Abstract
The classical visual and beta arrestins belong to a larger family of proteins that likely share structural similarity. Humans have an additional six related proteins sometimes termed the alpha-arrestins, whose functions are now emerging. Surprisingly, several alpha-arrestins play prominent roles in the regulation of metabolism and obesity. One alpha-arrestin, thioredoxin interacting protein (Txnip) has critical functions in regulating glucose uptake and glycolytic flux through the mitochondria. Another alpha-arrestin, Arrdc3, is linked to obesity in men and was recently identified in mice as a regulator of body mass, adiposity, and energy expenditure. Here we discuss recent evidence suggesting potential common themes for all arrestins, including physiological roles for classical arrestins in metabolism and functions of alpha-arrestins in receptor signaling and endocytosis.
Beta-Arrestins: Signaling Proteins that Control Metabolism
Arrestins were originally identified as intracellular proteins that bind phosphorylated G-protein coupled receptors and cause their desensitization, thus “arresting” their activation. There are two types of these classical arrestins: visual arrestins and beta-arrestins. Visual arrestins are characterized by their limited localization in the photoreceptor cells of the retina and function in transduction of light by specifically binding to and desensitizing active, phosphorylated opsins [1, 2]. Beta-arrestins (beta-arrestin-1 and beta-arrestin-2) were named for their role in the desensitization of the beta-adrenergic receptors, but unlike the visual arrestins, they can bind to and desensitize a wide range of G-protein-coupled receptors [2, 3]. Recruitment of a beta-arrestin to a G-protein coupled receptor is consistent enough that it is frequently used as a proxy for receptor activation [4].
The beta-arrestins have surprising promiscuity in regulating signaling, and the list of signaling systems that require beta-arrestins for downstream events continues to grow. Beta-arrestins can bind to and cause downregulation of unconventional seven-transmembrane-spanning receptors (7TMRs) such as the Smoothened and Frizzled receptors that mediate Hedgehog and Wnt signaling [5–8]. Importantly, beta-arrestins are not limited to functioning as endocytic adaptor proteins: they can be required for recruitment of a variety of downstream signaling proteins in response to receptor activation but independent of G-protein signaling [9–11]. Beta-arrestins can also bind to receptors that are not members of the 7TMR superfamily, including TGF-beta family receptors [12] and cytokine receptors such as TNF-α, where beta-arrestins appear to be primarily involved in activating MAP kinases signaling cascades rather than inducing clathrin-mediated endocytosis of the receptors [13].
Although this essential role for beta-arrestins in multiple types of receptor signaling is well established, important physiological roles in metabolism have only recently emerged. In mouse models of diabetes and obesity (db/db mice and high-fat feeding), the beta-arrestins were found to be downregulated in liver and skeletal muscle. A causal role for beta-arrestin-2 in metabolism was then identified by the demonstration that beta-arrestin-2 knock-out mice are insulin-resistant, and that this phenotype is rescued by gene transfer of beta-arrestin-2. [14].
Evidence from multiple laboratories suggests that beta-arrestins directly regulate insulin receptor signaling [14, 15]. Luan and colleagues have shown that upon insulin stimulation, beta-arrestin-2 binds to the insulin receptor and is required for recruiting the tyrosine-protein kinase Src and the serine/threonine protein kinase Akt to the activated receptor [14]. Although these studies suggest that beta-arrestins have specific signaling capabilities that play a role in metabolic dysfunction, it remains unclear how the beta-arrestins are able to coordinately affect physiology despite interacting with a broad set of receptors from multiple receptor families.
The Arrestin Superfamily
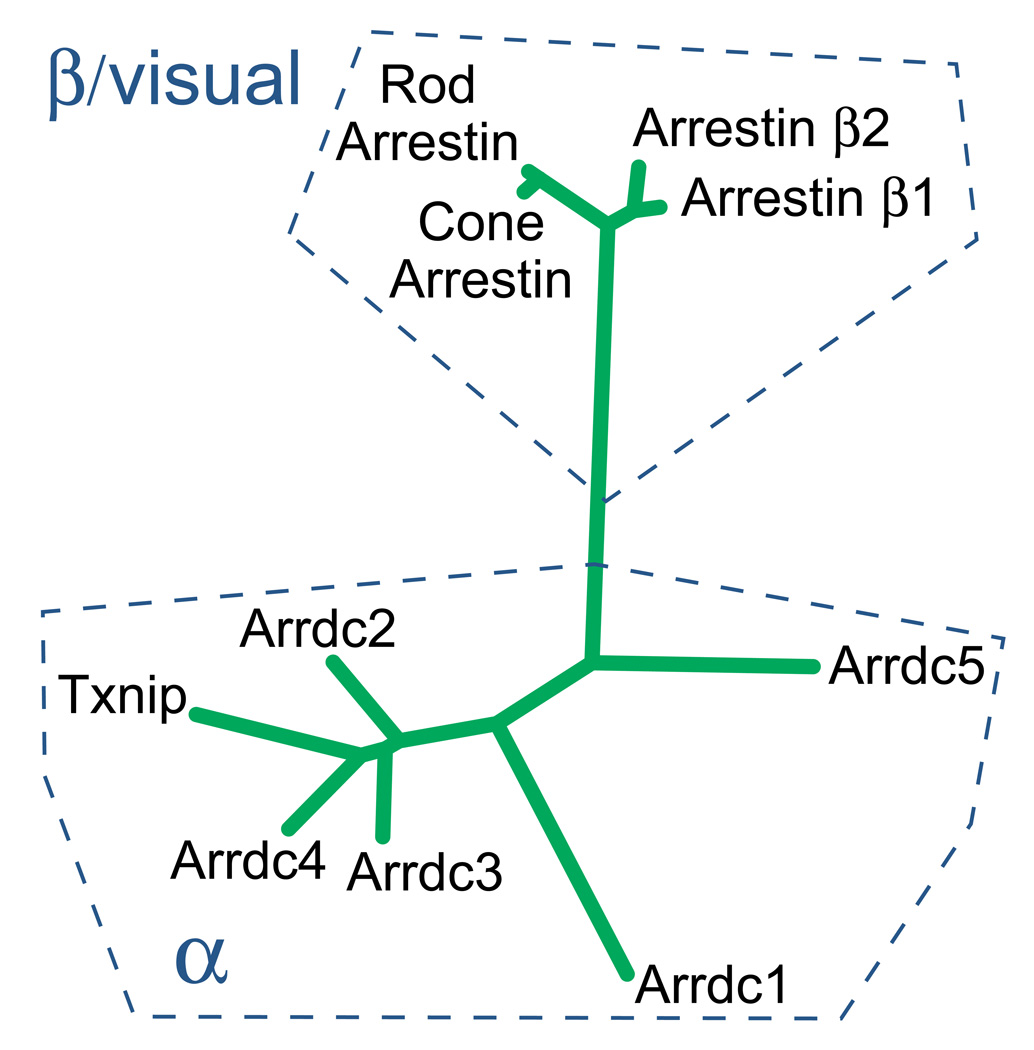
Beyond the well-studied beta arrestins and visual arrestins, there is a larger family of proteins that are predicted to share the overall arrestin fold structure [16]. These other proteins, sometimes termed the “alpha-arrestins”, are the more ancient branch, as these arrestins are present in filamentous fungi and the budding yeast as well as protists. Humans express six alpha-arrestins (Figure 1), but the molecular function of the arrestin domain in these proteins is not yet clear.
