Abstract
RNA virus population dynamics is complex, and sophisticated approaches are needed in many cases for therapeutic intervention. One such approach, termed lethal mutagenesis, is directed at targeting the virus population structure for extinction or error catastrophe. Previous studies have demonstrated the concept of this approach with human immunodeficiency virus type 1 (HIV-1) by use of chemical mutagens (i.e., 5-azacytidine) as well as by host factors with mutagenic properties (i.e., APOBEC3G). In this study, these two unrelated mutagenic agents were used concomitantly to investigate the interplay of these distinct mutagenic mechanisms. Specifically, an HIV-1 was produced from APOBEC3G (A3G)-expressing cells and used to infect permissive target cells treated with 5-azacytidine (5-AZC). Reduced viral infectivity and increased viral mutagenesis was observed with both the viral mutagen (i.e., G-to-C mutations) and the host restriction factor (i.e., G-to-A mutations); however, when combined, had complex interactions. Intriguingly, nucleotide sequence analysis revealed that concomitant HIV-1 exposure to both 5-AZC and A3G resulted in an increase of G-to-A viral mutagenesis at the expense of G-to-C mutagenesis. A3G catalytic activity was required for the diminution in G-to-C mutagenesis. Taken together, our findings provide the first demonstration for potentiation of the mutagenic effect of a cytosine analog by A3G expression, resulting in concomitant HIV-1 lethal mutagenesis.
Keywords: lentivirus, retrovirus, evolution, deamination, transversion
INTRODUCTION
The quasispecies model describes how rapidly evolving viruses exist within a mutation-selection balance 1; 2. The model asserts that high viral mutation rates drives the formation and maintenance of the quasispecies and allows for these viruses to readily adapt to a changing environment. However, the quasispecies also presents a significant obstacle in the successful treatment of rapidly evolving viruses such as human immunodeficiency virus type 1 (HIV-1). It has been predicted that a purposeful increase in viral mutation rate would cause an irreversible meltdown of the genetic information 3; 4; 5; 6. This process, termed extinction catastrophe, has been exploited as a novel therapeutic approach, yet findings from short-term cell culture based studies fail to predict the shortcomings observed clinically. For example, RNA viruses have been lethally mutagenized in vitro by various nucleoside analogs, including: ribavirin, 5-flurouracil, and 5-AZC 7; 8. Specifically, ribavirin was shown to lethally mutagenize poliovirus and hepatitis C virus 3; 9, while 5-flurouracil was shown to be an active viral mutagen against foot-and-mouth-disease virus 8. The compound, 5-AZC, was also demonstrated to lethally mutagenize HIV-1 in cell culture through induction of G-to-C mutations 7. A related compound, KP1212 was shown to lethally mutate HIV-1 in cell culture; however, the compound did not decrease viral loads or increase viral mutation loads in patient samples 10; 11. Similarly, abundant mutations were identified in patient-derived hepatitis C virus suggesting purposeful mutagenesis by the ribavirin-interferon regimen 12; however, a second study showed only a transient increase in mutation rate in patients on ribavirin monotherapy indicating that lethal mutagenesis may not be the sole antiviral mechanism 13.
Lethal mutagenesis can be induced not only by drugs, but also by the APOBEC3 (A3) family of proteins 14; 15; 16; 17; 18; 19; 20; 21; 22; 23. These proteins have emerged as innate restriction factors that induce targeted hypermutagenesis of viral genomes. While their importance is suggested by the rapid evolutionary expansion of the A3 locus, most APOBEC3 proteins seem to be active against retroviruses and retroelements. However, A3G and A3F exert potent anti-HIV-1 activity through lethal mutagenesis (reviewed in 24 and 25). Both A3G and A3F, along with A3B, possess the capacity to restrict other retroviral genera, including: murine leukemia virus (MLV, gammaretrovirus) 14; 16; 20, human T-lymphotrpic virus 1 (HTLV-1, deltaretrovirus) 21, foamy viruses (FVs, spumavirus) 26, as well as equine infectious anemia virus (EIAV, lentivirus) 16. In addition to retroviruses, hepatitis B virus (HBV, hepadnavirus), and adeno-associated virus (AAV, parvovirus), are also susceptible to members of the A3 family 18; 19.
The mechanism by which A3G hypermutates retroviral genomes has been well established (reviewed in 27, 28, and 29). Briefly, A3G is packaged into budding virions, after which the virion matures and binds to a target cell. In the target cell, A3G deaminates cytosines (C) present in the single-stranded negative-sense viral DNA during reverse transcription process. The deamination of C leads to uracil (U) and this pre-mutatgenic lesion can template for adenine (A) during plus strand DNA synthesis rather than guanine (G). The deamination of C by A3G during reverse transcription generates G-to-A mutation signatures in the resulting provirus14. However, the ability of A3G to mutate the viral genome depends on its ability to overcome viral countermeasures – such as the HIV-1 Vif protein. In a host-specific manner, Vif targets A3 proteins for proteosomal degradation. However, through saturating A3G levels or less-stringent Vif alleles, A3G proteins can gain access to the nascent virions and mutate the viral genome as described above. The ability of A3G to escape Vif is evident in patient samples where signature mutations indicative of A3G have been observed30; 31. A deaminase-independent mechanism has been proposed for HIV-1, but this model remains controversial 32; 33; 34; 35.
Many of the compounds that lethally mutagenize HIV-1 are C analogs including KP1212, 5-OH-dC, and 5AZC7; 10; 36. Competitive replacement by C mutagens could interfere with A3G-mediated deamination. For instance, the kinetics of 5-AZC- and A3G-generated mutations indicate that 5-AZC incorporation into viral DNA precedes the ability of A3G to catalyze cytosine deamination. Replacement of C with 5-AZC may remove potential sites that would otherwise be mutated by A3G. Therefore, compounds targeting C residues may not be the most efficient at inducing lethal mutagenesis in the presence of APOBEC3 proteins. To examine this type of interaction, we investigated mutagen-specific alterations to both mutation spectra as well as the mutational load. Interestingly, our results show that exposure of HIV-1 to both 5-AZC and A3G concomitantly increased the frequency of G-to-A mutations at the expense of G-to-C mutagenesis. Furthermore, the diminution of G-to-C mutations were dependent on A3G catalytic activity. This is the first demonstration for potentiation of the mutagenic effect of a cytosine analog by A3G expression, resulting in concomitant HIV-1 lethal mutagenesis.
