Abstract
Experimental studies showed that genomic imprinting is fundamental in fetoplacental development by timely regulating the expression of the imprinted genes to overlook a set of events determining placenta implantation, growth and embryogenesis.
We examined the expression profile of 22 imprinted genes which have been linked to pregnancy abnormalities that may ultimately influence childhood development. The study was conducted in a subset of 106 placenta samples, overrepresented with small and large for gestational age cases, from the Rhode Island Child Health Study.
We investigated associations between imprinted gene expression and three fetal development parameters: newborn head circumference, birth weight, and size for gestational age. Results from our investigation show that the maternally imprinted/paternally expressed gene ZNF331 inversely associates with each parameter to drive smaller fetal size while paternally imprinted/maternally expressed gene SLC22A18 directly associates with the newborn head circumference promoting growth. Multidimensional Scaling analysis revealed two clusters within the 22 imprinted genes which are independently associated with fetoplacental development. Our data suggest that cluster 1 genes work by assuring cell growth and tissue development while cluster 2 genes act by coordinating these processes. Results from this epidemiologic study offer solid support for the key role of imprinting in fetoplacental development.
Keywords: genomic imprinting, placenta, imprinted gene expression, fetal development
Introduction
The role of imprinted genes in fetoplacental development has been widely reported in the literature [1–2]. Genomic imprinting is predicted to involve only about 1% (~200 genes) of the expressed genome with only about 90 imprinted genes that are well characterized at present [3–5]. This gene set shares the unique characteristic of expressing from only one of two parental alleles in a parental specific fashion. Silencing of the inactive allele is thought be achieved by epigenetic mechanisms including DNA methylation, histone modification and long non-protein-coding RNAs (lncRNAs) that act upon specific imprinting control regions (ICRs) or ICR-like elements within promoters and enhancers [6–7]. Imprinted genes often aggregate in clusters under the control of specific ICRs; single imprinted genes regulated by dedicated ICR-like elements are however not infrequent [6]. Interestingly imprinted genes are functionally haploid in those tissues/organs that perpetuate the imprinting epigenetic signal while they otherwise behave as normal diploid genes.
Imprinted genes, as indicated by the Ingenuity Pathway Analysis (IPA) (Ingenuity® Systems, www.ingenuity.com), belong to gene networks critical for the proper cellular and organ development; perturbations of these networks have been associated with developmental, neurological, endocrine and muscular disorders as well as cancer (IPA) (Table 1). Consistently, by using animal models, imprinted gene have been shown to: 1) regulate the exchange of resources between mother and fetus; 2) program the metabolism in the early postnatal period to determine growth and metabolic phenotype; and 3) participate in the development of metabolically important organs such as the pituitary, pancreas, liver, fat, the hypothalamus and the placenta [8–9].
Table 1.
List of 22 candidate imprinted genes and their putative functions by network analysisa
| Gene Networkb | Networks | |
|---|---|---|
| Functional Areas | Associated Disease | |
| 1. DLK1, H19, HOXD10, IGF2, NNAT, TP73 | Cellular Growth and Proliferation | Developmental Disorders Genetic Disorders |
| 2. HOXA11, PEG3, SNRPN | Nervous System Development and Function | Cancer Skeletal and Muscular Disorders Reproductive System Diseases |
| 3. CD44, EPS15, ILK | Cellular Development Cell Cycle |
Cardiovascular Diseases |
| 4. CCDC86, CDKAL1 | Small Molecule Biochemistry | Gastrointestinal Diseases Hepatic System Diseases |
| 5. MEST, PLAGL1 | Gene Expression | Cancer Connective Tissue Disorders |
| 6. DHCR24, PEG10, ZNF331 | Cell Cycle Cellular Development |
Cancer |
| 7. PHLDA2 | Organ Development Respiratory System Development and Function Cellular Assembly and Organization |
– |
| 8. MEG3 | Connective Tissue Development and Function | Behavioral Syndromes |
| 9. SLC22A18 | – | Cancer, Developmental Disorders |
: elaborated by using the Ingenuity Pathway Analysis informatics tool.
: networks includes several other not imprinted genes which are not reported here for ease of reading.
Additionally, imprinted gene expression demonstrated low transcriptional noise, as shown in placenta [10]. This finding is in agreement with the imprinted genes functional importance that, as shown for other such genes [11–12], once altered, can lead to prominent phenotypic changes [13–14] such as lethality [15–16] that is further enhanced by their constitutional haploinsufficiency [17].
