Abstract
A method for the selective detection and quantification of peptide:oligonucleotide heteroconjugates, such as those generated by protein:nucleic acid cross-links, using capillary reversed-phase high performance liquid chromatography (cap-RPHPLC) coupled with inductively coupled plasma mass spectrometry detection (ICPMS) is described. The selective detection of phosphorus as 31P+, the only natural isotope, in peptide-oligonucleotide heteroconjugates is enabled by the elemental detection capabilities of the ICPMS. Mobile phase conditions that allow separation of heteroconjugates while maintaining ICPMS compatibility were investigated. We found that trifluoroacetic acid (TFA) mobile phases, used in conventional peptide separations, and hexafluoroisopropanol/triethylamine (HFIP/TEA) mobile phases, used in conventional oligonucleotide separations, both are compatible with ICPMS and enable heteroconjugate separation. The TFA-based separations yielded limits of detection (LOD) of ~40 ppb phosphorus, which is nearly seven times lower than the LOD for HFIP/TEA-based separations. Using the TFA mobile phase, 1-2 pmol of a model heteroconjugate were routinely separated and detected by this optimized capLC-ICPMS method.
Keywords: Inductively coupled plasma mass spectrometry, Protein-nucleic acid cross-links, Elemental phosphorus detection, Liquid chromatography-mass spectrometry, Quantitative analysis
Introduction
Protein:nucleic acid complexes are critical for cell survival and are involved in various cell functions including DNA replication, packaging, repair, and transcription as well as RNA maturation, transport, and translation [1-6]. Chemical and photochemical cross-linking has proven to be a useful approach for covalently linking proteins with nucleic acids to impose distance constraints on such complexes [7-13]. These constraints, when combined with data from biophysical techniques such as X-ray crystallography, nuclear magnetic resonance (NMR), and cryoelectron microscopy, allow researchers to develop more accurate three-dimensional structures, thereby improving our understanding of these biologically significant complexes [14-17].
While a number of biophysical approaches have been used for characterizing protein:nucleic acid cross-links, one of the more promising instrumental methods is the use of mass spectrometry (MS) [12, 18]. The potential advantages of MS for the analysis of cross-links include its ability to analyze high molecular weight complexes either intact or through analysis of enzymatic digestion products, the relatively high sensitivity of MS for peptides and oligonucleotides, and the compatibility of this method with a variety of different cross-linking reagents [8, 12, 19].
Although MS methods are especially useful in identifying the proteins involved in protein:nucleic acid complexes, identifying the specific sites of cross-linking within these complexes is not trivial because of the low abundance of cross-linked species and the high background of non-cross-linked components [20]. A large number of analytical developments to improve the sensitivity and reliability of cross-link detection have been pursued, primarily by Urlaub and co-workers [8, 11, 13, 21-23], along with some work by our group [24-26]. Urlaub and co-workers have utilized Edman degradation combined with matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) [7, 27] or alkaline phosphatase treatment with MALDI-MS [22] to identify cross-linking sites on the protein component of the protein:nucleic acid complex. Reversed-phase high performance liquid chromatography electrospray ionization mass spectrometry (RP-HPLC-ESI-MS) methods that have been explored include the use of multiple reaction monitoring (MRM) scans designed to identify the loss of a metaphosphoric acid group (HPO3, −80 Da), which serves as a signature for peptide:oligonucleotide cross-links present in a mixture of peptides [23], or LC-ESI-MS-based scans utilizing the unique mass defect of peptide:oligonucleotide cross-links to differentiate between peptides and cross-links [25]. Most recently, the use of electron-induced dissociation methods for complete sequencing of electrosprayed peptide: oligonucleotide heteroconjugates was presented [26].
While MALDI- and ESI-MS approaches have been used extensively for characterizing peptide:oligonucleotide heteroconjugates and protein:nucleic acid cross-links, to our knowledge, there have been no prior investigations into the use of inductively coupled plasma mass spectrometry (ICPMS) for such analyses. This discovery was surprising as ICPMS is a powerful analytical tool for element-specific analysis, as the signal response is directly proportional to the element concentration in the sample and this ionization source is not matrix or sequence dependent [28, 29].
Recent ICPMS developments allow for the detection of previously intractable elements such as phosphorus. Quantitation of post-translationally modified proteins (phosphoproteins and sulfated proteins) by phosphorus or sulfur monitoring during LC-ICPMS analysis of proteolytic digests is now commonly performed [28-31]. Similarly, LC-ICPMS monitoring of phosphorus or sulfur can also be used for the identification and quantification of oligodeoxynucleotides and phosphorothioate oligonucleotides [32, 33].
Here we have examined the applicability of LC-ICPMS for identifying and characterizing peptide:oligonucleotide heteroconjugates. This work is based on the premise that such heteroconjugates can be selectively detected by monitoring for elemental phosphorus, which is present in the heteroconjugate from the phosphodiester linkage of the oligonucleotide, using ICPMS. If possible on these heteroconjugates, this instrumental approach could then enable the selective detection and quantification of protein:nucleic acid cross-links. Before pursuing such cross-links, these initial studies have focused on the appropriate chromatographic and instrumental conditions required to detect phosphorus arising from the oligonucleotide component of the heteroconjugate, and to identify the analytical figures of merit for quantitative analysis of phosphorus from such heteroconjugates. Multiple chromatographic conditions were investigated, and a suitable analytical method for characterizing heteroconjugates is demonstrated using a model peptide: oligonucleotide construct.