Figure 1. A schematic phylogenetic tree for the human arrestin protein family with proposed nomenclature.
The classical human arrestins include two visual arrestins and two beta-arrestins. The visual arrestins desensitize the light-transducing opsins in the photoreceptor cells of the retina by binding specifically to the active, phosphorylated receptor. The ubiquitously expressed β-arrestins (β-arrestin 1 and β-arrestin-2) are required for desensitization and endocytosis of a variety of G-protein-coupled receptors. In addition, there are six new members of the arrestin superfamily, termed the “alpha-arrestins”. Alpha-arrestins, distantly related to the beta/visual arrestins but predicted to share the arrestin fold, have emerging roles in regulation of metabolism and obesity.
The classical visual and non-visual beta-arrestins have important structural similarities. For example, the structural basis for the binding specificity to phosphorylated receptors is through a conserved polar core. When an arrestin interacts with a receptor that has a negatively charged phosphate, positively charged residues in the polar core are destabilized and allow the arrestin to bind tightly to the receptor [17]. These polar core residues are conserved in all four classical visual and beta-arrestins but not in the larger family of related alpha-arrestins [16]. The clathrin-binding sites of the classical arrestins that facilitate their function in receptor endocytosis are also not present in alpha-arrestins. Instead, alpha-arrestins have highly conserved PPxY motifs, which allow binding to WW domains in proteins such as ubiquitin ligases [16].
There is, at the time of this writing, no solved structure of an alpha-arrestin, although predictions have been made from the protein sequence alone. Interesting evidence for conservation of the arrestin fold structure came from an even more distantly related protein, Vacuolar protein sorting-associated protein 26A (Vps26), which was found to have an arrestin fold (a two-lobe, immunoglobulin-like, β-strand sandwich) [18]. Vps26 is more closely homologous to the ancient alpha-arrestins than the visual/beta arrestins, suggesting that the arrestin fold is conserved through evolution of all arrestins [16]. Vps26 functions in the retromer complex, a large multimeric complex which is involved in the retrieval of cargoes from endosomes to the trans-Golgi network. Thus, evolutionary biology suggests conserved ancestral roles for arrestin-fold proteins in directing cargo through multiple stages of intracellular trafficking [19, 20].
Txnip: Link between redox stress and metabolism?
Thioredoxin interacting protein (Txnip) was initially identified as an inhibitor of thioredoxin, a small ubiquitous antioxidant protein. Txnip binds to thioredoxin through a disulfide bond, and overexpression of Txnip in vitro inhibits thioredoxin from performing its antioxidant functions [21]. Txnip has therefore been considered to act as an endogenous inhibitor of thioredoxin thus promoting oxidative stress [21].
Txnip’s metabolic functions were revealed when a nonsense mutation in the Txnip gene was identified as being responsible for the phenotype of the “hyplip” mouse [22]. The hyplip mouse has high triglyceride levels along with high ketone and lactate levels, consistent with decreased fatty acid flux through the tricarboxylic acid cycle in the mitochondria [22].
A central physiological role in glucose flux and glycolytic metabolism is now established for Txnip. In addition, recent studies have shown that Txnip transcription and translation is strongly induced by high glucose levels in multiple cell lines and primary cells [23–25]. In clinical studies, Txnip expression is increased in diabetic tissues, and is one of the genes in skeletal muscle that is most strongly repressed by insulin, during glucose clamp [26]. Therefore, Txnip has a profound effect on cellular glucose uptake [26]. In addition, Txnip inhibits basal glucose uptake, even in cell lines with minimal insulin-stimulated glucose uptake [26, 27].
These results are consistent with studies in Txnip-null mice that were generated by several different laboratories [28–30]. Txnip-null mice have low blood glucose and insulin levels and exhibit particularly profound fasting hypoglycemia. Glucose clamp studies revealed a 3-fold increase in glucose clearance in Txnip-null mice, and glucose uptake was enhanced in skeletal muscle and adipocytes [28]. Multiple lines of evidence now suggest that a major effect of Txnip is to enhance flux through the mitochondrial citric acid cycle, linking loss of Txnip to the Warburg cancer phenotype [28, 29, 31]. Txnip thus seems to act as a negative feedback regulator for glucose uptake: when glucose is in plentiful, Txnip is induced and then inhibits further glucose uptake [26].
Given Txnip’s clear functions in regulating glucose uptake and binding to thioredoxin, an obvious hypothesis emerges: could Txnip be the critical link connecting high glucose levels to oxidative stress and insulin resistance [21, 23, 32]? This hypothesis is likely an oversimplification. Despite efforts from multiple laboratories, little evidence has come to light that supports a direct link between Txnip and metabolism through increased redox stress from inhibition of thioredoxin. In addition, in vivo studies have not revealed differences in available thioredoxin in the liver or heart of Txnip-null animals [28, 33, 34]. Nevertheless, in the context of the larger family of arrestins, an alternative hypothesis might exist. It is likely that Txnip does not function primarily as an inhibitor of thioredoxin; instead it might be a special case of an alpha-arrestin that is regulated by its binding to thioredoxin [27, 35].