In this framework the placenta is considered as the main determinant of the fetal phenotype and therefore represents the appropriate tissue for analyzing the imprinted gene expression profile in relation to growth and developmental outcomes. The placenta indeed: 1) supplies the much needed fetomaternal interface, and functions as immune and endocrine organ [18–19]; 2) overlooks and coordinates the embryonic growth [20]; 3) synchronises with the brain development by the coordinated expression of many imprinted genes [21–22]; and 4) supplies serotonin to the developing brain to support neuronal differentiation [23–24]. The placenta is also of predominantly fetal origin as it originates from the outer layer of the blastocyst [25] and therefore provides a unique snapshot of the fetal epi/genetic status.
Expression profiling of the placenta has already provided hints about the activity of imprinted genes in correlation with pregnancy outcomes such as intrauterine growth restriction (IUGR) [10, 26] and preeclampsia (PE) [27]. These disorders of pregnancy have been independently linked to chronic and developmental abnormalities in children as mostly neurodevelopmental, consistent with the Barker hypothesis [28–30]. These findings are supportive of the theory that links perturbations of imprinting regulation in the placenta to chronic and developmental disorders through an altered fetal phenotype. Nevertheless the correlation between imprinted gene expression and fetal growth has been previously tested only in a small study by our group with limited information on the newborns [10]. In the current study, we analyzed the expression of a panel of imprinted genes (Table 1) in 106 human term placenta samples from a birth cohort of infants, the Rhode Island Child Health Study (RICHS) and examined the correlation between imprinted gene expression and fetal development.
Material and Methods
Study Population
Placenta samples were collected in the framework of the RICHS, which enrolls mother-infant pairs at Women and Infants Hospital of Rhode Island. Every term small for gestational age (SGA) (<10th percentile) and large for gestational age (LGA) (>90th percentile) infant enrolled, as calculated from the Fenton growth chart based on birthweight and gestational age [31], was matched for gender, gestational age (±3 days), and maternal age (±2 years) with one appropriate for gestational age (AGA) newborn.
Exclusion criteria adopted were: multiple pregnancies, maternal age <18 years, life-threatening medical complications of the mother, and congenital or chromosomal abnormalities of the infant. Data on maternal ethnicity, age and insurance were obtained through both a structured chart review and an interviewer-administered questionnaire. Data on gestational week, delivery method, infant gender, head circumference, birth weight and size for gestational age were abstracted from charts. For this study, the first 106 subjects enrolled between September 2009 and May 2010, were selected. All subjects were consented accordingly to the specific protocol approved by the Institutional Review Boards for Women and Infants’ Hospital and Brown University.
Placental Tissue Collection and RNA Isolation
Placental tissue was biopsied from each of the 4 placenta quadrants midway from the cord insertion and the placental rim, within 2 h from the delivery. The maternal decidua was then excised and the biopsies placed in RNAlater (Qiagen – Valencia, CA, USA) for 72 h at 4° C. Tissue was then blotted dry, snap-frozen in liquid nitrogen, homogenized with mortar and pestle and stored in ultrafreezer at −80° C. RNA was later extracted by using the RNeasy kit (Qiagen – Valencia, CA, USA) supplemented by double DNase I (Qiagen – Valencia, CA, USA) on column digestion in order to clear any DNA contamination. Extracted RNA was finally quantified with Nanodrop spectrophotometer and stored at −80°C.
Gene Expression Analysis
The list of imprinted genes was populated by consolidating data of 3 previous experiments: 1) the analysis of the expression of 52 imprinted genes expressed in placenta from pregnancies diagnosed with severe intrauterine growth restriction (IUGR) which returned 9 dysregulated genes [10]; 2) the investigation of the loss of genomic imprinting (LOI) profile at the RNA level in 22 severe IUGR placentas by using a quantitative assay that we developed [10, 32]; 3) a test we ran on the expression of 2 homeobox genes in a subset of 60 samples from the RICHS cohort (unpublished data) (see Table 1).
Gene expression was measured by the Mount Sinai School of Medicine Real-Time PCR facility using quantitative real-time PCR (qRT-PCR) with a robotized fluid handling system for 384 well plates. Expression values were cascade normalized against the three housekeeping genes RPS11, ACTB and TUBB. The RNA copy number for each imprinted gene for each sample was then calculated as:
where RCN is the RNA copy number, k is the equation constant, eff is the average replication efficiency, Ctcn are the cascade normalized (cn) Ct values from the 3 housekeeping genes and Ctgt are the Ct values for each gene tested (gt).
Statistical Analysis
Cascade normalized gene expression values for all imprinted genes tested were input into three separate regression models. RCNs were log transformed in order to have them normally distributed for such regressions. For infant head circumference and birth weight we used two separate multinomial linear regressions where the outcomes were modeled against the RCN of each gene with the following covariates: maternal age, ethnicity and insurance, gestational week and delivery method, and finally the infant gender. We used PASW statistical software (version 18.0.2) (SPSS Inc. – Chicago, IL, USA) to run a stepwise regression. The size for gestational age, as defined in the Study Population section, was modeled into a multinomial logistic regression using AGA (10th ≤ AGA ≤ 90th percentile) as reference category and a model otherwise overlapping the setting used for the linear regressions.