Materials and Methods
Materials
HPLC/Spectro grade acetonitrile and methanol (MeOH) were purchased from Tedia (Fairfield, OH, USA). HPLC grade triethylamine (TEA), trifluoroacetic acid (TFA), urea, and ammonium bicarbonate were from Fisher Scientific (Fairlawn, NJ, USA). Bovine serum albumin (BSA), 1,1,1,3,3,3 hexafluoro-2-propanol (HFIP), neurotensin, bradykinin, Substance P fragment 2-11, adrenocorticotropic hormone (ACTH), dithiothreitol, iodoacetamide, and dT10 were obtained from Sigma-Aldrich (St. Louis, MO, USA). RNase T1 (Roche Molecular Biochemicals, Indianapolis, IN, USA) was purified and used as described previously [34]. Sequence grade modified trypsin was purchased from Promega (Madison, WI, USA) and used per the manufacturer’s instructions. Pp60c-src was from American Peptide Company (Sunnyvale, CA, USA). The model heteroconjugate was an 11-amino acid reside peptide conjugated to a 5-mer oligonucleotide via a hexylaminolinker on the aspartic residue (Ac-GARGAD(agcca)RAVLA-NH2) obtained from BiomerTech (Hayward, CA, USA) [25].
HPLC
HPLC was carried out on an Agilent 1200 series system (Santa Clara, CA, USA) equipped with a vacuum degasser, binary pump, a thermostatically controlled well plate auto-sampler, a thermostatically controlled column compartment, a diode array detector, and a 500 nL flow cell. All lines within the capillary system were 50 μm i.d. PEEK fused silica tubing. An Agilent Zorbax 300 Extend C18 capillary column (0.3×100 mm) with a pore size of 300 Å and 3.5 μm particles was used.
The TFA-based mobile phase consisted of mobile phase A of water with 0.05% TFA and mobile phase B of 0.045% TFA in acetonitrile. Gradient conditions, at a flow rate of 10 μL min−1, including column equilibration, were 5 min at 0% B followed by a linear ramp to 60% B at 55 min. The gradient was held at 60% B for 5 min before ramping to 100% B in 5 min. After holding at 100% B for 5 min, the gradient was linearly decreased to 0% B in 5 min and held there for 20 min in preparation for the next run.
The HFIP/TEA mobile phase consisted of mobile phase A composed of 150 mM HFIP and 4 mM TEA in water, while mobile phase B was 150 mM HFIP and 4 mM TEA in methanol. Gradient conditions at a flow rate of 8 μL min−1 and column temperature of 50 °C were a linear ramp from 0% B to 10% B in 2 min followed by a ramp to 20% B in 13 min. After holding at 20% B for 5 min, the gradient was ramped to 60% B in 5 min and then 100% B in 5 min. The gradient was held at 100% B for 10 min before decreasing to 0% B in 5 min. The gradient was held at 0% B for 20 min in preparation for the next run.
Inductively Coupled Plasma Mass Spectrometer
ICPMS experiments were conducted on an Agilent 7700x ICPMS (Agilent Technologies, Santa Clara, CA), equipped with an octopole reaction system (ORS), which is composed of an on-axis rf-only octopole that produces a tight ion beam. The ICPMS included a shield torch, an octopole cell with pressurized helium gas (purity of 99.999%), a quadrupole mass analyzer and an electron multiplier. Interference removal is accomplished by the ORS through kinetic energy discrimination. The polyatomic spectral interferences are eliminated when pressurized gas fills the cell and collides or reacts with such molecules resulting in a loss of kinetic energy. The sample eluting from the HPLC was introduced into the plasma at a flow rate of 8–10 μL min−1 through a DS-5 micro-concentric nebulizer (MCN) purchased from CETAC Technologies (Omaha, NE, USA). Phosphorus was monitored at m/z 31. Instrumental parameters used were RF forward power, 1550, plasma Ar gas flow rate, 15.0 Lmin−1, carrier Ar gas flow rate, 1.10 Lmin−1, and helium gas flow rate, 4.0 ml min−1. The resulting data were analyzed using Agilent MassHunter ICPMS software.
Results and Discussion
The use of RP-HPLC-ICPMS for the detection of peptide: oligonucleotide heteroconjugates is quite distinct from MALDI- or ESI-MS approaches. ICPMS enables element-specific detection (e.g., phosphorus and/or sulfur), minimal matrix interferences, and quantitative analysis as the element response is directly proportional to the total mass of that element in the sample. In developing an LC-based ICPMS method for covalently linked heteroconjugate detection, the RP-HPLC conditions required to adequately retain a heteroconjugate within a complex sample that are compatible with the ICP source must be identified.
