Abstract
Although, the nucleolus has been observed for nearly 200 years in neurons, studies that directly address neuronal roles of this subnuclear structure have appeared only recently. The goal of this review is to discuss that recent progress and identify some critical questions which remain to be answered. As expected for the cellular center of ribosome biogenesis, the nucleolus is essential for the growth of developing neurons, including neurite morphogenesis and long-term maintenance of mature neurons. In addition, the nucleolus contributes to neuronal stress responses, including the regulation of apoptosis. Hence, disrupted neurodevelopment or neurodegeneration are among the likely consequences of nucleolar dysfunction. Conversely, the presence of active nucleoli may determine the potential for neurorepair.
Keywords: RNA-Polymerase-1, rRNA, neuronal stress, nucleus, transcription, chromatin
Introduction
Nearly two centuries ago, microscopic observations of neurons resulted in the identification of a subnuclear structure that in 1839 was named the nucleolus by Gabriel Valentin (reviewed in [1]). Subsequent work done mostly in transformed cells revealed that the nucleolus is an initiation site for ribosomal biogenesis and as such determines cellular translation capacity and regulates cell growth (for recent reviews see [2–5]). Additional nucleolar functions have also been identified, including various stress responses (reviewed in [6, 7]). Although neurons are post-mitotic cells, which upon reaching maturity have a limited growth potential, they often display prominent nucleoli. However, until recently their neuronal functions have not been addressed by direct experimentation. The goal of this review is to discuss the results of several studies from the past few years which have directly addressed the significance of neuronal nucleoli as well as to identify some of the important questions that remain to be addressed.
Biology of the nucleolus
The nucleolus is assembled around several hundred copies of the repeated 45S rRNA gene (ribosomal DNA or rDNA) that are located in several clusters at various chromosomal locations throughout the mammalian genomes (as reviewed in [2, 3]) (Fig. 1A). Ribosomal DNA is transcribed by a specialized nucleolar RNA polymerase, RNA polymerase-1 (Pol1) [3]. Transcription of rDNA initiates ribosomal biogenesis (Fig. 1A). Additionally, Pol1 activity promotes the formation and maintenance of the nucleolus including that of post-mitotic neurons [8, 9] (Fig. 1B). Hence, it is not surprising that the size and structural organization of the nucleolus is directly related to the rate of ribosomal biogenesis [2].
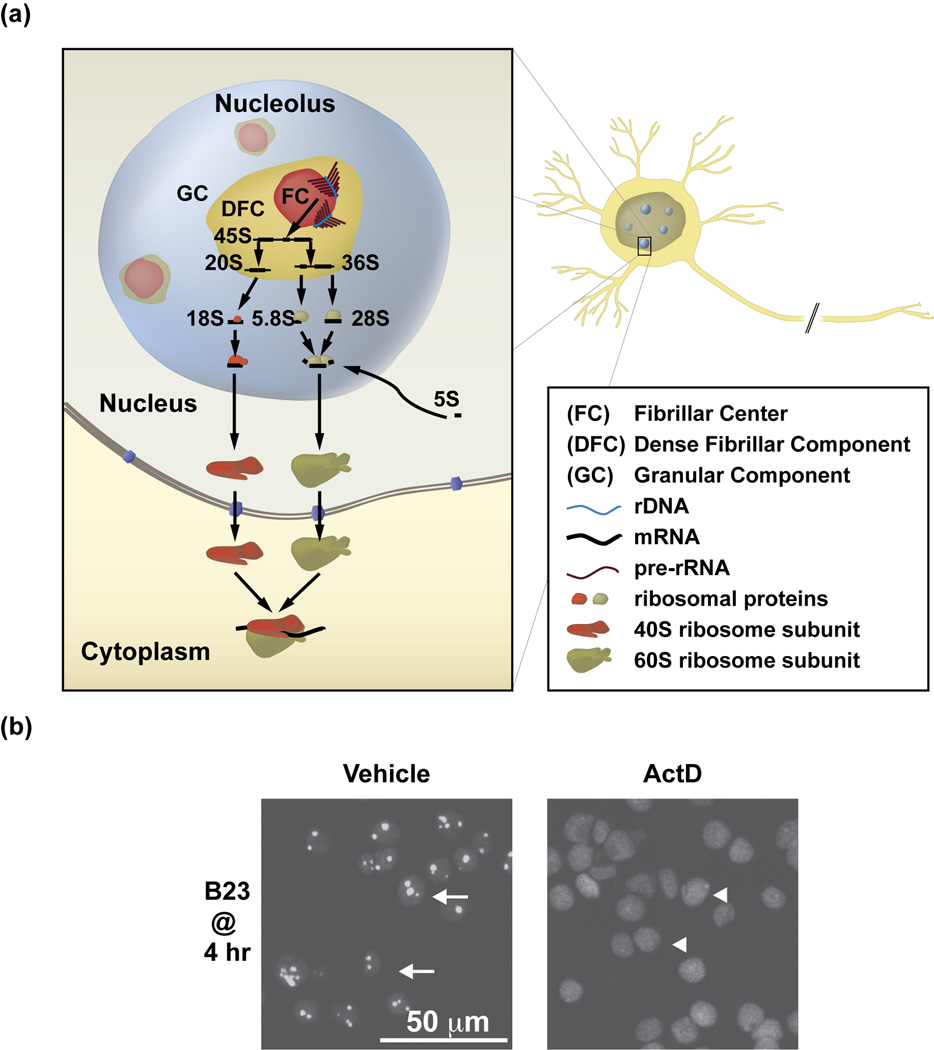
Figure 1. The nucleolus is a subnuclear structure, which carries out ribosomal biogenesis and is generated by the Pol1-dependent transcription of the nucleolar rRNA genes (rDNA).