Multidimensional scaling (MDS) and hierarchical clustering analysis were run by using the R 2.13.0 statistical package in order to obtain a visual bidimensional representation of the distances among studied genes. The distance is calculated as “1 – the Pearson correlation coefficients among the genes” as described by the log normalized RCN values.
The multinomial regressions carried out to test the inter-correlation of the genes in each cluster were run as follows. For infant head circumference, we first excluded genes SLC22A18 (cluster 1) and ZNF331 (cluster 2) contemporaneously and then separately from the model; for birth weight and SGA phenotype we excluded only ZNF331. The second cluster inter-correlation test was carried out by again excluding SLC22A18 and ZNF331 together with the genes that were found significant after the first exclusion round.
Results
Demographics and variable characteristics of the study population are presented in Table 2. The study population is a subpopulation of the RICHS study of which details have been published previously [33]. Because the study is designed to enroll one control AGA infant for every SGA and LGA cases, the cohort is overrepresented with SGA and LGA cases.
Table 2.
Population demographics (A) and variable characteristics (B) for the 106 samples from the RICHS cohort.
| A. Population demographics
| ||
|---|---|---|
| Variable | N. | % |
| Size per Gestational Age | ||
| SGA | 33 | 31 |
| AGA | 51 | 48 |
| LGA | 22 | 21 |
| Infant Gender | ||
| Female | 59 | 56 |
| Male | 47 | 44 |
| Delivery Method | ||
| Vaginal | 67 | 63 |
| Caesarean Section | 38 | 36 |
| Unknown | 1 | 1 |
| Maternal Ethnicity | ||
| White | 79 | 75 |
| African American | 7 | 7 |
| Asian | 3 | 3 |
| Other | 14 | 12 |
| Unknown | 3 | 3 |
| Maternal Insurance | ||
| Public | 45 | 42 |
| Private | 61 | 58 |
| B. Variable characteristics
| |||||
|---|---|---|---|---|---|
| Variable | N. | Mean | Std. Dev. | Min | Max |
| Maternal Age (years) | 106 | 29.4 | 6.0 | 18 | 40 |
| Gestational Age (weeks) | 106 | 38.9 | 1.1 | 37 | 41 |
| Birth Weight (g) | 106 | 3,310.0 | 744.9 | 1,705 | 5,080 |
| Head Circumference (cm) | 106 | 34.1 | 1.9 | 30.0 | 38.1 |
The set of imprinted gene selected for this study was determined by consolidating the following three lists of gene: 1) 9 imprinted genes that we found dysregulated in severe SGA (<5th percentile with associated Doppler findings) [10]; 2) 14 imprinted genes of which we previously reported the association between their LOI and severe IUGR [10, 32]; and 3) 2 homeobox genes that have been recently predicted to be imprinted and showed a high degree of correlation with fetal growth outcomes in a subset of samples from the RICHS cohort (unpublished data). Given that some genes were common in these lists, the final list contains a total of 22 imprinted genes shown in Table 1.
We tested the expression levels of the 22 imprinted genes on 106 samples from the RICHS cohort and subsequently analyzed their correlation with three main fetal growth indexes, i.e. infant head circumference, birth weight size for gestational age. The size for gestational age is a categorical variable used to classify babies as SGA, AGA or LGA depending on their birth weight percentile and gestational age. Expression data, calculated as RCN, were cascade normalized against the housekeeping genes RPS11, ACTB and TUBB and log transformed to assure normality.
We first analyzed the correlation of the expression levels of the imprinted genes tested with the three outcomes using multinomial linear regressions for the continuous outcomes (head circumference, birth weight) and multinomial logistic regression for the categorical outcome (size for gestational age) (Table 3). Multivariate models were adjusted for maternal age, ethnicity and insurance, together with gestational week, delivery method and infant gender. Stepwise selection procedures were used aiming at identifying genes or co-variates that predict study outcomes.
Table 3.