There are specific concerns that can arise when LC methods are coupled to ICPMS. As the ionization step in ICPMS first requires vaporization and atomization of the sample solution, the use of high amounts of organic solvents can lead to plasma instability and carbon deposition on the interface cones. Because the separation of biomolecules, such as peptides and oligonucleotides, using RP-HPLC typically requires high concentrations of organic solvent or buffer to elute the sample from the column, care must be taken to reduce the overall volume of LC eluent introduced into the ICP source. Thus, we first examined both peptide-based RP-HPLC conditions, where TFA is present in the mobile phase, and conventional oligonucleotide ion-pairing RP-HPLC conditions, where HFIP and TEA are the major components of the mobile phase, using cap-LC to ensure that the mobile phase conditions amenable to ICP-MS [31, 32] would adequately separate the biomolecules of interest. The acidic mobile phases and solution conditions investigated have been utilized previously with heteroconjugates, such as that used here as a model system, and were not found to affect the integrity of the heteroconjugate [13, 21, 25, 26].
HPLC Retention Characteristics
Initial investigations into the ability of various HPLC mobile phases to separate and retain a mixture of peptides and oligonucleotides in the same sample were performed using diode array, rather than ICPMS, detection. A TFA-based mobile phase and C18 stationary phase were used as typical peptide-like HPLC conditions. To initially investigate these conditions, several standard peptides including bradykinin, ACTH and neurotensin, along with a tryptic digest of bovine serum albumin and an oligodeoxynucleotide (dT10) were used as representative samples. Not surprisingly, under these HPLC conditions, all of the peptides investigated could be retained on the column while the oligodeoxynucleotide eluted in the void volume (data not shown). Similarly, when these same samples were analyzed using HPLC mobile phases typically used for oligonucleotide separation, only the oligodeoxynucleotide was significantly retained on the same C18 column (data not shown).
Consistent with previous results from LC-ESI-MS [25], the nature of the heteroconjugate will dictate the optimal chromatographic conditions. A heteroconjugate that is more “peptide-like,” that is, one where the peptide component is large and the oligonucleotide component is small, is best analyzed using conventional peptide-like HPLC conditions such as TFA. Peptide-like heteroconjugates should be retained on the stationary phase effectively under these conditions. Conversely, a heteroconjugate that is more “oligonucleotide-like,” that is one where the oligonucleotide component is large and the peptide component is small, is best analyzed using oligonucleotide HPLC conditions.
Phosphorus Detection and Matrix Effects
While the initial investigations into HPLC conditions were conducted to verify the applicability of such conditions for samples containing peptide- and oligonucleotide-like components, detection was performed using UV spectroscopy. The next series of experiments focused on the applicability of these mobile phases using ICPMS for element-specific detection of heteroconjugates. Here, phosphorus was chosen as the element to be monitored at m/z 31 (31P+) for heteroconjugate detection. Phosphorus is present from the oligonucleotide component of the heteroconjugate and, except when phosphopeptides are present, background interferences are expected to be minimal, provided the ORS is used. LC-ICPMS conditions were first optimized using Pp60, a phosphorylated peptide, as the model compound for peptide-like HPLC conditions and dT10 as the model compound for oligonucleotide-like HPLC conditions, after which the effects of background non-phosphorus-containing species on the identification of these model compounds was pursued.
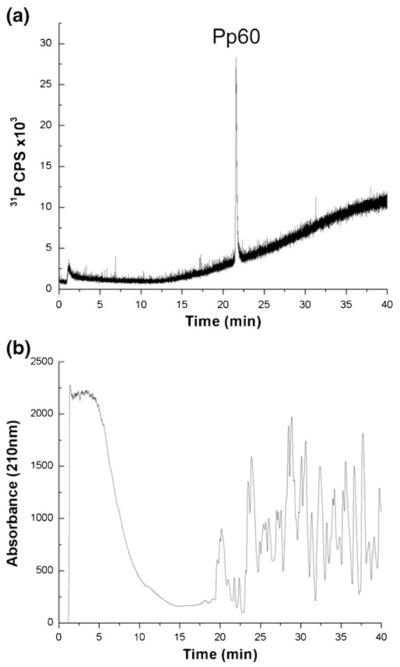
A tryptic digest of BSA was chosen as a representative sample matrix to examine whether a background of non-phosphorylated peptides would interfere with the detection of phosphorus-containing samples, such as a heteroconjugate, during ICPMS analysis. The interest here was to identify whether heteroconjugates would need to be purified extensively from uncross-linked protein samples prior to LC-ICPMS. BSA does not contain any sites of phosphorylation, and there was no ICPMS signal from 31P+ detected when running a tryptic digest of BSA as a control (data not shown). Using TFA mobile phase conditions, Pp60 was spiked into a tryptic digest of BSA, and detected using ICPMS. As shown in Figure 1a, the 31P chromatogram of the ICPMS gives rise to only the phosphorus signal attributable to the Pp60, eluting at 25 min. By way of comparison, the UV chromatogram (detection at 210 nm) of the same run (Figure 1b) reveals numerous eluting peaks arising from the tryptic peptides from BSA as well as the spiked Pp60.
Figure 1.
Separation of Pp60 and digested BSA using TFA-based mobile phase conditions. (a) Pp60 detected by monitoring m/z 31 (P+), (b) UV absorbance at 210 nm
Additional experiments were performed where the amount of Pp60 injected on column was held constant at 32.2 pmol, while the amount of the BSA tryptic digest was varied from 7.2 to 21 pmol of tryptic digest loaded on column. As shown in Table 1, the addition of the tryptic digest of BSA as a background had a negligible effect on the phosphorus response from Pp60 with a deviation of <10% for all amounts of the BSA digest investigated. Thus, we conclude that the use of the TFA-based mobile phase during LC-ICPMS enables the element-specific detection of phosphorus in the presence of a non-phosphorus-containing peptide background. Although the sample to matrix ratio investigated here is likely higher than what may be expected from isolation of cross-links from biological systems [13], these findings are consistent with other ICPMS results from a similar instrument for low abundance phosphopeptide detection [31].