RESULTS
Concomitant antiviral effects of 5-AZC and A3G
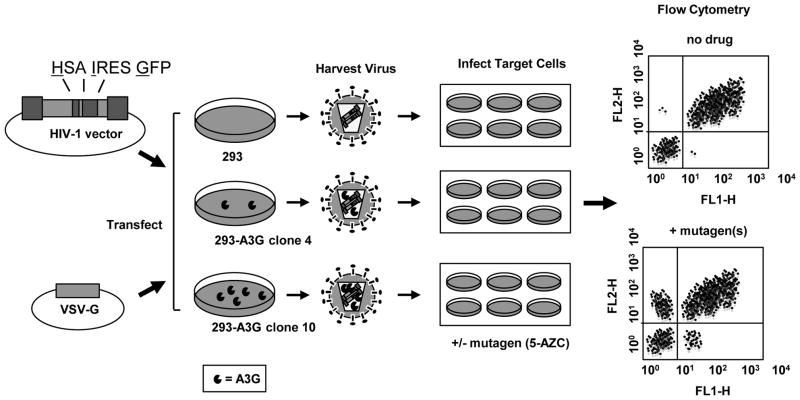
A single cycle vector assay was used to assess the concomitant antiviral effect of 5-AZC and A3G (Figure 1). This HIV-1 vector system utilizes a dual reporter cassette to monitor both infection efficiency as well as mutant frequency. Infectious vector virus was produced by transfection of a HIV-1 vector (pHIG) and VSV-G envelope expression plasmid into previously characterized 293 cell lines that stably express A3G at physiologically-relevant levels 37. These cell lines, previously characterized by our lab, were used in this study: 1) a 293-A3G null vector control which does not express A3G; 2) a cell line expressing low A3G levels (293-A3G clone 4); and 3) a cell line expressing high A3G levels (293-A3G clone 10)37. Virus produced from the A3G-expressing cells results in packaging of A3G into the virus particles. These virus particles are used to infect permissive target cells that have been treated with a range of 5-AZC (10–200 μM). Our assay allows for both HIV-1 mutagens to be present during reverse transcription. Any decrease in antiviral activity can be assessed by flow cytometry as a decrease in double-positive HSA+/GFP+-expressing cells.
Figure 1. HIV-1 vector assay for analysis of viral mutagens.
The HIV-1 vector (pHIG) 7 was co-transfected with a VSV-G envelope expression plasmid into previously characterized APOBEC3G (A3G)-expressing 293 cell lines 37. Cell culture supernatants were collected, filtered, and virus was used to infect U373-MAGICXCR4 target cells. Following normalization of virus titers, target cells were pre-treated for 2-hr with 5-AZC and infected with HIV-1 at an MOI of 0.30. Seventy-two hours post-infection, cells were collected for analysis. Abbreviations: MOI = multiplicity of infection; VSVG = vesicular stomatitis virus glycoprotein; HSA = murine heat stable antigen, CD24; IRES = internal ribosome entry site; GFP = green fluorescent protein; A3G = APOBEC3G. FL1-H = fluorescence channel 1, height of intensity; FL2-H = fluorescence channel 2, height of intensity.
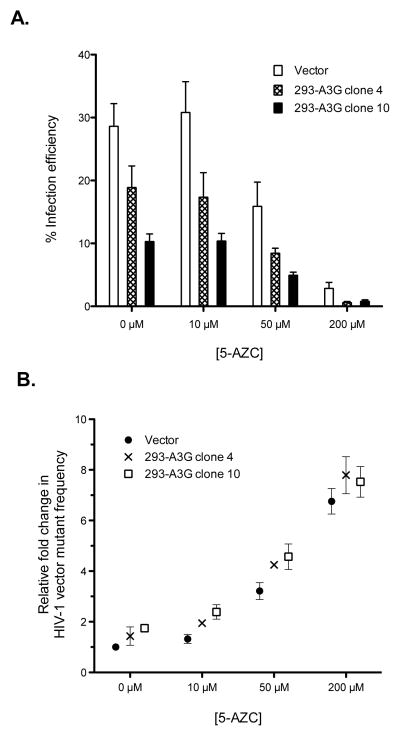
Similar to previous reports 14; 16; 17, A3G decreased HIV-1 replication (Figure 2A) and this decrease was greater as the expression of A3G increased (293-A3G clone 4 vs 293-A3G clone 10) 37. Specifically, vector virus produced from 293-A3G clone 4 reduced viral infectivity by one-third, while virus produced from 293-A3G clone 10 reduced viral infectivity by two-thirds. When permissive target cells were treated with various concentrations of 5-AZC (i.e., 10 μM, 50 μM or 200 μM), A3G increased 5-AZC’s antiviral effect (Figure 2A). For example, virus treated with 50 μM 5-AZC reduced infectivity by 2-fold compared to vector alone; however, this inhibitory effect was potentiated by 3.4- and 5.8-fold with concomitant exposure to A3G4 low and A3G10 high, respectively (Figure 2A). This same trend was observed with concomitant treatment of 200 μM 5-AZC and increasing amounts of A3G, indicating that A3G increased the inhibitory activity of 5-AZC (Figure 2A).
Figure 2. Concentration-dependent inhibition of 5-AZC of HIV-1 replication in the presence of A3G.
A. HIV-1 infectivity in the presence of 5-AZC and/or A3G. Virus stocks, produced from the A3G-expressing cell lines 293-A3G clone 4 and 293-A3G clone 1037, were normalized by transfection efficiency. U373-MAGICXCR4 cells were pretreated for 2 h with the indicated concentrations of 5-AZC prior to infection. Infection levels were determined by expression of both the HSA and GFP marker genes. B. Relative mutant frequency. The mutant frequency is shown relative to the vector + vehicle control. Data represent the mean fold change ± SD from at least 3 independent replicates. Mutant frequency was calculated as described in the Materials and Methods.
The concept of lethal mutagenesis suggests that decreased infectivity should correlate with an increase in mutation frequency. Therefore, we investigated if the loss of infectivity seen in Figure 2A correlated with an increase in mutant frequency. To do this, the single cycle assay was used, but the data were analyzed for expression of marker genes. For example, targets cells expressing a single marker gene (i.e. [HSA+/GFP−] or [HSA-/GFP+]) would likely indicate a loss-in-phenotype mutation that occurred during reverse transcription. As indicated in Figure 2B, A3G expression increased viral mutant frequency. Similarly, the mutant frequency was significantly increased when target cells were treated with either 50 μM or 200 μM 5-AZC. Again, the 5AZC-mediated increase in mutant frequency was further increased by A3G expression. For instance, vector virus treated with 50 μM 5-AZC increased the mutant frequency by 3.2-fold, while concomitant A3G exposure potentiated this effect by 4.3- and 4.6-fold, A3G4 low and A3G10 high, respectively (Figure 2B). Similar trends were found with concomitant A3G exposure in the presence of 200 μM 5-AZC (Figure 2B).