Multinomial regression statistics for the correlation between imprinted gene expression and the fetal growth parameters at birth
| Regression | Outcome | Regression Stats
|
Stepwise Variables | Regression Variables
|
|||
|---|---|---|---|---|---|---|---|
| p value | r square | Beta Coeff. | Std. Error | p value | |||
| Multinomial Linear | Head Circumference | <.001 | .324 | Constant | 10.675 | 5.835 | .071 |
| Gestational Week | 0.621 | 0.148 | <.001 | ||||
| logZNF331 | −1.771 | 0.441 | .001 | ||||
| logSLC22A18 | 1.498 | 0.448 | .003 | ||||
| Birth Weight | <.001 | .396 | Constant | −6,896.557 | 2,158.693 | .002 | |
| Gestational Week | 258.936 | 55.217 | <.001 | ||||
| logZNF331 | −594.526 | 187.361 | .002 | ||||
| Infant Gender | 346.630 | 123.795 | .006 | ||||
| Maternal Age | 24.502 | 10.441 | .021 | ||||
| Delivery Method | −291.248 | 130.762 | .028 | ||||
| Multinomial Logistic | Size per Gestational Age | .009 | .254 | SGA vs AGA | |||
| Intercept | −4.217 | 1.088 | <.001 | ||||
| logZNF331 | 3.130 | 0.927 | <.001 | ||||
| [Infant Gender = Female] | 1.034 | 0.555 | .062 | ||||
| [Infant Gender = Male] (1) | – | – | – | ||||
| LGA vs AGA | |||||||
| Intercept | −0.896 | 0.795 | .260 | ||||
| logZNF331 | 0.467 | 0.830 | .574 | ||||
| [Infant Gender = Female] | −0.905 | 0.570 | .112 | ||||
| [Infant Gender = Male] (1) | – | – | – | ||||
reference category, parameter set to zero
The maternally imprinted/paternally expressed gene ZNF331 was found inversely associated with newborn head circumference and birth weight while positively associated with the SGA phenotype. Contemporaneously, the paternally imprinted/maternally expressed gene SLC22A18 was positively correlated with newborn head circumference.
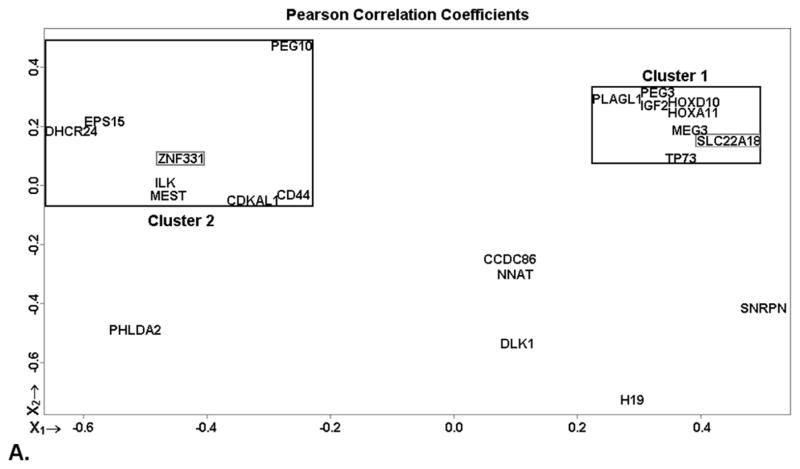
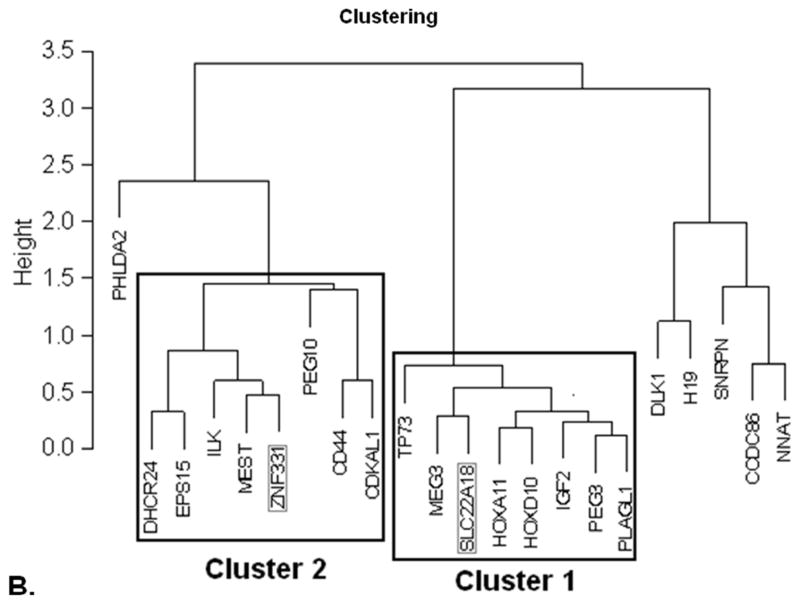
To account for the potential coordinated correlation of these key imprinted genes in fetoplacental growth and development, we applied MDS and hierarchical clustering analysis to the entire set of 22 genes based on expression data determined in this cohort (Figure 1). This approach identified 2 putative gene clusters, each composed of 8 genes. The first, cluster 1 (x1 > 0.2; x2 > 0.0), was very compact as shown by the close gene positioning in the MDS graph and the highly coherent clustering (Figure 1). This cluster includes genes HOXA11, HOXD10, IGF2, MEG3, PEG3, PLAGL1, SLC22A18 and TP73. Cluster 2 (x1 > −0.05; x2 < 0.2), was more broadly distributed and consisted of genes CD44, CDKAL1, DHCR24, EPS15, ILK, MEST, PEG10 and ZNF331. Interestingly, SLC22A18 and ZNF331, the genes significantly associated with the fetal outcomes tested, fell into different clusters.