Table 1.
Effect of Peptide Background, from Tryptic Digest of BSA, on Measured Phosphorus Response for Pp60 in TFA Mobile Phase Conditions and dT10 in HFIP/TEA Mobile Phase Conditions
| BSA (pmol) | Pp60 Area (cps) | dT10 Area (cps) |
|---|---|---|
| 21 | 16153 | 5239 |
| 14 | 16916 | 5474 |
| 10 | 19040 | 5595 |
| 8.5 | 17573 | 5363 |
| 7.2 | 20516 | 5467 |
| %RSD | 9.7 | 3.7 |
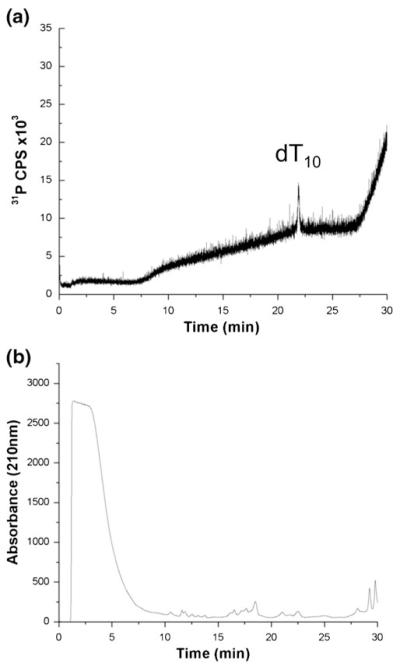
In a similar manner, LC-ICPMS with phosphorus-specific detection was investigated using HFIP/TEA mobile phase conditions. dT10 was spiked into a tryptic digest of BSA and detected using ICPMS. As shown in Figure 2a, the phosphorus chromatogram from the ICPMS only reveals the phosphorus signal attributable to dT10, which elutes at 12 min. However, the UV chromatogram with detection at 210 nm (Figure 2b) of the same run reveals a large background signal in the first 5 min, presumably attributable to unretained or slightly retained peptides, and only a few discernable peaks in the chromatogram after 10 min. These experiments confirm that LC-ICPMS can be used to selectively detect phosphorus (from dT10) even in a sample matrix composed of numerous other peptides.
Figure 2.
Separation of dT10 and digested BSA using HFIP/TEA mobile phase conditions. (a) dT10 detected by monitoring m/z 31 (P+), (b) UV absorbance at 210 nm
Next, the effect of sample-related matrix effects were investigated using the HFIP/TEA mobile phase. The amount of dT10 loaded on column was held constant at 3.6 pmol, and a variable amount of tryptic peptides from BSA ranging from 7.2 to 21 pmol were also loaded on column. As seen in Table 1, the addition of tryptic peptides as a background for dT10 detection had a minimal effect, with a deviation of <4% for the phosphorus signal. Thus, we conclude that the use of HFIP/TEA as a mobile phase during LC-ICPMS enables the element-specific detection of phosphorus irrespective of the amount of non-phosphorus-containing peptide background. Taken together, neither mobile phase conditions seemed adversely affected by sample matrix effects allowing either to be used for the LC-ICPMS detection of phosphorus from peptide:oligonucleotide heteroconjugates.
Quantitative Analysis of Phosphorus by HPLC-ICPMS
The final evaluation of HPLC-ICPMS conditions for heteroconjugate characterization focused on the quantitative measurement of phosphorus within the sample. The same standards for the TFA and HFIP/TEA mobile phases as before were used for these investigations. The experimental procedure involved using known amounts of Pp60 or dT10 loaded on column and measuring the ICPMS response for phosphorus. Response curves were generated from peak area measurements of phosphorus from which the limits of detection (LOD) and quantification (LOQ) for phosphorus could be determined. The LODs and LOQs were calculated using IUPAC-based calculations of 3σ and 10σ, respectively, where σ is the calculated standard deviation of the measurements [35].
Response curves for Pp60 analyzed using the TFA mobile phase and dT10 analyzed using the HFIP/TEA mobile phase were generated (data not shown). Using the TFA mobile phase, the LOD determined for the amount of phosphorus signal is 41 ppb P, which correlates to 1.3 pmol Pp60 loaded on column, and a corresponding LOQ of 137 ppb P equivalent to 4.4 pmol Pp60 loaded on column. Using the HFIP/TEA mobile phase, the LOD determined for the amount of phosphorus was 280 ppb, which corresponds to 1.0 pmol of dT10 loaded on column, and a corresponding LOQ of 936 ppb P equivalent to 3.4 pmol of dT10 loaded on column. These data show that the TFA mobile phase yields lower LODs and LOQs for phosphorus than the HFIP/TEA mobile phase, which may arise from effects in the source region due to the differences in the mobile phase compositions. Because dT10 contains nine phosphorus atoms per molecule, the amount of sample required to load on column is reduced, thus explaining why the sample loading was equivalent between Pp60 and dT10 even though the LOD for phosphorus is lower for the TFA conditions. These results also suggest that a larger oligonucleotide component (i.e., more phosphorus per oligonucleotide component) would be advantageous for ICPMS detection when using the HFIP/TEA mobile phase conditions. The LOD values obtained here (≤ μmol concentration) are comparable to values previously reported for cross-link or heteroconjugate characterization by LC-ESI-MS [16-21], and additional investigation of alternative nebulization sources may improve these LOD values further [32].