Txnip and Arrdc4: Conserved functions for alpha-arrestins in glucose homeostasis
The identification of how Txnip binds to thioredoxin led eventually to the understanding that inhibition of glucose uptake by Txnip does not occur through inhibition of thioredoxin [27]. Biochemical analysis of the Txnip-thioredoxin interaction suggested that the proteins form a covalent interaction through a disulfide bond. Targeted mutagenesis then allowed identification of a point mutation of a Txnip cysteine (cysteine 247) that abolishes its binding to thioredoxin. However, the Txnip mutant C247S is still able to inhibit cellular glucose uptake as strongly as wild type (wt) Txnip, demonstrating that this function does not require binding of Txnip to thioredoxin.
Furthermore, the glucose uptake function is intrinsic to the arrestin domains of Txnip. Txnip has a proline-rich C-terminal tail with conserved PPxY motifs. In yeast, the C terminal tail is responsible for recruiting E3 ligase activity. However, a truncated Txnip without the C terminal tail retains undiminished ability to inhibit glucose uptake [27]. Consistent with Txnip functioning through its arrestin domains, the ability to inhibit glucose uptake is not limited to Txnip. Another alpha-arrestin, Arrdc4, is an equally potent inhibitor of glucose uptake in vitro despite its inability to bind thioredoxin [27].
Remarkably, not only is the glucose uptake function conserved between Txnip and Arrdc4, but transcriptional regulation of both proteins is controlled through carbohydrate response elements (ChREs) [24, 25]. Txnip and Arrdc4 are in fact the most induced transcripts regulated by the MondoA transcription factor (related to the carbohydrate response element binding protein ChREBP) [25]. Thus both alpha arrestin proteins appear to be central to the cell’s glucose sensing program.
The molecular mechanisms for how arrestin domains regulate glucose uptake remain unknown. Do alpha-arrestins bind to and directly affect glucose transporters? It is tempting to speculate that Txnip and Arrdc4 regulate endocytosis, trafficking, or degradation of glucose transport through GLUT1, since they inhibit glucose transport under baseline conditions and in relatively insulin-insensitive cell lines [26, 27]. In budding yeast, two distant relatives of the alpha-arrestins (Art4 and Art8) have been shown to cause endocytosis of a high-affinity glucose transporter (Hxt6) in response to high glucose levels [36]. One of these proteins (Art4) is also required for linking altered glucose levels to endocytosis of the lactate transporter Jen1 through the action of the yeast AMPK homolog, Snf1 kinase. Finally, another alpha-arrestin-related protein (Art1) was recently identified as a key effector linking nutrient sensing through the TORC1 kinase signaling complex to endocytosis of amino acid transporters [37]. Thus very recent studies in yeast have provided substantial precedent for direct regulation of glucose transporter endocytosis by alpha-arrestins in response to changes in nutrient availability through conserved nutrient sensing pathways. However, investigations in the Txnip-null animals have yet to identify changes in membrane glucose transporter levels. Txnip does not regulate levels of GLUT4 on the plasma membrane in heart [34], nor GLUT1 levels in either heart or muscle [38]. It thus remains a major open question how Txnip and Arrdc4 affect glucose transport at the molecular level.
Understanding the mechanisms of Txnip and Arrdc4 function may yield insights not only into diabetes but also into how tumor cells manipulate metabolism to work in their favor. Txnip functions as a tumor suppresor gene in multiple types of cancers, and the mechanism may be directly related to its metabolic functions [39, 40]. Loss of Txnip inhibits mitochondrial oxidation while promoting glycolysis [28, 29], potentially favoring tumor cell growth through the “Warburg effect” [41, 42].
A Paradigm Twist: Thioredoxin Regulates Txnip in Adipogenesis
Although Txnip does not require binding to thioredoxin for its function in glucose metabolism, Txnip could still regulate other metabolic functions in part through inhibition of thioredoxin. Overexpression of Txnip in 3T3-L1 cells inhibits adipogenesis, and primary embryo-derived fibroblasts from mice lacking Txnip show enhanced adipogenesis [27]. This function of Txnip may be related to an intriguing feature of the Txnip-null mouse: while it maintains lower glucose levels and higher insulin responsiveness even when subjected to a high-fat diet, the Txnip-null mouse gains more body mass than wt mice do [27, 43]. Thus, high-fat diet has divergent effects on adiposity and insulin sensitivity in Txnip-null mice, consistent with the hypothesis that fat expansion can be protective against the metabolic syndrome.
Notably, while overexpression of Txnip inhibits adipogenesis, a Txnip mutant protein that is unable to bind thioredoxin (Txnip C247S) is much less effective in this regard [35]. This identifies a role for thioredoxin in the function of Txnip in preadipocytes, but it turns out to be indirect. Normal adipogenesis in 3T3-L1 cells requires rapid degradation of Txnip at the onset of differentiation [35]. This occurs through a conserved alpha-arrestin feature, namely the C-terminal tail PPxY motifs that allow recruitment of the E3 ubiquitin ligase Itch [35, 44]. However, the Txnip-thioredoxin complex appears to be resistant to such degradation [35]. Thus, without the ability to bind thioredoxin, Txnip C247S is more rapidly degraded and has reduced ability to inhibit adipogenesis.
These results illustrate a new paradigm for Txnip action: Txnip is not always functioning through inhibition of thioredoxin; rather, thioredoxin regulates some of Txnip’s intrinsic alpha-arrestin functions by enhancing its stability (Figure 2). It is therefore likely that multiple Txnip functions will be affected by its ability to bind thioredoxin, but this still does not imply that these functions work mechanistically through inhibition of thioredoxin. There are other functions of Txnip that are affected by thioredoxin binding, including nutrient sensing in the hypothalamus and apoptosis in vascular endothelial cells [45, 46]. In addition, Txnip reportedly affects cell cycle progression by binding directly to the cell cycle regulator Jab1 [47], and it also affects Histone deacetylase (HDAC) function by recruiting thioredoxin to the heat shock protein Dnajb5 [48]. For both Jab1 and Dnajb5, it is not yet clear whether these interactions require thioredoxin. It will therefore be interesting to determine whether these represent further examples of thioredoxin regulating Txnip, rather than Txnip regulating thioredoxin.