a, Ultrastructurally, the nucleolus has a tripartite organization that reflects spatial separation of various steps of ribosomal biogenesis (as reviewed in [2]). That process is initiated in the fibrillar centers (FC), where the 45S rRNA (pre-rRNA) is transcribed by Pol1 from rDNA. Note the “Christmas tree” structure of the transcribed rDNA loci/elongating pre-rRNA molecules at the edges of the FC. Further steps of ribosomal biogenesis including pre-rRNA modifications and processing to mature rRNAs (18S, 5.8S and 28S) as well as rRNA assembly with ribosomal proteins and the 5S rRNA occur in the dense fibrillar component (DFC) and granular component (GC) of the nucleolus. Final products of this process, the 40S and 60S ribosome subunits are exported out of the nucleus, where, following final maturation, they may form ribosomes. b, The nucleolus is a transcription-dependent structure. Upon 1 hr treatment with Actinomycin D to block Pol1 (0.05 µg/ml), the nucleoli of cultured rat cortical neurons disintegrate as demonstrated by nucleoplasmic translocation of the immunofluorescent nucleolar chaperone B23 [9]. Arrows (left panel) indicate neurons with intact nucleoli as suggested by predominantly nucleolar presence of B23; arrowheads (right panel) depict neurons with nucleolar disruption as indicated by diffusion of B23 throughout the nucleoplasm. Reproduced, with permission, from [9].
Numerous additional steps of the maturation of precursors of the small- and large ribosomal subunits (SSU, and LSU, respectively) are also nucleolar (as reviewed in [2, 4, 5]). The primary Pol1 transcript, 45S pre-rRNA undergoes extensive co-transcriptional modifications including ribose methylation and uridine isomerization to pseudouridine that may be required for the proper structural organization and activity of mature rRNA [4, 5]. Ribosomal proteins begin to assemble on the nascent transcript, which together with a vast array of accessory factors, stimulate subsequent steps of ribosomal subunit maturation including pre-rRNA processing and nuclear export. The pre-LSU assembly in the nucleolus also involves incorporation of 5S rRNA that is transcribed from non-nucleolar genes by RNA-Polymerase-3 (reviewed in [10]). Eukaryotic ribosomal biogenesis is a highly complex process that involves 80 ribosomal proteins, more than 200 non-ribosomal proteins and 75 small nucleolar RNAs (snoRNAs) [4, 5]. In a rapidly dividing cell, several thousand ribosomes are generated every minute [4]. Ribosomal RNA transcription accounts for at least half of the total transcriptional output of a cell during active proliferative growth [3, 4]. Hence, ribosomal biogenesis represents a major item on the energy budget of proliferating cells [11]. High rates of ribosomal biogenesis are also expected in cells that do not divide but, instead, rapidly increase their volume such as maturing oocytes, neurite-extending neurons or myelinating oligodendrocytes.
Multiple mechanisms are employed to adjust the rate of ribosomal production to cellular demand (as reviewed recently in [3]). The primary target for such regulation is rDNA transcription. Several studies have shown that stimulation of rDNA transcription is sufficient to increase the entire process of ribosomal biogenesis accelerating proliferation [12, 13]. Transcription of rDNA is regulated at the level of Pol1 activity and also by epigenetic mechanisms which affect the number of transcriptionally active rDNA genes (reviewed in [3, 14]). Thus, growth factors, nutrients, or stressors increase or decrease Pol1 activity, respectively. Many of their effects on Pol1 are mediated by signaling kinases including positive regulation by the mammalian target of rapamycin (mTor) and extracellular signal related kinases-1/2 (ERK1/2) or negative regulation by the c-Jun N-terminal kinase (JNK) or AMP-activated protein kinase (AMPK) [12, 15–20]. In addition, acetylation of rDNA transcriptional regulators provides another positive input to Pol1 activity [21–23]. Finally, epigenetic regulation is believed to play a role in reduction of the ribosomal biogenesis when differentiation follows rapid growth such as that during embryonic development (reviewed in [14]) or when cells are depleted of energy [11]. Epigenetic silencing of the nucleolus includes CpG methylation of rDNA promoters [14].