Model Heteroconjugate
The preceding experiments demonstrated that both TFA and HFIP/TEA conditions are suitable for LC-ICPMS analysis of phosphorus-containing compounds. As expected, TFA-based conditions would be applicable for characterizing heteroconjugates that contain a larger peptide coupled to a smaller oligonucleotide, while HFIP/TEA conditions would be preferred when the heteroconjugate contains a larger oligonucleotide and only one or a few amino acids. As neither mobile phase composition is affected by non-phosphorus-containing background ions and the quantitative capabilities are similar, the optimal set of LC-ICPMS experimental conditions can be dictated by the heteroconjugate of interest, thereby providing significant experimental flexibility for analytical use.
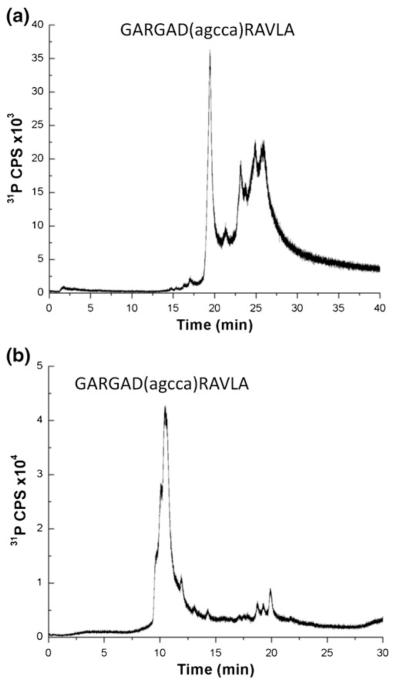
Because the optimization and characterization studies were conducted on simple non-heteroconjugate standards, to confirm that these findings hold for heteroconjugate analysis, the next experiments were performed using the unpurified synthetic reaction mixture of an 11-amino acid residue peptide conjugated to a 5-mer oligonucleotide via a hexlyaminolinker on the aspartic residue (Ac-GARGAD (agcca)RAVLA-NH2) [25, 26] as the demonstration system. The reaction mixture was analyzed by LC-ICPMS using TFA (Figure 3a) and HFIP/TEA (Figure 3b) mobile phases with all other experimental conditions equivalent to those used in the initial optimization studies.
Figure 3.
LC-ICPMS analysis of unpurified heteroconjugate (Ac-GARGAD(agcca)RAVLA-NH2 with peptide residues in upper case and nucleotide resides in lower case) using (a) TFA and (b) HFIP/TEA mobile phase conditions with detection by monitoring phosphorus at m/z 31 (P+)
As expected, both HPLC conditions were appropriate for retaining the heteroconjugate components, although the chromatographic profiles differ between the two mobile phases used. In Figure 3a, the major peak corresponding to the heteroconjugate elutes near 20 min, followed by several additional later-eluting peaks in the RP ion-pairing chromatogram. These additional peaks arise from incomplete deprotection of the peptide, which has previously been shown to yield a similar reversed-phase HPLC during UV detection [25], and thus represent different peptide components, each containing the same oligonucleotide. As the protecting groups increase the hydrophobicity of the peptide, these additional species are detected as later eluting components.
In contrast, using HFIP/TEA conditions, which separate based on the oligonucleotide characteristics of the heteroconjugate, only one major peak is detected eluting around 11 min (Figure 3b). This peak is not well resolved, with leading shoulders. Unlike the chromatogram obtained using TFA conditions in Figure 3a, here the different peptide components cannot be resolved by the HFIP/TEA mobile phase. The smaller set of peaks eluting around 20 min may be due to aggregation of the heteroconjugate due to the amount of sample loaded, although these peaks were not investigated further.
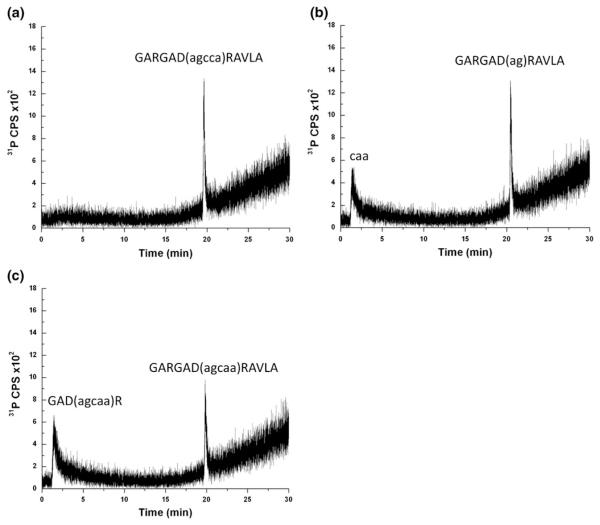
Finally, to further examine the influence of oligonucleotide size on peptide-based separations, the heteroconjugate sample was purified as described previously [25], digested with either trypsin or RNase T1, which cleaves oligonucleotides at unmodified guanosine residues, to generate a mixture of analytes where either the peptide or oligonucleotide component differs in length from the purified heteroconjugate. These samples were then analyzed by LC-ICPMS using the TFA mobile phase conditions. Figure 4a is the ICPMS data obtained when monitoring for phosphorus. As expected for the purified heteroconjugate, only a single peak eluting at 19.6 min was detected, consistent with the data obtained from the unpurified heteroconjugate sample analyzed in Figure 4a. Integrating the phosphorus signal over this peak area yielded a value of 2331 cps, which, based on the response curve for phosphorus under these chromatographic conditions, corresponds to ~175 ppb P or ~2.2 pmol of heteroconjugate.