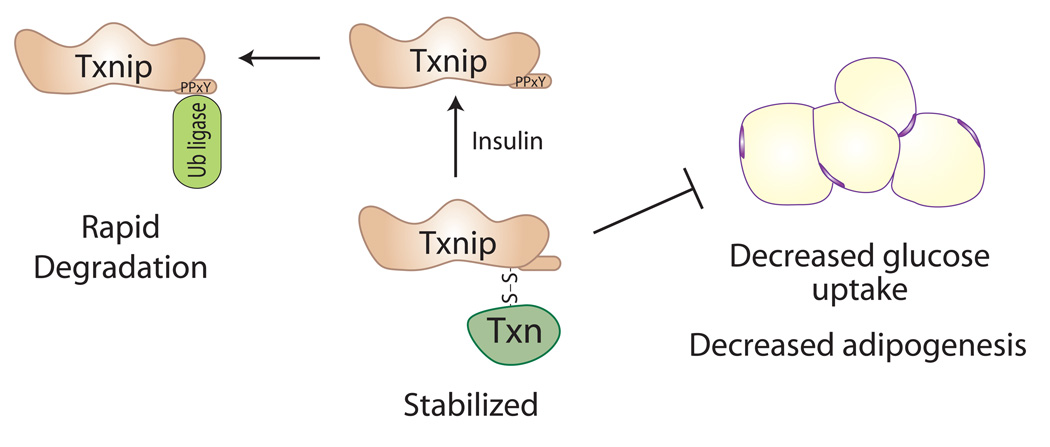
Figure 2. Regulation of Txnip by Thioredoxin.
Txnip was originally identified as a regulator of redox stress that binds thioredoxin and thereby inhibits its oxidoreductase activity. Txnip also has critical roles in blocking cellular glucose uptake and glycolytic metabolism, and in inhibiting adipogenesis. This led to the hypothesis that Txnip regulates metabolism by inhibiting thioredoxin and increasing oxidative stress. However, based on recent results, we now propose the reverse: that thioredoxin enhances Txnip’s function by increasing its stability [27, 35]. In this model, Txnip is subject to rapid protein degradation through recruitment of E3 ubiquitin ligases to its C-terminal PPxY motifs (for example, by treatment with insulin). In contrast, binding of Txnip to thioredoxin, perhaps by reducing accessibility of the PPxY motifs, leads to a Txnip-thioredoxin complex that resists degradation and thereby enhances Txnip functions.
In general, the arrestins appear to act as molecular scaffolds and signal integrators, and we anticipate multiple binding partners for alpha-arrestins such as Txnip will be discovered. Ultimately, the Txnip arrestin domains are likely to affect metabolism through other binding partners as well. As an example of such an unanticipated mechanism, a surprising new interaction has been reported for Txnip with the inflammasome protein NLRP3 that may link metabolism with innate immune response [49].
Arrdc3 Regulates Obesity
While only Txnip and Arrdc4 inhibit glucose uptake, do the other alpha-arrestins regulate metabolism? It has now emerged that Arrdc3 is linked to obesity — but interestingly, only in males. A genome-wide linkage study performed by DeCode and Merck scientists first linked Arrdc3 to body-mass index (BMI), in a population of over 8000 Icelanders [50]. Although expressed in multiple tissues, Arrdc3 transcript levels were found to be relatively high in adipose tissue samples. Further analysis of gene expression in human adipose tissue provided more support for a role of Arrdc3 in obesity. For example, whereas Arrdc3 expression levels in subcutaneous fat were not correlated with BMI, higher Arrdc3 expression levels in the more metabolically active omental fat were correlated with higher BMI, again only in males [51]. The mechanism behind the gender difference is not known.
The function of Arrdc3 has been further analyzed by experiments in mouse models that establish Arrdc3 as a causal regulator of obesity [51]. As predicted by the correlation of higher Arrdc3 with higher BMI in humans, mice with genetic deficiency of Arrdc3 remain lean and are resistant to obesity. Furthermore, although mice of both genders are resistant to obesity, the effect is more pronounced in male mice. The physiological mechanism for the resistance to obesity in the Arrdc3-deficient mice appears to be through increased energy expenditure, as heterozygous mice have both increased heat production and increased activity levels, compared to wt mice with similar weights [51].
The mechanism for increased energy expenditure in Arrdc3-null mice may be through not only increased thermogenesis in brown fat, but also through “browning” of the white fat depots. Arrdc3-null mice show increased thermogenic response to cold when placed at 4° C, which can be a result of either increased shivering or non-shivering (brown fat) thermogenic function. The role of brown fat in this regard is supported by an observed significant increase in type II deiodinase expression and activity, an enzyme that is required for the thermogenic function in brown fat. Interestingly, Arrdc3 expression in mice is highest in visceral fat and in the absence of Arrdc3, a striking increase in the expression of the brown fat uncoupling protein 1(Ucp1) in visceral white fat is seen.
Full circle: Functions of the Alpha-arrestins in signaling and endocytosis
The critical question about the molecular function of the alpha-arrestins remains: are they capable of acting to cause endocytosis of activated receptors, as classical beta and visual arrestins do? Evidence is rapidly accumulating that alpha-arrestins also have a conserved role in receptor signaling and vesicular trafficking.
It now seems clear that a conserved function of the ancestral arrestins is to regulate trafficking of diverse plasma membrane proteins in part through promoting endocytosis, by recruiting ubiquitin ligases. In the budding yeast Saccharomyces cerevisiae, multiple arrestins bind specific transporters and recruit the Nedd4-like ubiquitin ligase Rsp5, resulting in the ubiquitination of both proteins, and induction of endocytosis [36, 52, 53]. Similar functions have been observed in filamentous fungi [54, 55]. More recently, fungal arrestins have been shown to direct trafficking through other vesicular transport pathways as well [56]. Although distant relatives, these fungal arrestins bear closer homology to the alpha-arrestin branch of the family, than the visual/beta arrestin branch [16]. These ancestral alpha-arrestin functions suggest that regulation of diverse vesicular trafficking pathways has always been a function of the arrestins.