Figure 1.
Multidimensional scaling (MDS) and hierarchical clustering analysis. By using the Pearson correlation coefficients to describe the distances between the tested samples as returned by the RCNs for the 22 imprinted gene tested, 2 gene clusters emerged. They appear boxed in black in the MDS plot (A) and are confirmed by clustering analysis (B). Cluster 1 is particularly compact, while cluster 2 appears more scattered, but still consistently distributed. Significant genes of these clusters, ZNF331 and SLC22A18, are boxed in gray. A third cluster, with only 5 genes also emerged from the hierarchical clustering analysis; it, however, presents as the least cohesive (see hierarchical value scale) and never showed any correlation with the investigated outcomes. Imprinted gene PHLDA2 shows no tendency to cluster with any other gene.
In order to find out if these two genes exerted independent effect on fetal growth or instead they behaved as markers for their respective gene clusters, we proceeded to exclude them from the regression models. As expected from the clustering analysis, this procedure confirmed the cluster effect. In each regression model SLC22A18 and ZNF331 were replaced by genes belonging to the same clusters of the excluded genes (Table 4). This hypothesis was further confirmed by conducting a second regressions’ round by excluding SLC22A18, ZNF331 and their substitutes from the first round. Other genes of the clusters of those excluded became significantly correlated to each outcome or the SGA phenotype (data not shown).
Table 4.
Effect of the inter-correlation of the genes in each cluster as shownf by excluding the significant genes from the regression models.
| Outcome | Full Regressions
|
Excluded Genes | Restricted Regressions
|
||||
|---|---|---|---|---|---|---|---|
| Significant Genes | Correlation | Cluster | Significant Genes | Correlation | Cluster | ||
| Head Circumference | SLC22A18 | + | 1 | SLC22A18 | IGF2 | + | 1 |
| ZNF331 | − | 2 | ZNF331 | PEG10 | − | 2 | |
| SLC22A18 | PEG3 | + | 1 | ||||
| ZNF331 | − | 2 | |||||
| ZNF331 | SLC22A18 | + | 1 | ||||
| CDKAL1 | − | 2 | |||||
| Birth Weight | ZNF331 | − | 2 | ZNF331 | EPS15 | − | 2 |
| Size per Gestational Age | ZNF331 | SGA | 2 | ZNF331 | MEST | SGA | 2 |
This cluster effect is even more evident when considering that: 1) after excluding SLC22A18 and ZNF331, the newly significant genes conserved the correlation direction; 2) for the infant head circumference, when separately excluding SLC22A18 and ZNF331, the one gene that is not excluded, as the best representative of that cluster, is confirmed as significantly correlated to the outcome by the model; and 3) none of the 6 genes not clustered showed any significant correlation at any level for any of the regressions run.
Discussion
The existing data on the correlation between imprinted gene expression and the most relevant pregnancy outcomes IUGR [10, 26, 34] and PE [27], together with the association of these with a plethora of chronic and developmental disorders in children [27, 29, 35], calls for a deeper understanding of the role of imprinted genes in fetal development. The importance of identifying early biomarkers of adverse embryonic growth becomes even more evident when considering the steady increase of disorders like asthma [36–37], obesity [38], neurodevelopmental syndromes [39–40], learning disabilities [41], birth defects [42–43] and cancer [44] affecting children, and the cost they bring about for the health care system [45–46].
We herein analyzed the expression profile of 22 imprinted genes, which have been previously shown to be involved with several growth phenotypes [10, 27, 34], in 106 placental samples from the RICHS cohort. We focused on 3 birth outcomes that have widely been used as a proxy for fetal growth and development. The infant head circumference provides indications about the neural development and the risk of manifestation of neural syndromes [35]. While clearly linked to head circumference, the birth weight and its trajectory had been associated with other chronic disorders like asthma and obesity [47–48]. Finally the size for gestational age is based on birth weight and gestational age and has been initially elaborated as a parameter for maternal-fetal medicine clinical purposes [31] and it often overlaps with the birth weight in its ability to predict childhood diseases [35].