Figure 4.
LC-ICPMS analysis using TFA mobile phase conditions with detection by monitoring phosphorus at m/z 31 (P+). (a) Sample injected is purified heteroconjugate (Ac-GARGAD(agcca)RAVLA-NH2 with peptide residues in upper case and nucleotide resides in lower case). (b) Sample injected is RNase T1 digest of purified heteroconjugate. (c) Sample injected is tryptic digest of purified heteroconjugate. Approximately 10-fold less purified heteroconjugate was used for these analyses compared to the analyses in Figure 3
Digestion of the heteroconjugate using RNase T1 is predicted to generate two products: the trinucleotide 5′-CpApA-3′ and the intact peptide now containing a dinucleotide component [(Ac-GARGAD(ag)RAVLA-NH2]. LC-ICPMS analysis of the RNase T1 digest of the heteroconjugate is presented in Figure 4b. The major peak, corresponding to the heteroconjugate now containing a dinucleotide, elutes at 20.4 min, and a small peak elutes at 1.4 min, which is near the void volume for the column. In this case, the new heteroconjugate elutes at a statistically significant later elution time, likely due to the increase in hydrophobicity of this heteroconjugate as the oligonucleotide component has been reduced in size. The additional peak eluting near the void volume is consistent with the trinucleotide that results after RNase T1 digestion. As discussed earlier, the trinucleotide would not be retained under these RP-HPLC conditions due to the polarity of the sample and the lack of ion pairing reagent in the mobile phase. A quantitative characterization of the RNase T1 digest of the heteroconjugate can be done by integrating both phosphorus peak areas. The phosphorus signal for the trinucleotide eluting at 1.4 min yielded a value of 1063 cps (~80 ppb P), and the heteroconjugate with the dinucleotide remaining attached yielded a value of 1157 cps (~87 ppb P, ~1.9 pmol of digested heteroconjugate). Not unexpectedly, given that both peaks containing two phosphorus atoms per oligonucleotide, the detected ICPMS response is approximately equivalent and half that from the intact heteroconjugate.
Digestion of the heteroconjugate with trypsin is predicted to generate a smaller peptide still conjugated to the oligonucleotide component [GAD(agcaa)R] along with two smaller peptides that would not be conjugated to the oligonucleotide (GAR and AVLA), which cannot be detected by the LC-ICPMS method described here. Analysis of the tryptic digest of the heteroconjugate by LC-ICPMS (Figure 4c) yields two peaks. The major peak, eluting at 19.8 min, corresponds to the starting heteroconjugate that was not digested by trypsin. Interestingly, the additional peak also elutes very early in the chromatographic method, 1.4 min, which is near the void volume of the column. Because this method can only detect phosphorus, this early eluting peak is a tryptic peptide that remains conjugated to the oligonucleotide. The identity of this tryptic product cannot be determined by ICPMS analysis, although several possibilities exist: GAD(agcaa)R, GARGAD(agcaa)R, and GAD(agcaa)RAVLA. Based on the early retention time for this product using the TFA mobile phase, the most likely product is that with the smallest peptide component [GAD (agcaa)R], although further analysis would be required to confirm that identification.
A possible interpretation for the tryptic peptide-oligonucleotide component eluting so early is that the chromatographic behavior of the heteroconjugate is no longer governed primarily by the peptide component; thus, the overall polarity and low hydrophobicity of such heteroconjugates leads to a lack of retention with the TFA mobile phase. Again, because ICPMS detection allows for a quantitative comparison of the phosphorus signal from both samples, the two peak areas were integrated, yielding a value of 1263 cps (~ 95 ppb P, ~1.2 pmol digested heteroconjugate) for the peak eluting at 1.4 min and 1181 cps (~ 88 ppb P, ~1.1 pmol undigested heteroconjugate) for the peak eluting at 19.8 min. Overall, analysis of the purified heteroconjugate and various enzymatic digests illustrates that the LC-ICPMS method can be used for the qualitative identification of peptide:oligonucleotide heteroconjugates with high selectivity and provides useful semiquantitative information relating to the abundance of each component in the sample (assuming the number of phosphates per heteroconjugate is known).