Several studies now strongly suggest that the mammalian alpha-arrestins also function in directing endocytosis through recruitment of ubiquitin ligases [44, 57, 58]. The localization of the alpha-arrestins, other than Txnip, which co-localize with the plasma membrane and vesicles carrying early endosomal markers, has b een suggestive [27, 57, 59]. A fascinating new finding is that one of the human alpha-arrestins, Arrdc1, interacts with proteins comprising the ESCRT (endosomal sorting complex required for transport) machinery to direct a new form of microvesicle budding from the cell membrane into the extracellular space [57, 60]. Thus, mammalian alpha-arrestins may retain ancient arrestin functions directing multiple stages of vesicular trafficking.
The beta-arrestins were first identified as proteins required for down-regulation of the beta-2 - adrenergic receptor (β2-AR) [61, 62]. Recently, surprising results from Nabhan and colleagues suggest that the alpha-arrestins may fundamentally change this story. In an RNA-knockdown screen for transcripts affecting β2-AR levels, Arrdc3 was identified as one of the transcripts [58]. Increased Arrdc3 caused a reduction in β2-AR on the membrane, through ligand-induced ubiquitination of the receptor. Continuing a theme for the alpha-arrestins, recruitment of the ubiquitin ligase requires the PPxY motifs in the C-terminal tail of Arrdc3 [58]. Further studies have shown that an interaction with β2-AR is not unique to Arrdc3 — other alpha-arrestins (but not Txnip) bind to the receptor as well [51].
Evidence that adrenergic regulation by Arrdc3 has physiological importance comes from studies in the Arrdc3-null mouse. Isolated adipose tissue from the Arrdc3-null mouse shows strongly enhanced adrenergic response. Stimulation by norepinephrine results in enhanced cAMP signaling and enhanced lipolysis [51]. Thus, Arrdc3 may share adrenergic receptor regulation functions paralleling those of beta-arrestins.
However, the physiological mechanism of resistance to obesity in the Arrdc3-null mouse is more consistent with enhanced signaling through the beta-3 adrenergic receptor (β3-AR), which is required for non-shivering thermogenesis [63]. Beta-3 specific agonists are also known to enhance brown fat-like function in white fat [63, 64], as seen in the Arrdc3-null mouse. But unlike other adrenergic receptors, the β3-AR is not phosphorylated after activation, does not bind beta-arrestins, and is relatively resistant to downregulation [63, 65]. Thus it is especially surprising that Arrdc3 does bind to β3-AR [51], potentially providing a direct mechanism for its ability to regulate energy expenditure, and a new explanation for functional desensitization of the β3-AR in brown fat (Figure 3).
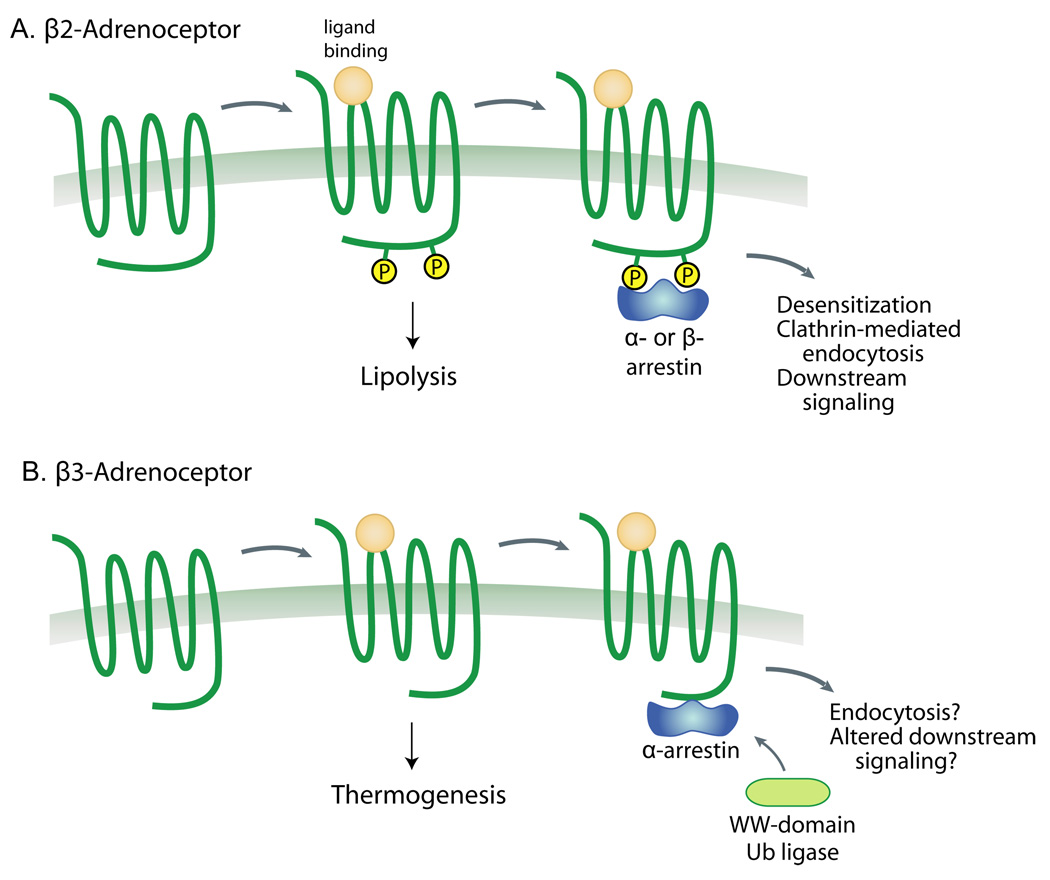
Figure 3. Schematic representation of open hypotheses for alpha and beta arrestin actions on beta-adrenergic receptors in adipose tissue.