In addition to ribosomal biogenesis, maturation of tRNAs and small nuclear RNAs (snRNAs) that participate in mRNA splicing has been shown to occur in nucleoli (reviewed in [24, 25]). Further, nucleolar disruption is a rapid consequence of Pol1 inhibition following DNA damage or oxidative injury which has allowed the nucleolus to evolve as a sensor of cellular stress [19, 26, 27]. Thus, the disintegrating nucleolus releases several proteins that activate the stress response transcription factor p53 resulting in apoptosis (reviewed in [6, 7]). Other proteins with dedicated functions in stress response such as several DNA repair enzymes are localized to the nucleolus in the absence of genotoxic stress and released upon DNA damage to restore genome integrity [28, 29]. Nucleolar sequestration is also a regulatory mechanism for several proteins that serve as molecular switches during cell cycle progression or differentiation (reviewed in [30]). In addition, the nucleolus is involved in nuclear export of ubiquitinated nuclear proteins such as p53 enabling their degradation by the proteasome system in the cytoplasm [31, 32]. Finally, at least in yeast, the inherent instability of rDNA repeats determines the life span by linking the aging-associated loss of rDNA to the activation of DNA damage response and cell senescence (reviewed in [33]). Taken together, the highly dynamic nucleolus is the key controller of cell growth and an important sensor of cellular stress.
Role of the nucleolus in neuronal growth
Despite being non-dividing cells, neurons are able to grow. During development, neurotrophic factors stimulate neuronal growth including increases in the size of the perikarion as well as length, caliber and complexity of neurites (reviewed in [34–36]). The large increase in the cell volume that is associated with such growth suggests that its dependence on ribosome production is similar to that observed for growth of dividing cells. This view is supported by observations from various regions of the nervous system that indicate an increasingly active morphology of neuronal nucleoli as neurons are progressing through development until they reach their final dimensions [8, 37, 38]. Some growth abilities are also retained in the mature nervous system such as the growth of axons which occurs during peripheral nerve regeneration after injury (reviewed in [36]). There is a good correlation between regenerative growth of injured neurons and increased nucleolar activity in these cells [39, 40]. Taken together, morphological observations suggest a link between neuronal growth and the dynamic changes of the nucleolus indicative of increased ribosomal biogenesis. A recent work suggests that such a relationship is likely due to a requirement of ribosomal production for neuronal growth [41] (Fig. 2).
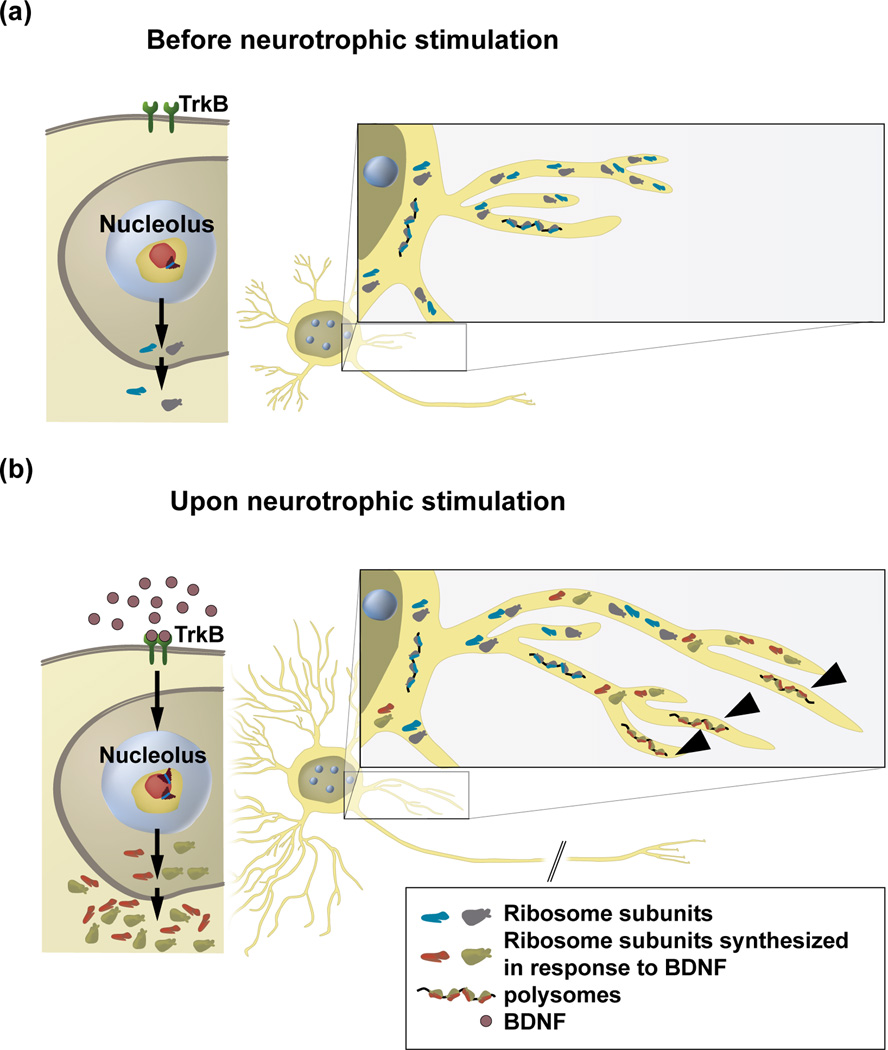
Figure 2. A hypothetical model of a nucleolar mechanism that supports neurite growth in response to extracellular signals.
a, Without growth-promoting signals, basal activity of Pol1 maintains neuronal homeostasis by replacing ribosomes that were damaged by oxidation. b, Upon growth-promoting signals (e.g. BDNF), Pol1 is stimulated to produce more ribosome subunits. In BDNF-treated rat forebrain neurons, Pol1-driven transcription is activated by the TrkB-ERK1/2 signaling pathway [41]. The newly made ribosome subunits (marked in red and green) may be preferentially transferred to the growing portions of neurites supporting their growth. Such growth support may include local protein synthesis as indicated by the contribution of the newly generated ribosomes to the translating polysomes (arrowheads).