Results of our investigation lend strong support to the fundamental role of genomic imprinting in fetal growth. We specifically showed that 16 of the 22 imprinted genes investigated can be grouped into two clusters that independently drive fetoplacental growth, further demonstrating the molecular interplay between imprinted genes in this process (see Figure 1).
Among genes in cluster 1, SLC22A18 demonstrated the strongest positive correlation with the head circumference. SLC22A18 is a transporter of organic cations involved in the transport of chloroquine and quinidine-related compounds [49] which are known to regulate cholinergic receptors [50–51]. SLC22A18 is located in the chromosomal region 11p15.5 which is associated with the Beckwith-Wiedemann Syndrome (BWS) [1]. BWS is characterized by overgrowth, increased risk of cancer and developmental delay [1], in substantial agreement with an increased head circumference. In agreement with our data, genes that populated cluster 1 are thought to be mostly involved in cell growth as well as nervous and connective tissue development linked to behavioral and disorders of development (IPA networks 1 and 2 – see Table 1).
Among the genes in cluster 2, ZNF331 showed the strongest negative correlation with infant head circumference and birth weight, which are linked to the SGA phenotype. It is interesting to point out that we previously found perturbations of ZNF331 expression linked to the IUGR phenotype, together with other 4 (CDKAL1, DHCR24, ILK and PEG10) of the 8 imprinted genes of cluster 2 [10]. These findings further support the role of cluster 2 genes in limiting fetal growth as opposed to cluster 1. ZNF331 is in fact a KRAB zinc-finger protein with a theorized gene silencing power in line with the activity of other such proteins [52]. Genes in cluster 2 mostly belonged to networks dedicated to cell cycle control, cell development and metabolism setting linked to metabolic disorders (IPA networks 3 and 6 – see Table 1).
Despite these novel findings, our study is limited by several factors. The limited sample size and lack of detailed lifestyle/environmental information from the mothers limits our ability to study other influence on fetal growth or imprinting dysregulation. In addition, the three measured outcomes (i.e. head circumference, birth weight and size for gestational age) are only indicative of possible childhood developmental abnormalities and follow-up of the newborn would be needed to further substantiate these results.
In summary clusters 1 and 2 appear to be independently associated with fetoplacental development by exerting different functions. Imprinted genes belonging to these two clusters are represented by proteins and lncRNAs that work to ensure a proper fetal development by assuring cell growth and tissue development (Cluster 1) and promoting an orderly growth (Cluster 2). Our data support the hypothesis that imprinted genes are critical for fetal growth and development; the imprinting expression profile has a potential to be developed into an early biomarker of suboptimal embryonic or fetal growth so that early intervention procedure may be carried out.
Acknowledgments
Statement of Financial Support
This work was supported by the Venture Capital Research Funding Program of the Mount Sinai Children’s Environmental Health Center and Mount Sinai Child Health and Development Institute and by the National Institutes of Health (NIH) grants from the NIEHS [P42ES013660] and the NCRR [P20 RR018728]
Abbreviations
- AGA
Appropriate per Gestational Age
- ICR
Imprinting Control Region
- IPA
Ingenuity Pathway Analysis
- IUGR
Intrauterine Growth Restriction
- LGA
Large per Gestational Age
- MDS
Multidimensional Scaling
- RCN
RNA Copy Number
- RICHS
Rhode Island Child Health Study
- SGA
Small per Gestational Age
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.American Journal of Medical Genetics. Special Issue: Imprinted Genes and Human Disease. Am J Med Genet C Semin Med Genet. 2010;154C (3):317–86. doi: 10.1002/ajmg.c.30268. [DOI] [PubMed] [Google Scholar]
- 2.Sasaky H, Ishino I, editors. Genomic Imprinting: Cytogenet Genome Res. 2006. [Google Scholar]
- 3.Geneimprint. The Genomic Imprinting Website. [database on the Internet] Available from: http://www.geneimprint.com/site/home.
- 4.Catalogue of Parent of Origin Effects [database on the Internet] Available from: http://igc.otago.ac.nz/home.html.
