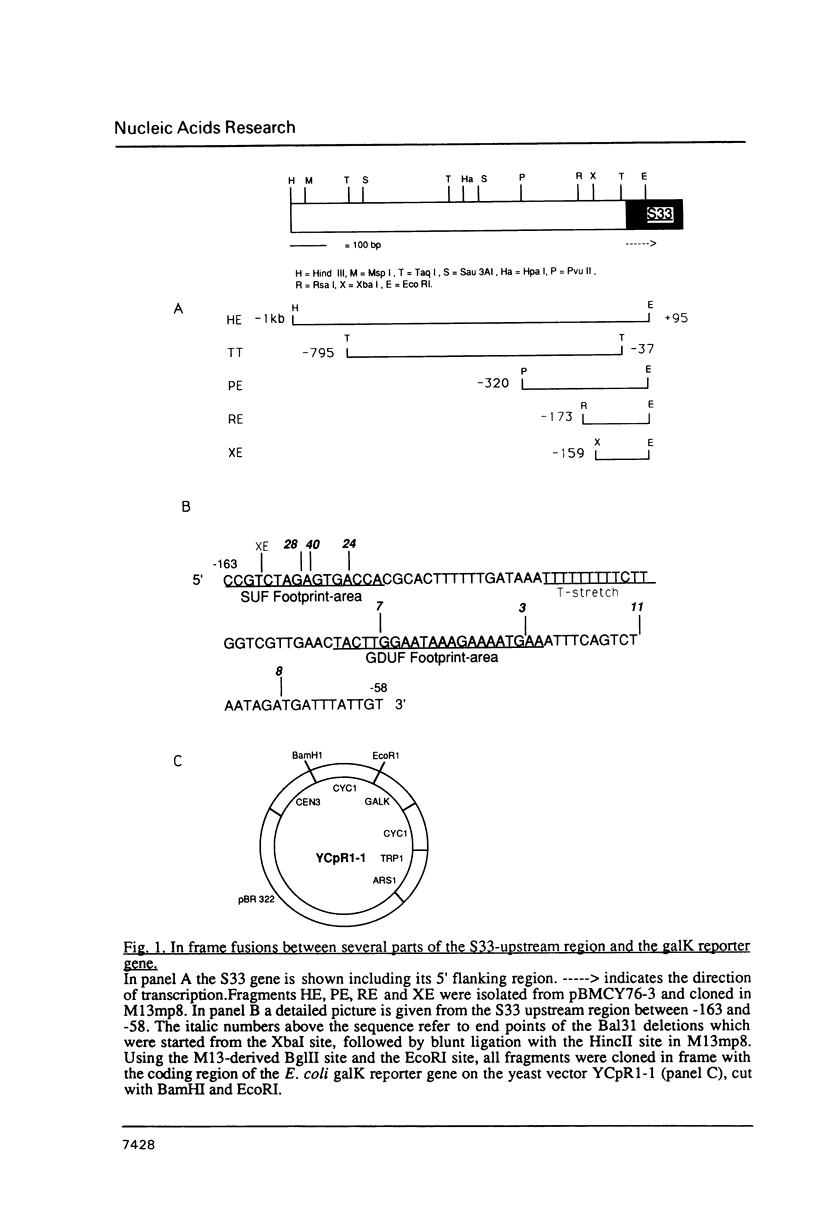
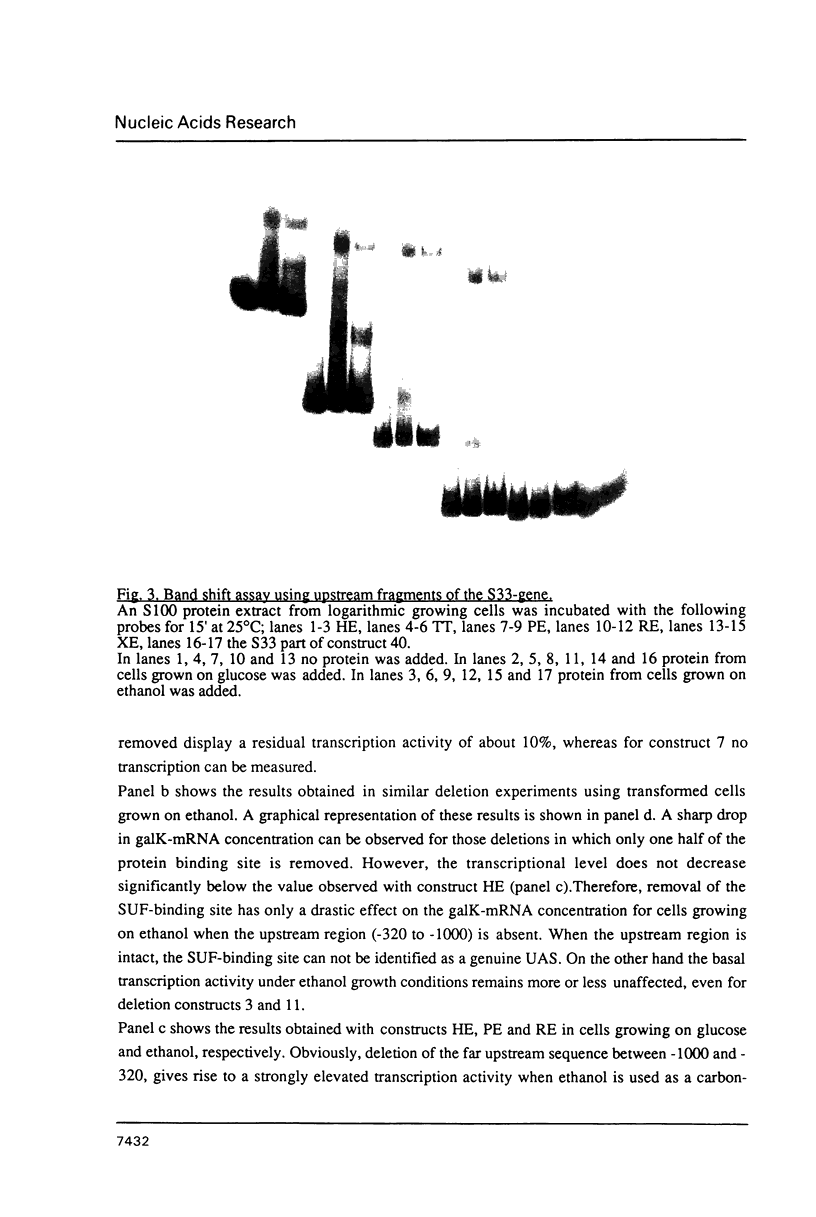
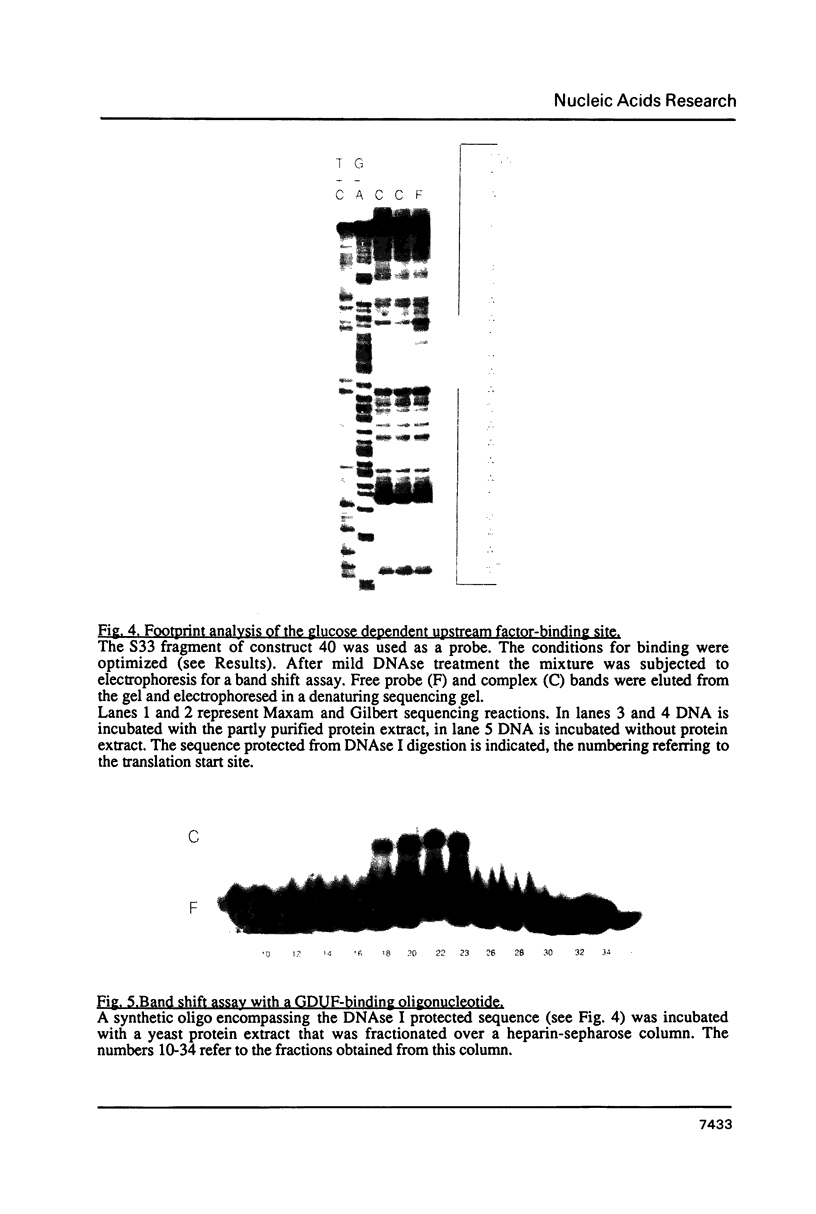
At least 4 different, protein binding cis-acting elements are present in the upstream region of the S33-gene. The major protein binding site is situated between positions -148 and -163 relative to the ATG start codon. It binds a trans-acting factor designated SUF (S33 Upstream Factor). When yeast cells are growing on glucose, deletion of this site results in a decrease of transcription of 50%. Using ethanol as a carbon-source, deletion of the SUF-responsive site lowers the transcription as much as 80%. A second protein binding site is found between positions -85 and -105. Only extracts from glucose-grown cells contain a factor that is able to bind to this site in vitro. A third protein binding site was found using a protein extract from ethanol-grown cells. This site, which is located quite close to the transcriptional start site, is probably responsible for the 20% residual transcription when the SUF binding site is removed. Finally, a site far upstream was found, which binds a protein from both glucose-grown and ethanol-grown cells. This site may function as an upstream repression site which is only functional when a non-fermentable carbon-source is used. Taking these findings into account, we present a model for the carbon-source dependent transcription activation of the gene encoding S33.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berman J., Tachibana C. Y., Tye B. K. Identification of a telomere-binding activity from yeast. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3713–3717. doi: 10.1073/pnas.83.11.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Kimmerly W. J., Rine J., Kornberg R. D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jan;8(1):210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan D. M., Pearson N. J. Transcriptional regulation of ribosomal proteins during a nutritional upshift in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jul;6(7):2429–2435. doi: 10.1128/mcb.6.7.