Brain-derived neurotrophic factor (BDNF) is a major growth stimulus for developing neurons of the cerebral cortex and the hippocampus (reviewed in [34, 35]). In addition to inducing growth of their somas, BDNF is also a powerful stimulant of neuritic morphogenesis. Such effects result from the interaction of BDNF with its high affinity receptor, the Tropomyosin-Related Kinase B (TrkB), and subsequent activation of several signaling pathways including the well established regulators of Pol1, mTor and ERK1/2 (eg. [42, 43])).
BDNF has been shown to stimulate Pol1 activity in primary cultures of cortical or hippocampal neurons from newborn rats and in whole rat pup hippocampi and cortices [41]. At least in cultured neurons, that effect required the ERK1/2 pathway. When Pol1 was inhibited by an shRNA-mediated knockdown of its specific co-activator, transcription initiation factor-1A (TIF1A), BDNF failed to stimulate growth of cultured hippocampal neurons. Thus, in neurons with normal TIF1A levels, a 24 hr treatment with BDNF increased perikarial volume, total length of neurites and number of secondary neuritic branches. None of these effects were observed after TIF1A knockdown suggesting requirement of Pol1 for BDNF-induced neuronal growth. In addition, the neuron growth stimulation with the constitutively active mutant form of the ERK1/2 activator, Map Kinase Kinase-1 (MKK1/MEK1) was as sensitive to Pol1 inhibition as that of BDNF.
In non-neuronal cells, TIF1A is phosphorylated at the serine residues 633 and 649 by ERK2 and the ERK1/2-activated RSK2, respectively, to stimulate Pol1 activity [12]. Overexpression of a mutant TIF1A with phosphomimetic substitutions at those phosphorylation sites (Ser to Asp) promoted robust growth of cultured hippocampal neurons including increases of soma volume, total neuritic length and neuritic branching. A similar growth response was observed if wild type TIF1A was overexpressed together with low levels of active MKK1, either of which were insufficient to stimulate neuronal growth. Collectively, these results suggest that Pol1 is both necessary and sufficient for perikarial growth and neuritic morphogenesis in developing neurons under neurotrophic stimulation. In addition, they identify Pol1 as a major target for the growth-stimulating ERK1/2 pathway.
These neuronal growth studies with newborn rat hippocampal neurons were carried out 7–9 days after cell isolation (in vitro days 6–8). At that time, axons are already well established while dendrites grow and branch out [44]. Therefore, most of the aforementioned neuritic growth effects of manipulating Pol1 are dendritic. Hence, an obvious question emerges whether axonal growth during development or during regeneration is also dependent on Pol1. Another question that needs to be addressed is whether pro-morphogenic signaling pathways like mTor, calcium/calmodulin-dependent kinases (CaMKs) or cAMP ([42, 43] and references therein) also rely on Pol1 to transduce the extrinsic signals for morphogenesis. Conversely, Pol1 may be a target for the intrinsic neuronal growth regulators such as transcription factors Runx1 or SnoN [45, 46].
As increased ribosomal production follows Pol1 activation, one could expect that neuronal growth requires greater number of ribosomes. However, inhibition of Pol1 activity for 48 h did not reduce general protein synthesis despite antagonizing neuronal growth [41]. One interpretation of such results could be that in a growing neuron, a continuous local delivery of the newly generated ribosomes would be indispensable for the growth even without an overall increase in protein translation (Fig. 2). Such a local ribosome delivery hypothesis is supported by the observations that neuronal ribosomal turnover is fairly slow with an estimated ribosome half life time of at least 8 days [47]. Hence, the inhibition of Pol1 activity for 24–72 h that is sufficient to affect neuronal growth [41] would not be long enough to alter the total pool of neuronal ribosomes. In growing dendrites, the local ribosome delivery hypothesis fits well with the well established ribosomal presence in this type of neurites [48]. If our ongoing studies confirm that axonal growth also requires Pol1, a similar local ribosome delivery hypothesis may also apply to axonal growth, at least in these neuronal population that contain axonal ribosomes [49]. Imaging ribosomes in live neurons using inducible fluorescent protein tags will help answer the question as to whether newly made ribosomes are enriched in expanding neurites.