Concomitant Exposure to 5-AZC and A3G alters HIV-1 Mutation Spectra
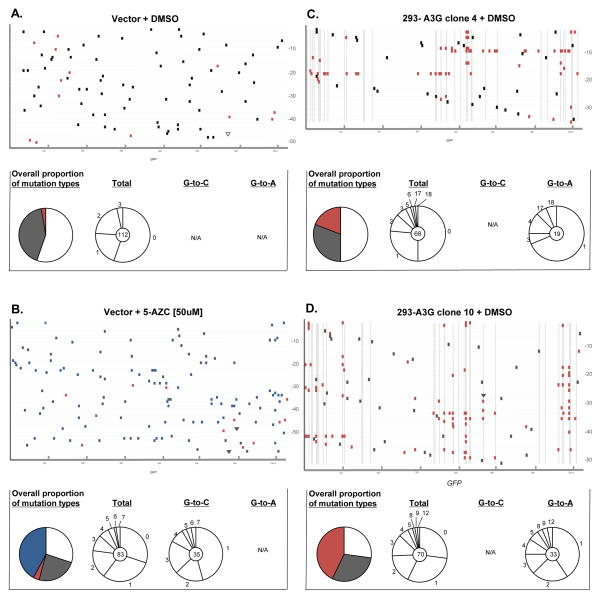
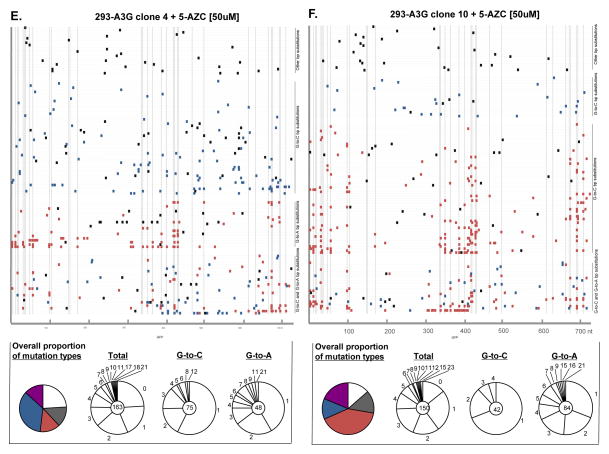
The increase in mutant frequency suggests that viral genomes were heavily mutated by the action of A3G and 5-AZC. Since both mutagens target cytosines and induce distinct mutations, we examined proviruses that had been exposed to either A3G, 5AZC (EC50 concentration), or to both. Since the resulting proviruses were expected to have a very high mutational load, primers were designed to minimize PCR bias caused by heavily mutated sequences (as described in the Methods and Materials). Table 1 shows the cumulative mutation spectra with each mutation type denoted as the percentage of total number of mutations. As expected, A3G levels correlated to an increase in the percentage of G-to-A mutations. As an example, 65% of all mutations identified were G-to-A when sequencing proviruses exposed to low A3G levels (i.e., 293-A3G clone 4) whereas 70% of mutations were G-to-A when sequencing provirus exposed to high A3G levels (293-A3G clone 10). The lack of a more dramatic difference in the percentage of G-to-A mutations may be due to the presence HIV-1 Vif protein, and its ability to partially restrict A3G activity in these cells. Another possibility could be due to the unexpectedly high number of G-to-A mutations at non-A3G signature sites (5′-GG-3′) found in virus produced from the 293-A3G clone 4 cells (Figure 3C). In contrast, 5-AZC induced a robust increase (61%) in the number of G-to-C transversions compared to untreated cells (Table 1) as previously reported 7.
TABLE 1.
Mutation spectra in the GFP gene of vector proviral sequences
| Mutation from: | Mutation to % (±SD):
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vector + vehicle controla |
Vector + 5-AZC [50μM]b |
A3G clone 4 + vehiclec |
A3G clone 10 + vehicled |
A3G clone 4 + 5-AZC [50μM]e |
A3G clone 10 + 5-AZC [50μM]f |
|||||||||||||||||||
| A | T | C | G | A | T | C | G | A | T | C | G | A | T | C | G | A | T | C | G | A | T | C | G | |
| A to | - | 0 | 2 (2) | 39 (5) | - | 0 | 0 | 13 (9) | - | 0 | 0 | 16 (5) | - | 0 | 1 (2) | 14 (1) | - | 0 | 1 (1) | 11 (5) | - | 0 | 0 | 6 (3) |
| T to | 2 (3) | - | 24 (8) | 0 | 0 | - | 8 (4) | 2 (3) | 0 | - | 10 (5) | 0 | 0 | - | 9(4) | 1 (1) | 0 | - | 5 (1) | 0 | 0 | - | 3 (2) | 0 |
| C to | 0 | 13 (5) | - | 0 | 0 | 6 (3) | - | 1 (1) | 0 | 9 (8) | - | 0 | 0 | 2 (2) | - | 1 (2) | 1 (1) | 7(4) | - | 1 (1) | 0 | 5 (1) | - | 0 |
| G to | 19 (6) | 0 | 0 | - | 4 (4) | 5 (2) | 61 (12) | - | 65 (12) | 0 | 0 | - | 70 (5) | 1 (2) | 0 | - | 28 (9) | 4(2) | 41 (16) | - | 64 (6) | 1 (1) | 20 (10) | - |
Total no. of sequences, 112; total no. of mutations, 81; no. of nucleotides sequenced, 80640; mutation frequency × 10−3, 1.035 (±0.136)
Total no. of sequences, 83; total no. of mutations, 132; no. of nucleotides sequenced, 59760; mutation frequency × 10−3, 1.953 (±0.459)
Total no. of sequences, 68; total no. of mutations, 95; no. of nucleotides sequenced, 48960; mutation frequency × 10−3, 1.602 (±0.368)
Total no. of sequences, 70; total no. of mutations, 127; no. of nucleotides sequenced, 50400; mutation frequency × 10−3, 2.203 (±0.335)
Total no. of sequences, 158; total no. of mutations, 363; no. of nucleotides sequenced, 113760; mutation frequency × 10−3, 3.255 (±0.383)
Total no. of sequences, 155; total no. of mutations, 468; no. of nucleotides sequenced, 111600; mutation frequency × 10−3, 4.272 (±0.305)
Figure 3. Mutation spectra analysis from recovered proviruses.
Total genomic DNA from infected cells was purified, proviral HIV-1 DNA amplified, cloned, and analyzed by DNA sequencing. A. Vector + DMSO, B. vector + 5-AZC, C. 293-A3G clone 4 + DMSO, D. 293-A3G clone 10 + DMSO, E. 293-A3G clone 4 + 5-AZC, F. 293-A3G clone 10 + 5-AZC. Each gray horizontal line represents an individual GFP gene sequence (720 bp) from a recovered provirus (5′-to-3′). Mutation locations are indicated by colored rectangular boxes (G-to-A mutations, orange; G-to-C mutations, blue; other base substitution types, black) or by triangles (insertion mutations, black; deletions, white). Dashed vertical lines indicate base locations in the GFP gene sequences that are predicted to be A3G hotspots, as determined using CLUSTERM (as described in the Materials and Methods). To the right of each of experimental set (A–F) of GFP gene sequences are summary pie charts that indicate total numbers of G-to-A, G-to-C, and other mutation types characterized in experiments. Below each of experimental set (A–F) of GFP gene sequences are mutation summaries – shown in pie charts. The first column shows colored pie charts and the proportion of recovered clones with no mutations, white; without either G-to-A or G-to-C mutations, grey; with G-to-A mutations, red; with G-to-C mutations, blue; with G-to-A and G-to-C mutations, purple. The other 3 columns show pie charts indicating the number of mutations per proviral sequence (i.e., 1, 2, 3, etc.) from the total recovered clones (2nd column), G-to-C only mutated clones (3rd column), or G-to-A only mutated clones (4th column). The total number of proviral clones is indicated by the number at the center of the pie chart. N/A = not applicable.