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsman J. C., van Heeswijk W. C., Grivell L. A. Identification of two factors which bind to the upstream sequences of a number of nuclear genes coding for mitochondrial proteins and to genetic elements important for cell division in yeast. Nucleic Acids Res. 1988 Aug 11;16(15):7287–7301. doi: 10.1093/nar/16.15.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamil K. G., Nam H. G., Fried H. M. Constitutive transcription of yeast ribosomal protein gene TCM1 is promoted by uncommon cis- and trans-acting elements. Mol Cell Biol. 1988 Oct;8(10):4328–4341. doi: 10.1128/mcb.8.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herruer M. H., Mager W. H., Woudt L. P., Nieuwint R. T., Wassenaar G. M., Groeneveld P., Planta R. J. Transcriptional control of yeast ribosomal protein synthesis during carbon-source upshift. Nucleic Acids Res. 1987 Dec 23;15(24):10133–10144. doi: 10.1093/nar/15.24.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet J., Cottrelle P., Cool M., Vignais M. L., Thiele D., Marck C., Buhler J. M., Sentenac A., Fromageot P. A general upstream binding factor for genes of the yeast translational apparatus. EMBO J. 1985 Dec 16;4(13A):3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmerly W., Buchman A., Kornberg R., Rine J. Roles of two DNA-binding factors in replication, segregation and transcriptional repression mediated by a yeast silencer. EMBO J. 1988 Jul;7(7):2241–2253. doi: 10.1002/j.1460-2075.1988.tb03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., Van Raamsdonk-Duin M. M., Mager W. H., Planta R. J. Conserved sequences upstream of yeast ribosomal protein genes. Curr Genet. 