A) Activation of the adipose tissue β1 and β2 adrenergic receptors causes a Gs-mediated increase in cAMP levels, stimulating lipolysis. This receptor has a classical paradigm for desensitization in which beta-arrestins are recruited to the active phosphorylated receptor, leading to ubiquitination and clathrin-mediated endocytosis [11]. We hypothesize that alpha-arrestins such as Arrdc3 may serve similar roles in this process. B) In contrast, the β3-adrenergic receptor is not phosphorylated and is largely not downregulated, at least in vitro. Furthermore, signaling is through Gi activation of ERK and through G-protein-independent recruitment of Src [63]. Whereas beta-arrestins are not recruited to the β3-adrenergic receptor, we hypothesize that alpha-arrestins may be able to either promote receptor ubiquitination or alter downstream signaling, thereby regulating thermogenesis in adipose tissue.
Concluding remarks
The fascinating story of the beta and visual arrestins may be the tip of the iceberg, given the paucity of information on their older relatives, the alpha-arrestins. Over the past decade, one alpha-arrestin, Txnip, has been established as a critical regulator of glycolytic metabolism, through clinical studies as well as genetic manipulation in mice. The molecular mechanism for Txnip’s metabolic regulation remains elusive, but it is clear that the concept that Txnip functions to inhibit thioredoxin is incomplete. It is interesting that Txnip is the only alpha-arrestin that binds thioredoxin, placing Txnip in a unique position between arrestin-mediated signal control and redox state. The alternative concept of regulation of Txnip by thioredoxin is likely to yield significant new insights into how redox biology controls metabolism.
The recent identification that the alpha-arrestin Arrdc3 regulates obesity and energy expenditure highlights that alpha-arrestins have evolved to play important roles in metabolism, despite a diversity of function among specific family members. Equally important may be the connection of Arrdc3 to classical arrestin-like functions in regulating signaling through the beta-adrenergic receptors. Important mechanistic questions remain to be addressed: can alpha-arrestins functionally replace beta-arrestins, or are there differences? A consistent theme for alpha-arrestins across evolution is the importance of the C-terminal tail domain PPxY motifs for recruiting ubiquitin ligases that lead to turnover of transmembrane transporters and receptors. However, as the more ancient group of proteins, recent reports also suggest that the alpha-arrestins may simply function in a wider diversity of vesicular transport pathways than the beta-arrestins [20, 56, 57, 60].
The unexpected overlap of alpha and beta arrestins in regulation of adrenergic signaling suggests that the two subfamilies are not functioning in an entirely independent manner. As noted previously, beta and visual arrestins can associate to form heterodimers, suggesting alpha/beta heterodimers as a possible explanation for an overlap in function [16].
In addition, alpha-arrestins may have an expanded range of receptor affinities, such as the ability of Arrdc3 to bind the beta-3 adrenergic receptor. With the identification of Arrdc3 as a new regulator of obesity, it will be critical to explore the mechanism of action of Arrdc3 further. Is the phenotype driven specifically by Arrdc3 action in adipose tissue or are there other tissues involved? Much will depend on this fundamental question — what allows Arrdc3 to bind to receptors that beta-arrestins can not? Confirmation that this interaction is relevant for the endogenous proteins and the in vivo phenotype will be crucial for understanding Arrdc3 function and its potential insights into the development of obesity in humans.
Outstanding Questions.
Which functions of Thioredoxin-interacting protein (Txnip) work by increasing redox stress through inhibition of Thioredoxin, and which functions are instead working directly through the arrestin domains of Txnip but regulated by Thioredoxin?
Two alpha-arrestins (Txnip and Arrdc4) inhibit cellular glucose uptake, but the molecular mechanism is not known. Are these alpha-arrestins able to bind glucose transporters and direct their endocytosis or intracellular trafficking, as suggested by the functions of other ancestral arrestins?
Several alpha-arrestins appear to share functions with beta-arrestins in directing receptor downregulation and endocytosis. Can alpha-arrestins truly substitute for classical beta-arrestin functions, or will there be differences in the mechanisms of action?
Some G-protein-coupled receptors, such as the beta-3-adrenergic receptor, do not recruit beta-arrestins. Are alpha-arrestins capable of binding to these non-classical receptors and regulating their downstream signaling if not their endocytosis?
Acknowledgments
Supported in part by grants from the NIH (HL103582, HL048743, and HL081523)
Glossary
- Visual arrestins
arrestins specifically expressed in retinal photoreceptor cells that cause desensitization of rod and cone photoreceptors. Rod arrestin (Sag), also known as S-antigen or arrestin-1, binds specifically to activated rhodopsin; cone arrestin (Arr3), also known as both arrestin-3 (from its official gene name) and arrestin-4 (as the last cloned of the four classical arrestins) regulates photoreceptors in the cone cells.
- Beta-arrestins
The two non-visual arrestins (β-arrestin-1 and β-arrestin-2) are ubiquitously expressed and bind to a broad range of GPCRs including the β2-adrenergic receptor. They function in desensitization, endocytosis, and recycling of the receptors as well as directing G-protein-independent downstream signaling.
- Alpha-arrestins
A term recently coined to describe the six human proteins predicted to contain an arrestin fold besides the classical visual and beta arrestins. Alpha-arrestins are the larger, more ancient proteins, with alpha-arrestin-like proteins characterized in fungi.
- Thioredoxin-interacting protein (Txnip)
An alpha-arrestin that binds to thioredoxin. It has critical functions in glucose metabolism.
- Arrestin domain containing 4 (Arrdc4)
An alpha-arrestin that, like its homolog Txnip, is induced by glucose levels and inhibits cellular glucose uptake, but does not bind thioredoxin.
- Arrestin domain containing 3 (Arrdc3)
An alpha-arrestin linked to obesity that regulates energy expenditure and binds to adrenergic receptors.
- Arrestin fold
a two-lobe, immunoglobulin-like, β-strand sandwich structure. The first lobe (the N-terminal arrestin domain) is connected to the second lobe (the C-terminal arrestin domain) by a short hinge loop in between. The structure is stabilized by a “polar core” buried at the interface of the two lobes.
- Thioredoxin (Txn1)
A small, ubiquitously expressed protein that functions as one of the major anti-oxidant defense systems of the cell. In conjunction with thioredoxin reductase, it reduces free radical oxygen species, other oxidants, and protein disulfides.