Since both A3G and 5-AZC induce mutations by targeting C residues, we next examined viral mutation spectra of combined expression of A3G with 5-AZC treatment. As shown in Table 1, viral genomes exposed to the 5-AZC EC50 with increasing levels of A3G significantly decreased the percentage of G-to-C transversion mutations. In particular, 61% of the mutations in viruses that were exposed to 5-AZC alone were G-to-C mutations, whereas 41% of the mutations in viruses exposed to both 5-AZC and a low level of A3G were G-to-C mutations. Finally, only 20% of mutations in viruses exposed to both 5-AZC and a high level of A3G were G-to-C mutations. The general increase in mutational loads suggests that A3G and 5-AZC significantly increased the viral mutation frequency, which is consistent with lethal mutagenesis.
The hotspot prediction algorithm CLUSTERM was used to determine whether 5-AZC-induced G-to-C mutations or A3G-induced G-to-A mutations were influenced by preference sites (i.e., hotspots). As expected by the random substitution of 5-AZC for C bases, the hotspot prediction algorithm found no G-to-C hotspots within GFP. Alternatively, as described in previous A3G studies, the clustering of G-to-A mutations at GG dinucleotides was predicted by CLUSTERM 38; 39. Of the 51 di-G nucleotide positions in the plus-strand DNA of GFP, 26 bases were found to represent A3G preference sites.
Disparate Mutational Load with Concomitant 5-AZC and A3G Exposure
While mutational spectra analysis offers a population view of the 5-AZC and A3G exposed virus, analysis of mutational load can discern differences among individual viral clones. The mutational load was determined for each mutation type of interest (i.e. G-to-C and G-to-A) by finding the mean number of mutations per viral clone (Table 2). As expected, the G-to-A mutational load increased from 0.03% to 0.35% and 0.37%, when comparing viral clones unexposed to A3G to those exposed to A3G low and A3G high levels, respectively (Table 2). The G-to-C mutational load was also increased to 0.31% relative to no drug controls (Table 2). Unexpectedly, the G-to-A mutational load was 18% higher (p = 0.026) when A3G and 5-AZC exposure was combined, compared to this amount of A3G alone (Table 2). Moreover, the concomitant exposure of A3G and 5-AZC also diminished the G-to-C mutational load by 36% (p = 0.013) compared to drug alone. These results suggest that 5-AZC and A3G an overlap of the mutagenic effects of these distinct viral mutagens, which ultimately influence the mutation load of G-to-A and G-to-C mutations.
TABLE 2.
Mutational load in the GFP gene of vector proviruses
| Vector + 5-AZC [50μM]
|
A3G clone 4 + DMSO
|
A3G clone 4 + 5-AZC [50μM]b |
A3G clone 10 + DMSO
|
A3G clone 10 + 5-AZC [50μM]b |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| # G-to-A mutations | 12 | 0.03% | 48 | 0.35% | 148 | 0.43% | 87 | 0.37% | 287 | 0.48% |
| Total # of G-to-A mutable positionsa | 41760 | 13680 | 34560 | 23760 | 59760 | |||||
| % change in G-to-A mutational load | + 12% (p > 0.2) | + 18% (p = 0.026) | ||||||||
| # G-to-C mutations | 78 | 0.31% | - | 0.00% | 157 | 0.27% | - | 0.00% | 63 | 0.20% |
| Total # of G-to-C mutable positionsa | 25200 | - | 58320 | - | 30960 | |||||
| % change in G-to-C mutational load | − 13% (p > 0.3) | − 36% (p = 0.013) | ||||||||
Number of clones multiplied by the 720 nucleotide positions in the GFP target sequence.
p values determined by χ2 analysis.
A3G catalytic activity is required for reductions in 5-AZC-mediated G-to-C mutations
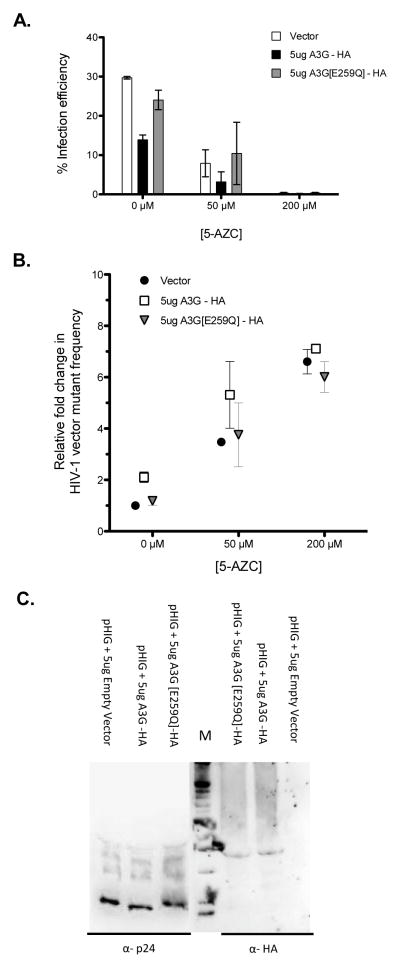
Since A3G has been shown to possess catalytic-independent antiviral activity, we next examined the effect of a catalytically inactive A3G on the infection efficiency. It was observed that the A3G_E259Q mutant had no effect on either the efficiency of infection or mutant frequency compared to that of vector only (Figure 4A-B). Importantly, there was no difference in G-to-C mutation spectra under concomitant exposure to 5-AZC and A3G_E259Q compared to that of 5-AZC alone – i.e., 57% vs 59%, respectively (Table 3). Immunoblot analysis of A3G virion incorporation demonstrated that A3G_E259Q was as efficiently packaged into HIV-1 particles as wt A3G, indicating that the lack of efficient incorporation of A3G_E259Q was not responsible for these observations (Figure 4C). Taken together, these data indicate that A3G catalytic activity is necessary in order to observe the decrease in infection efficiency as well as the potentiation of the mutagenic effect of 5-AZC by A3G expression, which results in concomitant HIV-1 lethal mutagenesis.
Figure 4. Requirement of APOBEC3G catalytic activity for reducing the frequency of 5-AZC-mediated G-to-C mutations.
293T cells were transfected with the HIV-1 vector (pHIG), VSV-G envelope expression plasmid and either a A3G-expression vector, the A3G(E259Q) catalytically-inactive mutant expression vector, or vector only. Twenty-four hours post-transfection, cell culture supernatants were harvested and used to either infect 5-AZC treated target cells with viral infectivity (A) or viral mutant frequency (B) determined or used for immunoblot analysis of HIV-1 Gag or A3G (C).
Table 3.