The local ribosome delivery hypothesis implies that increased local protein synthesis may be critical for neuronal growth. In fact, local protein synthesis is well documented in dendrites and axons and is believed to contribute to their growth (reviewed in [48–51]). Thus, newly made ribosomes might be needed to satisfy demand for local protein synthesis that is required for growth. In axons, those newly made ribosomes could populate the periaxoplasmic plaques that were proposed as centers of axonal protein synthesis [52]. In addition, it is conceivable that the local protein synthesis that is required for growth employs a distinct type of ribosomes whose biogenesis is stimulated by growth promoting signals. Such a scenario fits well with the hypothesized functional heterogeneity of cellular ribosomes (reviewed in [53]). However, local ribosome delivery may have additional effects that would add to the growth support. For instance, in addition to residing at the endoplasmic reticulum membranes or in the cytosol, ribosomes may associate with, and, possibly regulate the cytoskeleton, which, in turn, is critical for neuronal growth (reviewed in [54]). A recent genetic screen in yeast also identified a ribosomal requirement for activation of the mTor-Akt signaling pathway [55]. Hence, in addition to support of local protein synthesis, local ribosomes could promote neurite morphogenesis by regulating cytoskeleton and/or contributing to the intracellular signaling pathways for the extracellular signals that promote growth.
While this review focuses on the neuronal nucleolus, the glial nucleolus may also be important for growth and/or maintenance of neurites. For instance, in regenerating axons of sensory neurons, ribosome transfer from Schwann cells has been proposed as a mechanism contributing to the local protein synthesis [56]. Likewise, desomatized axons were shown to be recipients of Schwann cell ribosomes [57]. Further studies are needed to determine the role of ribosomal biogenesis in glia for neuritic growth and maintenance.
As neuronal growth is critical for the development of the nervous system, nucleolar impairment may contribute to neurodevelopmental disorders (Table 1). For instance, mutations reducing activity of two nucleolar proteins including the essential for mitotic growth-1 (EMG1/NEP1) and RNA binding motif protein-28 (RBM28) lead to microcephaly and mental retardation in Bowen-Conradi- and Alopecia, Neurological deficits and Endocrinopathy (ANE) syndromes, respectively [58, 59]. In non-neuronal cells, EMG1 and RBM28 function as transacting factors of ribosomal biogenesis. However, future studies are necessary to test whether neurological consequences of EMG1- and/or RBM28 deficiency are due to reduced ribosomal biogenesis in developing neurons and the subsequent impairment of their growth. Cockayne syndrome (CS) that is caused by inactivating mutations of several components of the DNA nucleotide excision repair (NER) pathway is associated with microcephaly, cognitive impairment and progressive degeneration of various neuronal populations (eg. [60]). It has been proposed that its pathogenesis may be due to reduced nucleolar transcription as products of 3 major CS-mutated genes play a direct role as Pol1 co-factors (eg. [61]). An autism-spectrum disorder, Prader-Willi syndrome (PWS), is caused by paternal deficiency within the 15q11–q13 chromosomal region. While this region contains a number of genes whose deficient expression contributes to the complex PWS phenotype, insufficiency of SNORD116 is critical for key characteristics of this disorder including obesity, hyperphagia and hypogonadism [62, 63]. SNORD116 (HBII-85) encodes a brain-specific C/D box snoRNA that forms a complex with the nucleolar protein fibrillarin [64]. In contrast to most C/D box snoRNAs, which contribute to ribosomal biogenesis, SNORD116 is not complementary to rRNA but to several mRNAs suggesting its role in mRNA splicing [65]. Nevertheless, nucleolar atrophy has been reported in cerebellar Purkinje neurons of PWS patients or transgenic mice with a PWS-like paternal deficiency of gene expression indicating that SNORD116 may, at least, indirectly regulate ribosomal biogenesis [66]. In Rett syndrome, which is caused by reduced activity of a transcriptional regulator methyl-CpG-binding protein-2 (MeCP2), dendritic atrophy without neuronal loss occurs in early childhood [67]. In a mouse model of Rett syndrome (Mecp2 knockout), this pathology is accompanied by reduced size of neuronal nucleoli [68]. It is tempting to speculate that ribosomal biogenesis may become a novel target for therapeutic interventions at least in some neurodevelopmental disorders.
Table 1.