- Retromer
a protein complex important in recycling transmembrane receptors from endosomes to the trans-Golgi network.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yarfitz S, Hurley JB. J. Biol. Chem. 1994;269:14329–14332. [PubMed] [Google Scholar]
- 2.Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. J. Biol. Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 3.Lefkowitz RJ, Rajagopal K, Whalen EJ. Mol Cell. 2006;24:643–652. doi: 10.1016/j.molcel.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Kocan M, Pfleger KDG. Methods Mol Biol. 2011;746:357–371. doi: 10.1007/978-1-61779-126-0_20. [DOI] [PubMed] [Google Scholar]
- 5.Chen W, Ren X-R, Nelson CD, Barak LS, Chen JK, Beachy PA, de Sauvage F, Lefkowitz RJ. Science. 2004;306:2257–2260. doi: 10.1126/science.1104135. [DOI] [PubMed] [Google Scholar]
- 6.Wilbanks AM, Fralish GB, Kirby ML, Barak LS, Li Y-X, Caron MG. Science. 2004;306:2264–2267. doi: 10.1126/science.1104193. [DOI] [PubMed] [Google Scholar]
- 7.Kovacs JJ, Whalen EJ, Liu R, Xiao K, Kim J, Chen M, Wang J, Chen W, Lefkowitz RJ. Science. 2008;320:1777–1781. doi: 10.1126/science.1157983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li S, Chen Y, Shi Q, Yue T, Wang B, Jiang J. PLoS Biol. 2012;10 doi: 10.1371/journal.pbio.1001239. e1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurevich VV, Gurevich EV, Cleghorn WM. Handb Exp Pharmacol. 2008:15–37. doi: 10.1007/978-3-540-72843-6_2. [DOI] [PubMed] [Google Scholar]
- 10.Noor N, Patel CB, Rockman HA. J. Mol. Cell. Cardiol. 2011;51:534–541. doi: 10.1016/j.yjmcc.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shenoy SK, Lefkowitz RJ. Trends Pharmacol. Sci. 2011;32:521–533. doi: 10.1016/j.tips.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mythreye K, Blobe GC. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8221–8226. doi: 10.1073/pnas.0812879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawamata Y, Imamura T, Babendure JL, Lu J-C, Yoshizaki T, Olefsky JM. J. Biol. Chem. 2007;282:28549–28556. doi: 10.1074/jbc.M705869200. [DOI] [PubMed] [Google Scholar]
- 14.Luan B, Zhao J, Wu H, Duan B, Shu G, Wang X, Li D, Jia W, Kang J, Pei G. Nature. 2009;457:1146–1149. doi: 10.1038/nature07617. [DOI] [PubMed] [Google Scholar]
- 15.Hupfeld CJ, Olefsky JM. Annu. Rev. Physiol. 2007;69:561–577. doi: 10.1146/annurev.physiol.69.022405.154626. [DOI] [PubMed] [Google Scholar]
- 16.Alvarez CE. BMC Evol Biol. 2008;8:222. doi: 10.1186/1471-2148-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurevich VV, Gurevich EV. Pharmacol. Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, Rojas R, Bonifacino JS, Hurley JH. Nat. Struct. Mol. Biol. 2006;13:540–548. doi: 10.1038/nsmb1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reider A, Wendland B. J. Cell. Sci. 2011;124:1613–1622. doi: 10.1242/jcs.073395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Donnell AF, Apffel A, Gardner RG, Cyert MS. Mol. Biol. Cell. 2010;21:3552–3566. doi: 10.1091/mbc.E10-07-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaimul AM, Nakamura H, Masutani H, Yodoi J. Free Radic. Biol. Med. 2007;43:861–868. doi: 10.1016/j.freeradbiomed.2007.05.032. [DOI] [PubMed] [Google Scholar]
- 22.Bodnar JS, Chatterjee A, Castellani LW, Ross DA, Ohmen J, Cavalcoli J, Wu C, Dains KM, Catanese J, Chu M, Sheth SS, Charugundla K, Demant P, West DB, de Jong P, Lusis AJ. Nat Genet. 2002;30:110–116. doi: 10.1038/ng811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulze PC, Yoshioka J, Takahashi T, He Z, King GL, Lee RT. J. Biol. Chem. 2004;279:30369–30374. doi: 10.1074/jbc.M400549200. [DOI] [PubMed] [Google Scholar]
- 24.Minn AH, Hafele C, Shalev A. Endocrinology. 2005;146:2397–2405. doi: 10.1210/en.2004-1378. [DOI] [PubMed] [Google Scholar]
- 25.Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, Ayer DE. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6912–6917. doi: 10.1073/pnas.0712199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, Saxena R, Ladd C, Schulze PC, Mazzini MJ, Jensen CB, Krook A, Björnholm M, Tornqvist H, Zierath JR, Ridderstråle M, Altshuler D, Lee RT, Vaag A, Groop LC, Mootha VK. PLoS Med. 2007;4:e158. doi: 10.1371/journal.pmed.0040158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patwari P, Chutkow WA, Cummings K, Verstraeten VLRM, Lammerding J, Schreiter ER, Lee RT. J. Biol. Chem. 2009;284:24996–25003. doi: 10.1074/jbc.M109.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chutkow WA, Patwari P, Yoshioka J, Lee RT. J. Biol. Chem. 2008;283:2397–2406. doi: 10.1074/jbc.M708169200. [DOI] [PubMed] [Google Scholar]
- 29.Hui STY, Andres AM, Miller AK, Spann NJ, Potter DW, Post NM, Chen AZ, Sachithanantham S, Jung DY, Kim JK, Davis RA. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3921–3926. doi: 10.1073/pnas.0800293105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oka S, Liu W, Masutani H, Hirata H, Shinkai Y, Yamada S, Yoshida T, Nakamura H, Yodoi J. FASEB J. 2006;20:121–123. doi: 10.1096/fj.05-4439fje. [DOI] [PubMed] [Google Scholar]
- 31.Peterson CW, Ayer DE. Front. Biosci. 2011;17:2206–2223. doi: 10.2741/3848. [DOI] [PubMed] [Google Scholar]