Effect of catalytically-inactive A3G expression in virus-producing cells on mutation spectra in the GFP gene of vector proviruses
| Mutation from: | Mutation to % (±SD):
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vector + 5-AZC [50μM]a |
A3G E259Q + DMSOb |
A3G E259Q + 5-AZC [50μM]c |
|||||||||||
| A | T | C | G | A | T | C | G | A | T | C | G | ||
| A to | - | 0 | 0 | 19 (3) | - | 2 (2) | 2 (1) | 19 (0) | - | 0 | 0 | 14 (3) | |
| T to | 0 | - | 3 (2) | 5 (2) | 0 | - | 29 (5) | 2 (2) | 0 | - | 7 (1) | 0 | |
| C to | 0 | 3 (2) | - | 0 | 0 | 17 (10) | - | 0 | 0 | 10 (3) | - | 1 (1) | |
| G to | 10 (4) | 5 (3) | 59 (5) | - | 26 (3) | 2 (2) | 0 | - | 10 (3) | 2 (2) | 57 (10) | - | |
Total no. of sequences, 36; total no. of mutations, 59; no. of nucleotides sequenced, 25920; mutation frequency × 10−3, 2.276 (±0.346)
Total no. of sequences, 53; total no. of mutations, 42; no. of nucleotides sequenced, 38160; mutation frequency × 10−3, 1.101 (±0.371)
Total no. of sequences, 90; total no. of mutations, 103; no. of nucleotides sequenced, 64800; mutation frequency × 10−3, 1.590 (±0.312)
DISCUSSION
Lethal mutagenesis as a therapeutic strategy has gained momentum, especially after ribavirin and KP-1212 gained clinical use 9; 11. Most of the exploratory HIV-1 mutagens target C analogs including the triazole base, 5-AZC. Since the APOBEC3 protein subfamily also targets C bases for deamination, these mutagens may exert redundant mechanisms when combined. This would suggest that compounds targeting C bases could be less effective mutagens in the presence of A3G. Alternatively, incorporation of 5-AZC could have secondary effects by altering A3G substrate specificity or target site architecture. In this study, we examined concomitant exposure of HIV-1 to 5-AZC and A3G. The ability of A3G to induce G-to-A hypermutagenesis provides an example of an evolutionary conserved mechanism that eliminates HIV-1 infectivity by lethal mutagenesis. Editing of retroviral genomes by A3G in the face of concomitant mutagen exposure has not been previously explored.
5-AZC is a first-in-class hypomethylating agent as substitution of N-5 can no longer be methylated 40. The ribonucleoside 5-AZC undergoes anabolic metabolism to the corresponding triphosphate, but a fraction is converted to the nucleotide triphosphate, 5-aza-2′-deoxycytidine (5-AZdC), and is substituted for dCTP in DNA 41; 42. Because aza-pyrimidines are chemically unstable, ring-opened intermediates may have the potential for non-Watson-Crick base pairing, leading to base mispairing 43. In fact, 5-AZC has been implicated in several studies as a mutagen, specifically introducing a rare GC-to-CG transversion type 7; 44; 45; 46. Experimentation with an HIV-1 vector system found that 5-AZC increased the preponderance of only G-to-C mutations, suggesting that the mutagenic product of azacytosine is able to template for C:C mispairings when incorporated during minus-strand DNA synthesis 7.
Intriguingly, the majority of retroviral minus-strand DNA is also transiently single-stranded during replication, and ssDNA is the preferential substrate of the APOBEC3 subfamily 14. During HIV-1 reverse transcription, A3G gains access to ssDNA, targeting cytosine residues for deamination to pre-mutagenic lesions 47. These kinetics, of A3G and 5-AZC-based mutagenesis, suggest that access to minus-strand ssDNA by A3G is subsequent to RT incorporation of 5-AZdC during minus-strand DNA synthesis.
The interaction between A3G and the non-canonical 5-azacytosine base could result in two possible obvious outcomes: 1) A3G is unable to engage 5-azacytosine or its ring-opened decomposition products. This would be observed as a decrease in G-to-A mutations with no difference in the number of G-to-C mutations; 5-azacytosine would effectively antagonize A3G deaminase activity. 2) A3G is able to catalyze deamination of 5-azacytosine to 5-azauracil and this uracil analog, results in increased G-to-A mutations at the expense of G-to-C mutation types. However, other mechanisms cannot be excluded at this time, such as: the ability of 5-azacytosine (or its ring-opened products) to alter local DNA secondary structure and subsequent ssDNA availability, or the influence of 5-azacytosine on A3G processivity or substrate specificity due to close-proximity base interactions, as suggested by Rausch et al. 48.
In order to better understand mutation type-specific differences of concomitant mutagen exposure, proviral DNAs were sequenced. The G-to-A mutational load (i.e., # of G-to-A mutations/proviral clone) was increased up to 18% in virus exposed to both 5-AZC and A3G, as compared to A3G exposure alone. This effect was at the expense of a 36% decrease to the 5-AZC-induced mutational load (i.e., # of G-to-C mutations/sequence). These results suggest a model whereby 5-AZC incorporation into retroviral DNA causes complex interactions with A3G, to inversely shift the G-to-A and G-to-C mutational loads. Furthermore, the E259Q catalytically inactive A3G, in conjunction with 5-AZC, showed no difference in G-to-C transversions compared to drug alone, suggesting a requirement for fully functional A3G in order to observe the increase of G-to-A mutations at the expense of G-to-C mutations.
One potential concern regarding the sequencing data is biased amplification due to the oligonucletides selected. Particular attention to primer design helped ensure universal and unbiased PCR. For example, the location of each oligonucleotide was carefully adjusted such that no di-GG nucleotide motif was positioned in the forward primer (and no di-CC nucleotide motif in the reverse primer) in order to eliminate the potential for mis-annealing due to the creation of A3G signatures. Moreover, no more than two G nucleotides were positioned at the forward primer regions (no more than two C nucleotides in the reverse primer region), while these potentially mutable bases were substituted with the degenerate S (50% G and 50% C), to exclude 5-AZC biased amplification. These primers amplified heavily mutated sequences in each treatment group (23 mut/720 bp), indicating that the sequenced DNAs were not biased against any mutation type.
Presently, concomitant use of viral mutagens and chain terminators shows efficacy against RNA viruses 49; 50; 51; 52; 53; 54. For example, Perales et al. demonstrated that in foot-and-mouth disease virus, sequential treatment with a traditional antiviral inhibitor followed by the viral mutagen ribavirin was much more effective in extinguishing picornavirus replication than these compounds used together, or either one alone 53. Future progress of mutagen utilization hinges on understanding the mutational constraints within RNA virus population structure. For example, sub-restrictive editing of retroviral genomes by APOBEC3 proteins has the potential to negatively influence therapeutic intervention, by rapid generation of drug-resistant mutants 55; 56. Furthermore, quasispecies theory predicts that populations will evolve more robust genomes, termed survival of the flattest, in the face of increased mutational load 57; 58. Alternative adaptive strategies include selection of anti-mutator viral polymerases 59; 60; 61 or enhanced discrimination between correct and mutagenic nucleotides 62. Evolution of drug resistance, or increased robustness, of mutagens can threaten such therapeutic approaches; yet, discovery of novel viral mutagens, as well as optimal applications, may lead to alternative therapeutic strategies.