- 5.Maynard ND, Chen J, Stuart RK, Fan JB, Ren B. Genome-wide mapping of allele-specific protein-DNA interactions in human cells. Nat Methods. 2008;5(4):307–9. doi: 10.1038/nmeth.1194. [DOI] [PubMed] [Google Scholar]
- 6.Lewis A, Reik W. How imprinting centres work. Cytogenet Genome Res. 2006;113(1–4):81–9. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- 7.Wagschal A, Feil R. Genomic imprinting in the placenta. Cytogenet Genome Res. 2006;113(1–4):90–8. doi: 10.1159/000090819. [DOI] [PubMed] [Google Scholar]
- 8.Bressan FF, De Bem TH, Perecin F, Lopes FL, Ambrosio CE, Meirelles FV, et al. Unearthing the roles of imprinted genes in the placenta. Placenta. 2009;30(10):823–34. doi: 10.1016/j.placenta.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes. 2007;14(1):3–12. doi: 10.1097/MED.0b013e328013daa2. [DOI] [PubMed] [Google Scholar]
- 10.Diplas AI, Lambertini L, Lee MJ, Sperling R, Lee YL, Wetmur J, et al. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics. 2009;4(4):235–40. doi: 10.4161/epi.9019. [DOI] [PubMed] [Google Scholar]
- 11.Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–6. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 12.Zaitoun I, Downs KM, Rosa GJ, Khatib H. Upregulation of imprinted genes in mice: an insight into the intensity of gene expression and the evolution of genomic imprinting. Epigenetics. 2010;5(2):149–58. doi: 10.4161/epi.5.2.11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 14.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31(1):69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 15.Blake WJ, Kaern M, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–7. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 16.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2(6):e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batada NN, Hurst LD. Evolution of chromosome organization driven by selection for reduced gene expression noise. Nat Genet. 2007;39(8):945–9. doi: 10.1038/ng2071. [DOI] [PubMed] [Google Scholar]
- 18.Desforges M, Sibley CP. Placental nutrient supply and fetal growth. Int J Dev Biol. 2010;54(2–3):377–90. doi: 10.1387/ijdb.082765md. [DOI] [PubMed] [Google Scholar]
- 19.Fowden AL, Forhead AJ, Coan PM, Burton GJ. The placenta and intrauterine programming. J Neuroendocrinol. 2008;20(4):439–50. doi: 10.1111/j.1365-2826.2008.01663.x. [DOI] [PubMed] [Google Scholar]
- 20.Hu D, Cross JC. Development and function of trophoblast giant cells in the rodent placenta. Int J Dev Biol. 2010;54(2–3):341–54. doi: 10.1387/ijdb.082768dh. [DOI] [PubMed] [Google Scholar]
- 21.Davies W, Isles AR, Wilkinson LS. Imprinted gene expression in the brain. Neurosci Biobehav Rev. 2005;29(3):421–30. doi: 10.1016/j.neubiorev.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Keverne EB, Aparicio SA, Ishino F, Barton SC, Surani MA. Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science. 1999;284(5412):330–3. doi: 10.1126/science.284.5412.330. [DOI] [PubMed] [Google Scholar]
- 23.Bonnin A, Goeden N, Chen K, Wilson ML, King J, Shih JC, et al. A transient placental source of serotonin for the fetal forebrain. Nature. 2011;472(7343):347–50. doi: 10.1038/nature09972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeltser LM, Leibel RL. Roles of the placenta in fetal brain development. Proc Natl Acad Sci U S A. 2011;108(38):15667–8. doi: 10.1073/pnas.1112239108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolpert L, Tickle C, Lawrence P, Meyerowitz E, Robertson E, Smith J, et al. Principles of Development. 4. New York, NY – USA: Oxford University Press; 2010. [Google Scholar]
- 26.McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, et al. Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta. 2006;27(6–7):540–9. doi: 10.1016/j.placenta.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Tycko B. Imprinted genes in placental growth and obstetric disorders. Cytogenet Genome Res. 2006;113(1–4):271–8. doi: 10.1159/000090842. [DOI] [PubMed] [Google Scholar]
- 28.Barker DJ. Maternal nutrition, fetal nutrition, and disease in later life. Nutrition. 1997;13(9):807–13. doi: 10.1016/s0899-9007(97)00193-7. [DOI] [PubMed] [Google Scholar]
- 29.Barker DJ. Developmental origins of adult health and disease. J Epidemiol Community Health. 2004;58(2):114–5. doi: 10.1136/jech.58.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson C, Syddall H, Rodin I, Osmond C, Barker DJ. Birth weight and the risk of depressive disorder in late life. Br J Psychiatry. 2001;179:450–5. doi: 10.1192/bjp.179.5.450. [DOI] [PubMed] [Google Scholar]