- 32.Hui TY, Sheth SS, Diffley JM, Potter DW, Lusis AJ, Attie AD, Davis RA. J. Biol. Chem. 2004;279:24387–24393. doi: 10.1074/jbc.M401280200. [DOI] [PubMed] [Google Scholar]
- 33.Sheth SS, Bodnar JS, Ghazalpour A, Thipphavong CK, Tsutsumi S, Tward AD, Demant P, Kodama T, Aburatani H, Lusis AJ. Oncogene. 2006;25:3528–3536. doi: 10.1038/sj.onc.1209394. [DOI] [PubMed] [Google Scholar]
- 34.Yoshioka J, Imahashi K, Gabel SA, Chutkow WA, Burds AA, Gannon J, Schulze PC, MacGillivray C, London RE, Murphy E, Lee RT. Circ Res. 2007;101:1328–1338. doi: 10.1161/CIRCRESAHA.106.160515. [DOI] [PubMed] [Google Scholar]
- 35.Chutkow WA, Lee RT. J. Biol. Chem. 2011;286:29139–29145. doi: 10.1074/jbc.M111.267666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikko E, Pelham HRB. Traffic. 2009;10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacGurn JA, Hsu P-C, Smolka MB, Emr SD. Cell. 2011;147:1104–1117. doi: 10.1016/j.cell.2011.09.054. [DOI] [PubMed] [Google Scholar]
- 38.Andres AM, Ratliff EP, Sachithanantham S, Hui ST. FEBS Lett. 2011;585:1223–1230. doi: 10.1016/j.febslet.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elgort MG, O’Shea JM, Jiang Y, Ayer DE. Genes Cancer. 2010;1:893–907. doi: 10.1177/1947601910389604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masutani H, Yoshihara E, Masaki S, Chen Z, Yodoi J. J Clin Biochem Nutr. 2012;50:23–34. doi: 10.3164/jcbn.11-36SR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Kaadige MR, Looper RE, Kamalanaadhan S, Ayer DE. Proc. Natl. Acad. Sci. U.S.A. 2009 doi: 10.1073/pnas.0901221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen J, Hui ST, Couto FM, Mungrue IN, Davis DB, Attie AD, Lusis AJ, Davis RA, Shalev A. FASEB J. 2008;22:3581–3594. doi: 10.1096/fj.08-111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang P, Wang C, Gao K, Wang D, Mao J, An J, Xu C, Wu D, Yu H, Liu JO, Yu L. J. Biol. Chem. 2010;285:8869–8879. doi: 10.1074/jbc.M109.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blouet C, Schwartz GJ. J. Neurosci. 2011;31:6019–6027. doi: 10.1523/JNEUROSCI.6498-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.World C, Spindel ON, Berk BC. Arterioscler. Thromb. Vasc. Biol. 2011;31:1890–1897. doi: 10.1161/ATVBAHA.111.226340. [DOI] [PubMed] [Google Scholar]
- 47.Jeon J-H, Lee K-N, Hwang CY, Kwon K-S, You K-H, Choi I. Cancer Res. 2005;65:4485–4489. doi: 10.1158/0008-5472.CAN-04-2271. [DOI] [PubMed] [Google Scholar]
- 48.Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD, Vatner SF, Sadoshima J. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 49.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Nat. Immunol. 2010;11:136–140. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 50.Emilsson V, Thorleifsson G, Sainz J, Walters G, Gulcher J, Lamb J, Schadt E. Haplotypes in the human thioredoxin interacting protein homologue (ARRDC3) gene associated with obesity. WO/2005/111239. [Google Scholar]
- 51.Patwari P, Emilsson V, Schadt EE, Chutkow WA, Lee S, Marsili A, Zhang Y, Dobrin R, Cohen DE, Larsen PR, Zavacki AM, Fong LG, Young SG, Lee RT. Cell Metab. 2011;14:671–683. doi: 10.1016/j.cmet.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 53.Herrador A, Herranz S, Lara D, Vincent O. Mol. Cell. Biol. 2010;30:897–907. doi: 10.1128/MCB.00132-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boase NA, Kelly JM. Mol. Microbiol. 2004;53:929–940. doi: 10.1111/j.1365-2958.2004.04172.x. [DOI] [PubMed] [Google Scholar]
- 55.Herranz S, Rodríguez JM, Bussink H-J, Sánchez-Ferrero JC, Arst HN, Peñalva MA, Vincent O. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12141–12146. doi: 10.1073/pnas.0504776102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galindo A, Calcagno-Pizarelli AM, Arst HN, Jr, Peñalva MÁ. Journal of Cell Science. 2012 doi: 10.1242/jcs.098897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rauch S, Martin-Serrano J. J. Virol. 2011;85:3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nabhan JF, Pan H, Lu Q. EMBO Rep. 2010;11:605–611. doi: 10.1038/embor.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oka S, Masutani H, Liu W, Horita H, Wang D, Kizaka-Kondoh S, Yodoi J. Endocrinology. 2006;147:733–743. doi: 10.1210/en.2005-0679. [DOI] [PubMed] [Google Scholar]
- 60.Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Proceedings of the National Academy of Sciences of the United States of America. 2012 doi: 10.1073/pnas.1200448109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lohse M, Benovic J, Codina J, Caron M, Lefkowitz R. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 62.Ferguson SSG, Downey WE, Colapietro A-M, Barak LS, Menard L, Caron MG. Science. 1996;271:363–366. doi: 10.1126/science.271.5247.363. [DOI] [PubMed] [Google Scholar]
- 63.Collins S, Cao W, Robidoux J. Mol. Endocrinol. 2004;18:2123–2131. doi: 10.1210/me.2004-0193. [DOI] [PubMed] [Google Scholar]
- 64.Ishibashi J, Seale P. Science. 2010;328:1113–1114. doi: 10.1126/science.1190816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bengtsson T, Redegren K, Strosberg AD, Nedergaard J, Cannon B. J. Biol. Chem. 1996;271:33366–33375. doi: 10.1074/jbc.271.52.33366. [DOI] [PubMed] [Google Scholar]