Understanding molecular details of potential mutagens as well as sequence space limitations of specific viral pathogens may provide a rationale for tailored therapeutic intervention rather than lengthy small molecule screening. Generally, HIV-1 viral mutagens are stealth nucleosides directly utilized by RT during replication to induce site-specific mutations. These compounds are referred to as universal bases because they can mispair with more than one of the canonical Watson-Crick base pairs. For instance, the viral mutagen KP1212, can base pair with either G or A due to a tautormeric shift in the pyrimidine base. 5-AZC, a close derivative to KP1212, induces a unique G-to-C transversion mutational pattern during HIV-1 replication because of its ability to mispair with C bases 7; 43; 44. Even by understanding specific molecular details, viral mutagens still lack a reliable framework to help predict successful treatment outcomes. Many parameters involved in understanding virus population dynamics and genetic diversity are not fully understood, including: sequence space, mutational robustness, effective population size, as well as the natural fitness landscape. Similarly, since nucleotide base composition among viral genera is known to be quite distinct, purposeful alterations to the mutational bias of a particular virus may pose a greater defect to viral fitness. For example, since HIV-1, like other lentiviruses, has an unusually A-rich genome 63, it is not clear if mutagens that cause more N-to-A mutations (versus A-to-N) are more detrimental to viral fitness.
In summary, we describe concomitant HIV-1 mutagenesis using two unrelated classes of viral mutagens. Our findings indicate a combined, yet intricate, interaction between the 5-AZC and A3G. The combined antiviral effect observed indicated that A3G potentiated the mutagenic effect of 5-AZC. Sequencing analysis revealed that the combined mutagenic effect resulted in an increase in the frequency of G-to-A mutations at the expense of G-to-C mutations, suggesting a complex interaction between A3G and 5-AZC upon incorporation into viral DNA. Future studies will provide greater details into the molecular mechanisms involved in concomitant HIV-1 lethal mutagenesis by A3G and 5-AZC.
MATERIALS AND METHODS
Plasmids constructs and cell lines
The HIV-1 vector pHIG has been previously described 7. Briefly, a ~2.0 kbp dual reporter cassette comprised of the murine heat stable antigen (HSA), an IRES element, and eGFP was placed in frame and 3′ to the NL4-3 nef start codon. The G protein of vesicular stomatitis virus (VSV-G) envelope expression plasmid HCMV-G was used to pseudotype virions and was a kind gift from J. Burns (UCSD). The A3G-expressing cell lines 293-A3G clone 4 and 293-A3G clone10 were previously characterized in Sadler et al. 37. The APOBEC-expression plasmids pcDNA3.1+-A3G-3xHA and pcDNA3.1+-A3G_E259Q-3xHA were a kind gift from R. Harris (University of Minnesota). U373-MAGICXCR4 target cells lines were obtained from the AIDS Reagent Program (from M. Emerman).
Single-cycle transduction and mutant frequency assays
Vector virus was produced by transient transfection of 293 cell lines with pHIG. Producer cell lines were transfected with 10μg pHIG and 1μg VSV-G env plasmids using Genjet ver. II reagent (SignaGen Rockville, MD) as per manufacturer’s recommendations. Twenty-four hours post-transfection, viral supernatants were collected and filtered through a 0.2-μm filter. Viral stocks were normalized by transfection efficiency, measured by %GFP-expressing producer cells. Viral stocks were either used to infect target cells or stored at −80°C. The single-cycle infection efficiency for the control A3G null virus was set to 30%. U373-MAGICXCR4 target cells were plated in flat bottom 12-well dish at concentration of 6.25 ×106 cells per ml 24-hr prior to infection. Experiments with the viral mutagen 5-AZC (Sigma-Aldrich St. Louis, MO) included a 2 hr pre-treatment of 1:1000 dilution prior to addition of normalized viral supernatant up to 1ml total volume. At 24 hr post-infection, media was replaced with fresh cell culture media.
Flow cytometry analysis
Target cells were prepared for flow cytometry to quantify transduction efficiency (via both GFP and HSA reporter gene expression) and mutant phenotype (via lack of GFP or HSA reporter gene expression). At 72 hr post infection cells were collected and stained with 1:250 anti-HSA-PE (BD Pharmingen). Following 20-min incubation at 4°C, cells were washed 1× in PBS-2% FC3, and resuspended for flow cytometry. Mutant frequency analysis was determined from the % target cells expressing a single reporter gene relative to the total infected cell population (i.e., %[HSA+/GFP−] plus %[HSA−/GFP+] divided by % of total infected cells). Mutant frequencies were set relative to virus null for A3G in the absence of drug. Cells were analyzed with a FACScan (BD Biosciences) and CellQuest software. Cells were gated by morphology (FFS vs. SSC) by counting 10,000 cells for fluorescence analysis. Excitation was done at 488 nm; fluorescence channel 1 (FL1) detected GFP emission at 507nm, and FL2 detected PE emission at 578nm. Compensation was set based on single color controls to eliminate spill-over and re-verified based on the geometric mean of single-color positive to negative-detected populations.
Proviral DNA sequence analyses
Approximately 25,000 target cells from each treatment group (+/− A3G and +/− 5-AZC) were collected for total genomic DNA isolation (Roche High Pure PCR Template Preparation Kit) and subjected to nested PCR of the GFP gene in the HIV-1 vector proviral DNA. Primer sets were carefully designed as to remove any bias in amplicon pool considering mutagenic potential of the agents used. Specifically, forward primers did not contain any 5′-GG-3′ motifs while reverse primers were absent of 5′-CC-3′ motifs to remove the selection by A3G. Additionally, forward primers were located to regions containing only 2 Gs (same for reverse, however regions contained only 2 Cs) but these sites were designed to contain a G or C, denoted by S. This approach limited the 5-AZC-induced G-to-C bias that may be excluded from the amplicon pool. Within the pHIG vector these primer sets would amplify a 944pb region of GFP: outer primer pair, 5′-CTCAATSCCACASCCATA-3′ and 5′-GTSTTSTTTGGGAGTGAATTAG-3′; inner primer pair, 5′-CTCTCCTCAASCSTATTCAAC-3′ and 5′-GGTATGGSTGATTATGATSTAGAGT-3′. PCR reactions used the Platnium PCR Supermix (Invitrogen), and the initial PCR reaction was purified with GenElute PCR Clean-up Kit (Sigma). Amplicons were verified for correct size and purity by DNA gel imaging before prior to ligation into the pGEM-T vector (Promega). Plasmids were transformed into E. coli, and insert containing-vectors were purified (DirectPrep 96 Miniprep Kit, Qiagen) and sequenced. Sequence alignments were performed using SeqMan of Lasergene 7 (DNAStar; Madison, WI). Only DNA sequences covering the entire GFP gene (720bp) were used for sequence analysis. This region was chosen because it has been previously used for sequencing analysis of A3G and 5-AZC-mediated viral mutagenesis 7; 14; 64; 65.