Conclusions
An optimized capillary RP-HPLC-ICPMS method for the selective detection of peptide:oligonucleotide heteroconjugates has been developed. ICPMS enables the selective detection of phosphorus at m/z 31, which is present in the phosphodiester linkage of the oligonucleotide component in this heteroconjugate. Selective detection of heteroconjugates and, hence, protein:nucleic acid cross-links, is of particular interest owing to the typical low cross-link yields in a background of much more abundant uncross-linked protein and/or nucleic acid. Because we have shown that either TFA-based or HFIP/TEA-based mobile phases, conventionally used for peptide and oligonucleotide RP-HPLC, respectively, are effective at retaining and separating heteroconjugates when coupled with ICPMS, the experimental design and implementation of a capLC-ICPMS method can be adapted based on the anticipated cross-link properties to be analyzed. A variety of enzymatic digestions can be used to generate cross-links where the anticipated peptide component is larger than the oligonucleotide component, thereby allowing for a TFA-based separation of the crosslink, with uncross-linked oligonucleotides eluting in the void volume. Alternatively, digestions can be manipulated to generate cross-links where the anticipated oligonucleotide component is larger than the peptide component, allowing for HFIP/TEA-based separation of the cross-link, with the added benefit that the sensitivity of ICPMS detection is based on the phosphorus response, which will increase with increasing size of the oligonucleotide.
A limitation of the developed method is that LC-ICPMS, alone, cannot be used to identify the peptide or oligonucleotide components nor the site of cross-linking. For those identifications, additional characterization using MALDI- or ESI-MS based methods would be required. Instead, the LC-ICPMS method developed in this work is envisioned as being a new option for screening protein:nucleic acid cross-link mixtures to identify the number and relative abundance of cross-links that are present. The selectivity of ICPMS can be used to verify the presence of a cross-link (through the 31P response), and because ICPMS is a quantitative technique, the amount of cross-linked material in a sample can be estimated. In addition, while 31P was used here for detection because of the phosphodiester linkages that are present in oligonucleotides, one can envision incorporating other ICPMS-active elements into the cross-linking reagent (e.g., sulfur or platinum) to improve the sensitivity and selectivity for cross-link detection. Furthermore, by using the selective detection of ICPMS, this method could serve as a detector (via split flow) in the first step of the purification of cross-links away from any uncross-linked components, with subsequent identification of the peptide and oligonucleotide components, along with the site of cross-linking, by MALDI- or ESI-based methods. Finally, such a combined approach for protein:nucleic acid cross-link analysis would be equally applicable to either DNA- or RNA-based protein: nucleic acid complexes.
Acknowledgments
The authors acknowledge financial support for this work by the National Institutes of Health (GM58843 to P.A.L.). The authors thank Jessica Kotha for purifying the heteroconjugate sample used in this work and Agilent Technologies for their continued support and instrumentation.
References
- 1.Falkenberg M, Larsson N-G, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu. Rev. Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A, Lindsey-Boltz LA, Uensal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 3.Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 4.Staley JP, Woolford JLJ. Assembly of ribosomes and spliceosomes: Complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 2009;21:109–118. doi: 10.1016/j.ceb.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hager GL, McNally JG, Misteli T. Transcription Dynamics. Mol. Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson RJ, Hellen CUT, Pestova TV. The Mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urlaub H, Hartmuth K, Kostka S, Grelle G, Lührmann R. A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA-protein contacts in native U1 and U4/U6. U5 snRNPs. J. Biol. Chem. 2000;275:41458–41468. doi: 10.1074/jbc.M007434200. [DOI] [PubMed] [Google Scholar]
- 8.Urlaub H, Hartmuth K, Lührmann R. A two-tracked approach to analyze RNA-protein cross-linking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods. 2002;26:170–181. doi: 10.1016/S1046-2023(02)00020-8. [DOI] [PubMed] [Google Scholar]
- 9.Back JW, Notenboom V, de Koning LJ, Muijsers AO, Sixma TK, de Koster CG, de Jong L. Identification of cross-linked peptides for protein interaction studies using mass spectrometry and 18O labeling. Anal. Chem. 2002;74:4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 10.Back JW, de Jong L, Muijsers AO, de Koster CG. Chemical cross-linking and mass spectrometry for protein structural modeling. J. Mol. Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 11.Kühn-Hölsken E, Lenz C, Sander B, Lührmann R, Urlaub H. Complete MALDI-TOF MS analysis of cross-linked peptide-RNA oligonucleotides derived from nonlabeled UV-irradiated ribonucleoprotein particles. RNA. 2005;11:1915–1930. doi: 10.1261/rna.2176605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urlaub H, Kuhn-Holsken E, Luhrmann R. Analyzing RNA-protein cross-linking sites in unlabeled ribonucleoprotein complexes by mass spectrometry. Methods Mol. Biol. 2008;488:221–245. doi: 10.1007/978-1-60327-475-3_16. [DOI] [PubMed] [Google Scholar]