Evidence supporting a role for nucleolar dysfunction in neurological disorders
| DISORDER | EVIDENCE OF NUCLEOLAR DYSFUNCTION | ||
|---|---|---|---|
| Morphological | Biochemical | Genetic/Epigenetic | |
| Alzheimer’s Disease | Reduced nucleolar size in the cerebro-cortical, hippocampal and Meynert’s nucleus neurons [85–87, 99] | Increased oxidation of rRNA and ribosomal failure in the hippocampus and/or cerebral cortex [88–90] | CpG hypermethylation of the rDNA promoter in the cerebral cortex [91] |
| ANE Syndrome | –a | – | Inactivating mutation of the nucleolar LSU biogenesis factor RBM28 cause the disease [59] |
| Bowen-Conradi Syndrome | – | – | Inactivating mutation of the nucleolar SSU biogenesis factor EMG1 causes the disease [58] |
| Cockayne Syndrome | – | – | Inactivating mutations of the NER mediators and Pol1 co-factors CSB, XPBb and XPDc cause the disease ([61] and references therein). |
| Huntington’s Disease (HD) | – | – | Reduced nucleolar transcription and levels/activity of the nucleolar transcription factor UBFd in a mouse striatal cell line with an HD-associated mutation in the mouse Htte and in the striatum of transgenic mice overexpressing an HD mutant fragment of human HTT (R6/2 mouse line) [23]. |
| Parkinson’s Disease | Reduction in nucleolar size and the nucleolar marker B23 in nigral neurons of PD patients [75, 100] | – | Mouse nigral neuron degeneration following dopaminergic-neuron-specific knock out of the Pol1 co-activator TIF1A [75] |
| Prader-Willi Syndrome (PWS) | Reduced nucleolar size in neurons from PWS patients and transgenic mice with the PWS-like paternal deficiency of gene expression [66] | – | Microdeletions within the chromosome 15 PWS region that selectively disrupt the snoRNA gene cluster SNORD116 (HBII-85) are sufficient to causes the key features of the disease [62, 63] |
| Rett Syndrome | Reduced nucleolar size in the Mecp2 mouse knockout model of Rett syndrome [68] | – | – |
| Suicide with history of abuse in childhood | – | Reduced levels of 18S rRNA in the hippocampus of affected individuals [101] | CpG hypermethylation of the rDNA promoter in the hippocampus ofaffected individuals [101] |
no data available;
XPB, xeroderma pigmentosum group B;
XPD, xeroderma pigmentosum group D;
UBF, upstream binding factor;
HTT, huntingtin
Role of the nucleolus as a neuronal stress sensor
The requirement of the ongoing rDNA transcription for the maintenance of the nucleolus as a structure makes it a convenient sensor of cellular stress (reviewed in [6, 7]) (Fig. 1B). Indeed, the transcription-dependent nucleolar compartmentalization of various proteins offers a number of potential stress messengers and/or stress effectors which can be released upon cell injury [7] (Fig. 3). Conversely, nuclear export and inactivation of such stress signaling molecules as p53 may require an intact nucleolus [31]. Therefore, the stress response function of the nucleolus could remain active in many types of differentiated cells including those which have exited the cell cycle. Direct evidence for the role of the nucleolus in neuronal stress responses has only recently emerged.
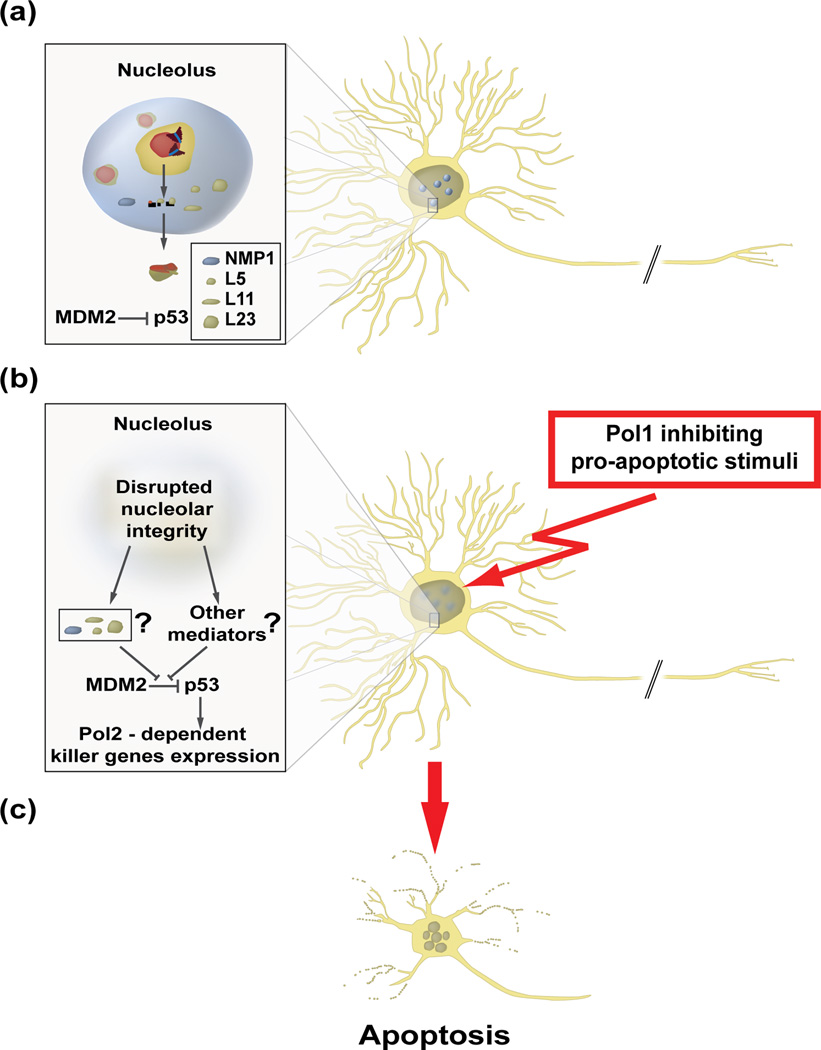
Figure 3. The nucleolus is a neuronal stress sensor that controls the pro-apoptotic activity of p53 in developing neurons.