1985;9(4):273–277. doi: 10.1007/BF00419955. [DOI] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Schoppink P. J., Cornelissen M. T., Cohen L. H., Mager W. H., Planta R. J. Yeast ribosomal protein S33 is encoded by an unsplit gene. Nucleic Acids Res. 1983 Nov 25;11(22):7759–7768. doi: 10.1093/nar/11.22.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager W. H. Control of ribosomal protein gene expression. Biochim Biophys Acta. 1988 Jan 25;949(1):1–15. doi: 10.1016/0167-4781(88)90048-6. [DOI] [PubMed] [Google Scholar]
- Planta R. J., Raué H. A. Control of ribosome biogenesis in yeast. Trends Genet. 1988 Mar;4(3):64–68. doi: 10.1016/0168-9525(88)90042-x. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Zitomer R. S., Schümperli D., Rosenberg M. The expression in yeast of the Escherichia coli galK gene on CYC1::galK fusion plasmids. Gene. 1983 Nov;25(2-3):249–262. doi: 10.1016/0378-1119(83)90229-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D., Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987 Dec 4;51(5):721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D., Stillman D. J., Brand A. H., Nasmyth K. A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987 Feb;6(2):461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais M. L., Woudt L. P., Wassenaar G. M., Mager W. H., Sentenac A., Planta R. J. Specific binding of TUF factor to upstream activation sites of yeast ribosomal protein genes. EMBO J. 1987 May;6(5):1451–1457. doi: 10.1002/j.1460-2075.1987.tb02386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudt L. P., Smit A. B., Mager W. H., Planta R. J. Conserved sequence elements upstream of the gene encoding yeast ribosomal protein L25 are involved in transcription activation. EMBO J. 1986 May;5(5):1037–1040. doi: 10.1002/j.1460-2075.1986.tb04319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]