- 13.Kramer K, Hummel P, Hsiao H-H, Luo X, Wahl M, Urlaub H. Mass spectrometric analysis of proteins cross-linked to 4-thiouracil and 5-bromouracil substituted RNA. Int. J. Mass Spectrom. 2011;304:184–194. [Google Scholar]
- 14.Campbell ID. Timeline: The march of structural biology. Nat. Rev. Mol. Cell Biol. 2002;3:377–381. doi: 10.1038/nrm800. [DOI] [PubMed] [Google Scholar]
- 15.Christodoulou J, Larsson G, Fucini P, Connell SR, Pertinhez TA, Hanson CL, Redfield C, Nierhaus KH, Robinson CV, Schleucher J, Dobson CM. Heteronuclear NMR investigations of dynamic regions of intact Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10949–10954. doi: 10.1073/pnas.0400928101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank J. Cryo-electron microscopy as an investigative tool: The ribosome as an example. BioEssays. 2001;23:725–732. doi: 10.1002/bies.1102. [DOI] [PubMed] [Google Scholar]
- 17.Bushnell DA, Westover KD, Davis RE, Kornberg RD. Structural basis of transcription: An RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms. Science (Washington, DC) 2004;303:983–988. doi: 10.1126/science.1090838. [DOI] [PubMed] [Google Scholar]
- 18.Jensen ON, Kulkarni S, Aldrich JV, Barofsky DF. Characterization of peptide-oligonucleotide heteroconjugates by mass spectrometry. Nucleic Acids Res. 1996;24:3866–3872. doi: 10.1093/nar/24.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 20.Rusconi F, Guillonneau F, Praseuth D. Contributions of mass spectrometry in the study of nucleic acid-binding proteins and of nucleic acid-protein interactions. Mass Spectrom. Rev. 2002;21:305–348. doi: 10.1002/mas.10036. [DOI] [PubMed] [Google Scholar]
- 21.Richter FM, Hsiao H-H, Plessmann U, Urlaub H. Enrichment of protein-RNA cross-links from crude UV-irradiated mixtures for MS analysis by on-line chromatography using titanium dioxide columns. Biopolymers. 2009;91:297–309. doi: 10.1002/bip.21139. [DOI] [PubMed] [Google Scholar]
- 22.Kühn-Hölsken E, Dybkov O, Sander B, Lührmann R, Urlaub H. Improved identification of enriched peptide RNA cross-links from ribonucleoprotein particles (RNPs) by mass spectrometry. Nucleic Acids Res. 2007;35:e95. doi: 10.1093/nar/gkm540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenz C, Kühn-Hölsken E, Urlaub H. Detection of protein-RNA cross-links by NanoLC-ESI-MS/MS using precursor ion scanning and multiple reaction monitoring (MRM) experiments. J. Am. Soc. Mass Spectrom. 2007;18:869–881. doi: 10.1016/j.jasms.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Brock JE, Pourshahian S, Giliberti J, Limbach PA, Janssen GR. Ribosomes bind leaderless mRNA in Escherichia coli through recognition of their 5′-terminal AUG. RNA. 2008;14:2159–2169. doi: 10.1261/rna.1089208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pourshahian S, Limbach PA. Application of fractional mass for the identification of peptide-oligonucleotide cross-links by mass spectrometry. J. Mass Spectrom. 2008;43:1081–1088. doi: 10.1002/jms.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krivos KL, Limbach PA. Sequence analysis of peptide:oligonucleotide heteroconjugates by electron capture dissociation and electron transfer dissociation. J. Am. Soc. Mass Spectrom. 2010;21:1387–1397. doi: 10.1016/j.jasms.2010.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urlaub H, Raker VA, Kostka S, Lührmann R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001;20:187–196. doi: 10.1093/emboj/20.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruger R, Kubler D, Pallisse R, Burkovski A, Lehmann WD. Protein and proteome phosphorylation stoichiometry analysis by element mass spectrometry. Anal. Chem. 2006;78:1987–1994. doi: 10.1021/ac051896z. [DOI] [PubMed] [Google Scholar]
- 29.Navaza AP, Encinar JR, Sanz-Medel A. Absolute and accurate quantification of protein phosphorylation by using an elemental phosphorus standard and element mass spectrometry. Angew. Chem. Int. Ed. 2007;46:569–571. doi: 10.1002/anie.200602517. [DOI] [PubMed] [Google Scholar]
- 30.Zinn N, Hahn B, Pipkorn R, Schwarzer D, Lehmann WD. Phosphorus-based absolutely quantified standard peptides for quantitative proteomics. J. Proteome Res. 2009;8:4870–4875. doi: 10.1021/pr900494m. [DOI] [PubMed] [Google Scholar]
- 31.Ellis J, Grimm R, Clark JF, Pyne-Gaithman G, Wilbur S, Caruso JA. Studying protein phosphorylation in low MW CSF fractions with capLC-ICPMS and nanoLC-CHIP-ITMS for identification of phosphoproteins. J. Proteome Res. 2008;7:4736–4742. doi: 10.1021/pr800294r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lokits KE, Limbach PA, Caruso JA. Interfaces for capillary LC with ICPMS detection: A comparison of nebulizers/spray chamber configurations. J. Anal. At. Spectrom. 2009;24:528–534. [Google Scholar]
- 33.Easter RN, Kroning KK, Caruso JA, Limbach PA. Separation and identification of oligonucleotides by hydrophilic interaction liquid chromatography (HILIC)-inductively coupled plasma mass spectrometry (ICPMS) Analyst. 2010;135:2560–2565. doi: 10.1039/c0an00399a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hossain M, Limbach PA. Multiple endonucleases improve MALDI-MS signature digestion product detection of bacterial transfer RNAs. Anal Bioanal Chem. 2009;394:1125–1135. doi: 10.1007/s00216-008-2562-2. [DOI] [PubMed] [Google Scholar]
- 35.McNaught AD, Wilkinson A, IUPAC . Compendium of Chemical Terminology. 2nd ed. Blackwell Scientific Publications; Oxford: 1997. p. 450. the “Gold Book”. [Google Scholar]