Immunoblot analysis of transiently transfected A3G-containing particles
293 cells were transfected with 10μg pHIG, 1μg HCMV-G env, and either 5μg pcDNA3.1+-A3G-3xHA or pcDNA3.1+-A3G_E259Q-3xHA or 5μg empty vector, using Genjet ver. II reagent as per manufacturer’s recommendations. Twenty-four hours after transfection, cell culture supernatants were collected and passed through a 0.2-μm filter. Next, 1.5 ml of cell culture supernatant was centrifuged at 16,000×g for 2 hr at 4°C. Virus particle pellets were resuspended in 2.5 μl of 10X RIPA buffer and 2.5 μl of a 10X protease inhibitor cocktail was added and incubated on ice for 15 min. Samples were then denatured by the addition of 6X loading dye and boiled for 10 min prior to SDS-PAGE with a 4–20% denaturing gel. Proteins were transferred to nitrocellulose and A3G and A3G_E259Q-containing was probed with a primary monoclonal antibody, HA.11 (Covance; Emeryville, CA), followed by a goat anti-mouse HRP conjugate (Invitrogen). To detect HIV-1 capsid (p24) protein, the nitrocellulose membrane was incubated with a rabbit anti-p24 antibody (Advanced Biotechnologies; Columbia, MD) followed by a goat anti-rabbit IgG (H+L)-HRP conjugate (BioRad). Detection of protein location on the nitrocellulose membranes was determined using a BioRad ChemiDoc Imager with Quantity One software (version 4.5.2, BioRad).
Statistical analyses
All statistical analyses and graphical representation were done using GraphPad Prism version 5.0 (GraphPad Software, La Jolla, CA). Bars graphs of infection efficiency and mutant frequency data were established with the mean +/− standard deviation. Statistical analysis of mutational load differences was performed using a chi-squared contingency table, comparing the number of mutated sites to the number of non-mutated sites with each mutation type (i.e., G-to-A and G-to-C). A3G hotspot prediction was determined using CLUSTERM (http://www.itba.mi.cnr.it/webmutation and ftp://ftp.bionet.nsc.ru/pub/biology/dbms/CLUSTERM.ZIP). This algorithm predicts a hotspot threshold for the number of mutations at a particular site. The threshold number is established by analyzing the frequency distribution of mutations among a pre-determined genomic region 39.
Acknowledgments
We thank Christine Clouser, Holly Sadler, Steve Patterson and Reuben Harris for helpful comments and suggestions. We also thank Igor Rogozin for help with the use of CLUSTERM for hotspot prediction analysis. This research was supported by NIH grant R01 GM56615. M.J.D. was supported by NIH grant T32DA007097 and C.M.H. by T32DE007288.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Eigen M. Viral quasispecies. Scientific American. 1993;269:42–9. doi: 10.1038/scientificamerican0793-42. [DOI] [PubMed] [Google Scholar]
- 2.Bull JJ, Meyers LA, Lachmann M. Quasispecies made simple. PLoS computational biology. 2005;1:e61. doi: 10.1371/journal.pcbi.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci U S A. 2001;98:6895–900. doi: 10.1073/pnas.111085598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graci JD, Cameron CE. Therapeutically targeting RNA viruses via lethal mutagenesis. Future virology. 2008;3:553–566. doi: 10.2217/17460794.3.6.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson JP, DR, Loeb LA. Viral error catastrophe by mutagenic nucleosides. Annu Rev Microbiol. 2004;58:183–205. doi: 10.1146/annurev.micro.58.030603.123649. [DOI] [PubMed] [Google Scholar]
- 6.Smith RA, LL, Preston BD. Lethal mutagenesis of HIV. Virus Res. 2005;107:215–28. doi: 10.1016/j.virusres.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Dapp MJ, Clouser CL, Patterson S, Mansky LM. 5-Azacytidine can induce lethal mutagenesis in human immunodeficiency virus type 1. Journal of virology. 2009;83:11950–8. doi: 10.1128/JVI.01406-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sierra S, Davila M, Lowenstein PR, Domingo E. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J Virol. 2000;74:8316–23. doi: 10.1128/jvi.74.18.8316-8323.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Contreras AM, Hiasa Y, He W, Terella A, Schmidt EV, Chung RT. Viral RNA mutations are region specific and increased by ribavirin in a full-length hepatitis C virus replication system. Journal of virology. 2002;76:8505–17. doi: 10.1128/JVI.76.17.8505-8517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris KS, Brabant W, Styrchak S, Gall A, Daifuku R. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antiviral Res. 2005;67:1–9. doi: 10.1016/j.antiviral.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 11.Mullins JI, Heath L, Hughes JP, Kicha J, Styrchak S, Wong KG, Rao U, Hansen A, Harris KS, Laurent JP, Li D, Simpson JH, Essigmann JM, Loeb LA, Parkins J. Mutation of HIV-1 genomes in a clinical population treated with the mutagenic nucleoside KP1461. PloS one. 2011;6:e15135. doi: 10.1371/journal.pone.0015135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuevas JM, Gonzalez-Candelas F, Moya A, Sanjuan R. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. Journal of virology. 2009;83:5760–4. doi: 10.1128/JVI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutchman G, Danehower S, Song BC, Liang TJ, Hoofnagle JH, Thomson M, Ghany MG. Mutation rate of the hepatitis C virus NS5B in patients undergoing treatment with ribavirin monotherapy. Gastroenterology. 2007;132:1757–66. doi: 10.1053/j.gastro.2007.03.035. [DOI] [PubMed] [Google Scholar]
- 14.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–9. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 15.Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 16.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–8. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 19.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Current biology : CB. 2006;16:480–5. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 20.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Current biology : CB. 2004;14:1392–6. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 21.Sasada A, Takaori-Kondo A, Shirakawa K, Kobayashi M, Abudu A, Hishizawa M, Imada K, Tanaka Y, Uchiyama T. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology. 2005;2:32. doi: 10.1186/1742-4690-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 23.Zennou V, Bieniasz PD. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349:31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 24.Albin JS, Harris RS. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert reviews in molecular medicine. 2010;12:e4. doi: 10.1017/S1462399409001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg BR, Papavasiliou FN. Beyond SHM and CSR: AID and related cytidine deaminases in the host response to viral infection. Advances in immunology. 2007;94:215–44. doi: 10.1016/S0065-2776(06)94007-3. [DOI] [PubMed] [Google Scholar]