a, Under normal conditions, RNA-polymerase-1 (Pol1)-dependent transcription ensures nucleolar sequestration of proteins that may disrupt murine double minute- 2 (MDM2)-dependent ubiquitination of the killer transcription factor p53 in the nucleoplasm, enabling its proteasome-mediated degradation in the cytoplasm. While the identity of p53 regulators in the neuronal nucleolus is not known at present, nucleophosmin/B23 as well as ribosomal proteins L5, L11 and L23 play that role in non-neuronal cells (reviewed in [7]). In non-neuronal cells, the negative regulation of p53 may also involve its nucleolus-dependent nuclear export [31]. b, Upon an insult by a pro-apoptotic agent that blocks Pol1 (e.g. inducers of DNA single strand breaks and/or large DNA adducts [9, 27]), the nucleolus disintegrates and p53 degradation is inhibited. The accumulated p53 induces Pol2-transcribed killer genes which activate neuronal apoptosis [9].
It has been known for some time that the morphological appearance of the neuronal nucleolus is sensitive to various types of injuries including DNA damaging anti-cancer drugs, ionizing irradiation, hypoxia or oxidants [69–75]. Also, in the case of pro-neurodegenerative mouse mutations such as Klotho or Pcd, nucleolar disruption in the sensitive neuronal populations has been associated with oxidative DNA damage [76–78]. Since most of those nucleolar stress conditions involve DNA damage, it is likely that nucleolar disruption is a consequence of DNA lesions that directly interfere with rDNA transcription (for review see [79]). Alternatively, stress-activated signaling cascades including hydrogen peroxide-activated JNK2 may inhibit Pol1-driven transcription to induce the nucleolar stress response [19].
The first report of the possible consequences of the nucleolar stress was focused on ischemia/hypoxia [80]. In this case, a search for binding partners of the activated death effector protein Bax that can help to localize it to mitochondria identified the nucleolar chaperone nucleophosmin/B23. B23 was released from the nucleolus upon hypoxia while B23 knockdown reduced apoptotic response to hypoxia in a cell line. These findings together with an observation that Bax is bound to B23 in a mouse model of stroke suggest that nucleolar stress contributes to neuronal loss in hypoxia [80].
A pro-apoptotic role of Pol1 inhibition and the subsequent nucleolar stress has been also proposed in developing neurons that are challenged with the DNA damaging anticancer drug camptothecin (CPT) [9] (Fig. 3). This compound inhibits DNA topoisomerase-1 (Topo1) inducing DNA single strand breaks (SSBs) and DNA-Topo1 adducts [81]. In cultured cortical neurons from newborn rats, CPT potently blocked Pol1 activity and disrupted integrity of their nucleoli [9] (Fig 1B). The CPT-induced nucleolar stress preceded activation of the DNA damage-regulated pro-apoptotic transcription factor p53 and apoptosis. Nucleolar disruption also occurred if p53 and/or apoptosis were blocked. In addition, Pol1 inhibition by knocking down TIF1A, was sufficient to induce p53-dependent neuronal apoptosis that required extranucleolar transcription. Also, neuronal apoptosis was induced by the transcriptional inhibitor Actinomycin D if it was applied at low concentrations which were relatively more potent against Pol1 than the mRNA-transcribing RNA-Polymerase-2 (Pol2). Finally, Pol1 inhibition, activation of p53 and apoptosis has been reported after TIF1A was knocked out in the neuroprogenitor cells of the developing mouse brain [82]. Hence, Pol1 inhibition in immature neurons triggers neuronal apoptosis that is dependent on a p53-mediated transcriptional response (Fig. 3).
In rat forebrain neurons, the nucleolar stress is specific to certain types of DNA damage [27]. While transcription inhibitory lesions such as single strand breaks (SSBs) and/or DNA protein adduct disrupted nucleoli, double strand breaks (DSBs) neither blocked Pol1 nor affected nucleolar integrity [27]. In addition, nucleolar stress is not a developmentally restricted response to DNA damage as it has been observed in both developing and mature neurons [27]. Such observations suggest that the nucleolus may serve as a neuronal sensor of genotoxic stress in both developing and mature nervous systems.
Apoptosis is a consequence of nucleolar stress that is restricted to developing neurons [9, 27, 82]. Conversely, adult mouse neurons with conditional knockout of TIF1A survive for months despite a loss of ribosomal biogenesis in the nucleolus, inhibition of mTor signaling, mitochondrial impairment and presence of oxidative stress [75, 82]. Hence, in adult neurons, nucleolar stress likely activates a survival program that allows these cells to cope with unfavorable conditions. In addition, in most cases, nucleolar stress is expected to be relatively transient as Pol1 will resume its activity after cellular homeostasis is restored [73, 83]. Therefore, one can speculate that many of nucleolar stress effectors will mediate compensatory responses that promote neuronal survival. Finally, under hypoxia or oxidative damage, a transient block of ribosomal biogenesis may be neuroprotective simply by conserving energy [11]. Taken together, while in developing neurons, nucleolar stress induces apoptosis, neuroprotection may be one of its major outcomes in the adult nervous system. In addition, even in developing neurons, transient- but not prolonged nucleolar stress may primarily support survival.