- 31.Fenton TR. A new growth chart for preterm babies: Babson and Benda’s chart updated with recent data and a new format. BMC Pediatr. 2003;3:13. doi: 10.1186/1471-2431-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambertini L, Diplas AI, Lee MJ, Sperling R, Chen J, Wetmur J. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Epigenetics. 2008;3(5):261–9. doi: 10.4161/epi.3.5.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maccani MA, Padbury JF, Marsit CJ. miR-16 and miR-21 expression in the placenta is associated with fetal growth. PLoS One. 2011;6(6):e21210. doi: 10.1371/journal.pone.0021210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambertini L, Diplas AI, Wetmur J, Lee MJ, Chen J. Evaluation of genomic imprinting employing the analysis of Loss Of Imprinting (LOI) at the RNA level: preliminary results. Eur J Oncol. 2009;14(3):161–9. [Google Scholar]
- 35.van Wassenaer A. Neurodevelopmental consequences of being born SGA. Pediatr Endocrinol Rev. 2005;2(3):372–7. [PubMed] [Google Scholar]
- 36.United States Environmental Protection Agency (US EPA) America’s children and the environment (ACE), measure D1: percentage of children with asthma. Washington (DC): EPA; Nov 19, 2010. Available from: http://www.epa.gov/economics/children/child_illness/d1-graph.html. [Google Scholar]
- 37.America’s children in brief: key national indicators of well-being. Merrifield (VA): The Forum [database on the Internet]; 2010. Available from: http://www.childstats.gov/americaschildren/health.asp. [Google Scholar]
- 38.Overweight and obesity: US obesity trends. Trends by state 1985–2009. Atlanta (GA): CDC; 2011. Mar 3, [database on the Internet]2011. Available from: http://www.cdc.gov/obesity/data/trends.html. [Google Scholar]
- 39.Centers for Disease Control and Prevention (CDC) Prevalence of autism spectrum disorders - Autism and Developmental Disabilities Monitoring Network, United States, 2006. MMWR Surveill Summ. 2009;58(10):1–20. [PubMed] [Google Scholar]
- 40.Boyle CA, Decoufle P, Yeargin-Allsopp M. Prevalence and health impact of developmental disabilities in US children. Pediatrics. 1994;93(3):399–403. [PubMed] [Google Scholar]
- 41.Pastor PN, Reuben CA. Diagnosed attention deficit hyperactivity disorder and learning disability: United States, 2004–2006. Vital Health Stat. 2008;10(237):1–14. [PubMed] [Google Scholar]
- 42.Paulozzi LJ, Erickson JD, Jackson RJ. Hypospadias trends in two US surveillance systems. Pediatrics. 1997;100(5):831–4. doi: 10.1542/peds.100.5.831. [DOI] [PubMed] [Google Scholar]
- 43.Vu LT, Nobuhara KK, Laurent C, Shaw GM. Increasing prevalence of gastroschisis: population-based study in California. J Pediatr. 2008;152(6):807–11. doi: 10.1016/j.jpeds.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 44.Surveillance epidemiology and end results (Homepage) Rockville (MD): 2011. [database on the Internet] Available from: http://seer.cancer.gov. [Google Scholar]
- 45.Trasande L. Quantifying the economic consequences of childhood obesity and potential benefits of interventions. Expert Rev Pharmacoecon Outcomes Res. 2011;11(1):47–50. doi: 10.1586/erp.10.86. [DOI] [PubMed] [Google Scholar]
- 46.Trasande L, Liu Y. Reducing the staggering costs of environmental disease in children, estimated at $76. 6 billion in 2008. Health Aff (Millwood) 2011;30(5):863–70. doi: 10.1377/hlthaff.2010.1239. [DOI] [PubMed] [Google Scholar]
- 47.Simmons R. Perinatal programming of obesity. Semin Perinatol. 2008;32(5):371–4. doi: 10.1053/j.semperi.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bush A. Asthma research: the real action is in children. Paediatr Respir Rev. 2005;6(2):101–10. doi: 10.1016/j.prrv.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 49.Tzortzaki EG, Yang M, Glass D, Deng L, Evan AP, Bledsoe SB, et al. Impaired expression of an organic cation transporter, IMPT1, in a knockout mouse model for kidney stone disease. Urol Res. 2003;31(4):257–61. doi: 10.1007/s00240-003-0318-1. [DOI] [PubMed] [Google Scholar]
- 50.Ballestero JA, Plazas PV, Kracun S, Gomez-Casati ME, Taranda J, Rothlin CV, et al. Effects of quinine, quinidine, and chloroquine on alpha9alpha10 nicotinic cholinergic receptors. Mol Pharmacol. 2005;68(3):822–9. doi: 10.1124/mol.105.014431. [DOI] [PubMed] [Google Scholar]
- 51.Sieb JP, Milone M, Engel AG. Effects of the quinoline derivatives quinine, quinidine, and chloroquine on neuromuscular transmission. Brain Res. 1996;712(2):179–89. doi: 10.1016/0006-8993(95)01349-0. [DOI] [PubMed] [Google Scholar]
- 52.Meiboom M, Murua Escobar H, Pentimalli F, Fusco A, Belge G, Bullerdiek J. A 3. 4-kbp transcript of ZNF331 is solely expressed in follicular thyroid adenomas. Cytogenet Genome Res. 2003;101(2):113–7. doi: 10.1159/000074165. [DOI] [PubMed] [Google Scholar]