- 26.Delebecque F, Suspene R, Calattini S, Casartelli N, Saib A, Froment A, Wain-Hobson S, Gessain A, Vartanian JP, Schwartz O. Restriction of foamy viruses by APOBEC cytidine deaminases. Journal of virology. 2006;80:605–14. doi: 10.1128/JVI.80.2.605-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nature reviews Immunology. 2004;4:868–77. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 28.Malim MH, Emerman M. HIV-1 accessory proteins--ensuring viral survival in a hostile environment. Cell host & microbe. 2008;3:388–98. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annual review of immunology. 2008;26:317–53. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 30.Pace C, Keller J, Nolan D, James I, Gaudieri S, Moore C, Mallal S. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. Journal of virology. 2006;80:9259–69. doi: 10.1128/JVI.00888-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez-Perez JA, Ormsby CE, Hernandez-Juan R, Torres KJ, Reyes-Teran G. APOBEC3G mRNA expression in exposed seronegative and early stage HIV infected individuals decreases with removal of exposure and with disease progression. Retrovirology. 2009;6:23. doi: 10.1186/1742-4690-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browne EP, Allers C, Landau NR. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–21. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shindo K, Takaori-Kondo A, Kobayashi M, Abudu A, Fukunaga K, Uchiyama T. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. The Journal of biological chemistry. 2003;278:44412–6. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- 34.Bishop KN, Holmes RK, Malim MH. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. Journal of virology. 2006;80:8450–8. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schumacher AJ, Hache G, Macduff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. Journal of virology. 2008;82:2652–60. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loeb LA, Essigmann JM, Kazazi F, Zhang J, Rose KD, Mullins JI. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:1492–7. doi: 10.1073/pnas.96.4.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sadler HA, Stenglein MD, Harris RS, Mansky LM. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. Journal of virology. 2010;84:7396–404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogozin IB, Babenko VN, Milanesi L, Pavlov YI. Computational analysis of mutation spectra. Briefings in bioinformatics. 2003;4:210–27. doi: 10.1093/bib/4.3.210. [DOI] [PubMed] [Google Scholar]
- 39.Rogozin IB, Pavlov YI. Theoretical analysis of mutation hotspots and their DNA sequence context specificity. Mutation research. 2003;544:65–85. doi: 10.1016/s1383-5742(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 40.Kaminskas E, Farrell AT, Wang YC, Sridhara R, Pazdur R. FDA drug approval summary: azacitidine (5-azacytidine, Vidaza) for injectable suspension. The oncologist. 2005;10:176–82. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 41.Paces V, Doskocil J, Sorm F. Incorporation of 5-azacytidine into nucleic acids of Escherichia coli. Biochimica et biophysica acta. 1968;161:352–60. [PubMed] [Google Scholar]
- 42.Li LH, Olin EJ, Buskirk HH, Reineke LM. Cytotoxicity and mode of action of 5-azacytidine on L1210 leukemia. Cancer research. 1970;30:2760–9. [PubMed] [Google Scholar]
- 43.Rogstad DK, Herring JL, Theruvathu JA, Burdzy A, Perry CC, Neidigh JW, Sowers LC. Chemical decomposition of 5-aza-2′-deoxycytidine (Decitabine): kinetic analyses and identification of products by NMR, HPLC, and mass spectrometry. Chemical research in toxicology. 2009;22:1194–204. doi: 10.1021/tx900131u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson-Grusby L, Laird PW, Magge SN, Moeller BJ, Jaenisch R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc Natl Acad Sci U S A. 1997;94:4681–4685. doi: 10.1073/pnas.94.9.4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pathak VK, Temin HM. 5-azacytidine and RNA secondary structure increase the retrovirus mutation rate. J Virol. 1992;66:3093–3100. doi: 10.1128/jvi.66.5.3093-3100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc Natl Acad Sci U S A. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nature structural & molecular biology. 2004;11:435–42. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 48.Rausch JW, Chelico L, Goodman MF, Le Grice SF. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. The Journal of biological chemistry. 2009;284:7047–58. doi: 10.1074/jbc.M807258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clouser CL, Patterson SE, Mansky LM. Exploiting drug repositioning for discovery of a novel HIV combination therapy. Journal of virology. 2010;84:9301–9. doi: 10.1128/JVI.01006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perales C, Agudo R, Domingo E. Counteracting quasispecies adaptability: extinction of a ribavirin-resistant virus mutant by an alternative mutagenic treatment. PloS one. 2009;4:e5554. doi: 10.1371/journal.pone.0005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pariente N, Airaksinen A, Domingo E. Mutagenesis versus Inhibition in the Efficiency of Extinction of Foot-and-Mouth Disease Virus. Journal of Virology. 2003;77:7131–7138. doi: 10.1128/JVI.77.12.7131-7138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pariente N, Sierra S, Lowenstein PR, Domingo E. Efficient virus extinction by combinations of a mutagen and antiviral inhibitors. J Virol. 2001;75:9723–30. doi: 10.1128/JVI.75.20.9723-9730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perales C, Agudo R, Tejero H, Manrubia SC, Domingo E. Potential benefits of sequential inhibitor-mutagen treatments of RNA virus infections. PLoS pathogens. 2009;5:e1000658. doi: 10.1371/journal.ppat.1000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perales C, Agudo R, Manrubia SC, Domingo E. Influence of mutagenesis and viral load on the sustained low-level replication of an RNA virus. Journal of molecular biology. 2011;407:60–78. doi: 10.1016/j.jmb.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 55.Mulder LC, Harari A, Simon V. Cytidine deamination induced HIV-1 drug resistance. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5501–6. doi: 10.1073/pnas.0710190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim EY, Bhattacharya T, Kunstman K, Swantek P, Koning FA, Malim MH, Wolinsky SM. Human APOBEC3G-mediated editing can promote HIV-1 sequence diversification and accelerate adaptation to selective pressure. Journal of virology. 2010;84:10402–5. doi: 10.1128/JVI.01223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Codoner FM, Daros JA, Sole RV, Elena SF. The fittest versus the flattest: experimental confirmation of the quasispecies effect with subviral pathogens. PLoS pathogens. 2006;2:e136. doi: 10.1371/journal.ppat.0020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanjuan R, Cuevas JM, Furio V, Holmes EC, Moya A. Selection for robustness in mutagenized RNA viruses. PLoS genetics. 2007;3:e93. doi: 10.1371/journal.pgen.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levi LI, Gnadig NF, Beaucourt S, McPherson MJ, Baron B, Arnold JJ, Vignuzzi M. Fidelity variants of RNA dependent RNA polymerases uncover an indirect, mutagenic activity of amiloride compounds. PLoS pathogens. 2010;6:e1001163. doi: 10.1371/journal.ppat.1001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfeiffer JK, Kirkegaard K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7289–94. doi: 10.1073/pnas.1232294100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arribas M, Cabanillas L, Lazaro E. Identification of mutations conferring 5-azacytidine resistance in bacteriophage Qbeta. Virology. 2011 doi: 10.1016/j.virol.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 62.Sierra M, Airaksinen A, Gonzalez-Lopez C, Agudo R, Arias A, Domingo E. Foot-and-mouth disease virus mutant with decreased sensitivity to ribavirin: implications for error catastrophe. Journal of virology. 2007;81:2012–24. doi: 10.1128/JVI.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berkhout B, van Hemert FJ. The unusual nucleotide content of the HIV RNA genome results in a biased amino acid composition of HIV proteins. Nucleic acids research. 1994;22:1705–11. doi: 10.1093/nar/22.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hache G, Liddament MT, Harris RS. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J Biol Chem. 2005;280:10920–4. doi: 10.1074/jbc.M500382200. [DOI] [PubMed] [Google Scholar]
- 65.Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Current biology : CB. 2004;14:1385–91. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]