Nucleolar malfunction and neurodegeneration
As neurons are dependent on translation for cell maintenance and synaptic plasticity, one can expect that if neuronal ribosome production is reduced by pathological factors, neurodegeneration and/or neuronal dysfunction will follow. In fact, a hypothesis that ribosomal biogenesis failure underlies aging dependent-neurodegeneration and/or neuronal dysfunction was proposed almost 40 years ago by Bernard Strehler [84]. More recently, relatively slow neurodegeneration has been reported following Pol1 inhibition by the neuron-specific knock out of TIF1A in adult mouse brain [75, 82]. However, while these results illustrate the importance of the nucleolus for the maintenance of adult neurons, their relevance to the common neurodegenerative diseases is uncertain.
While a complete inhibition of Pol1 activity such as in TIF1A knockout mice may be unlikely, partial nucleolar insufficiency may contribute to neurodegeneration (Table 1). For instance, reduced nucleolar size indicative of Pol1 inhibition has been reported in neurons from brain regions that are affected by AD [85–87]. Moreover, reduced numbers of ribosomes and/or extensive oxidation of rRNA have been demonstrated in hippocampal and cerebro-cortical samples from AD brains [88–90] (Fig. 4A, B). Such rRNA damage was associated with decreased protein synthesis [88–90]. Importantly, both rRNA oxidation and ribosomal defects have been found in mild cognitive impairment (MCI) which often represents an early stage AD [89, 90] (Fig. 4A, B). More recently, hypermethylation of the rDNA promoter has been reported in the cerebro-cortical- but not cerebellar samples from AD brains [91]. Such hypermethylation, which suggests epigenetic silencing of the nucleolar rRNA genes, was most pronounced at the early stages of AD. While rDNA promoter hypermethylation has been shown to reduce nucleolar transcription and ribosomal biogenesis, it may also be a consequence of lower transcriptional activity of Pol1 [92]. Irrespective of being a cause or the result of lower nucleolar transcription, hypermethylation of the rDNA promoter suggests nucleolar failure in AD. The resulting reduction of ribosomal biogenesis may lower the renewal rate of neuronal ribosomes resulting in accumulation of damaged ribosomes and the AD-associated translational insufficiency (Fig. 4C, D).
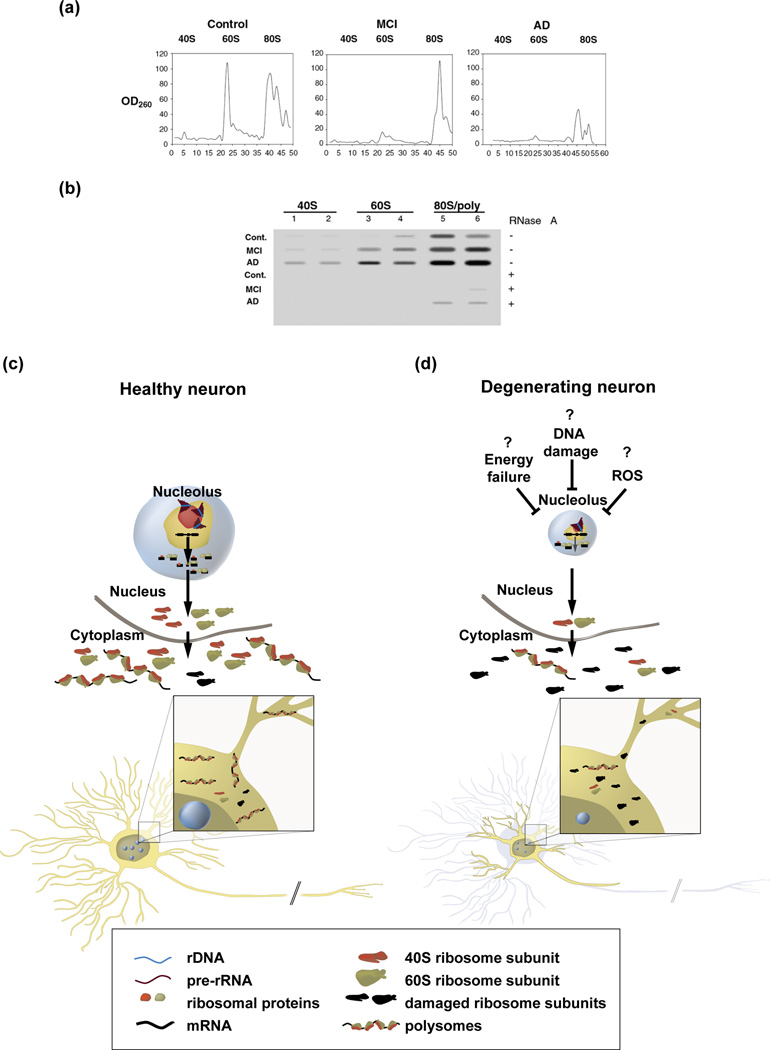
Figure 4. A hypothetical model for the role of nucleolar insufficiency in AD.
a–b, Reduced ribosomal content and increased rRNA oxidation in AD pathology-affected parietal cortex. a, Representative examples of ribosomal isolations from parietal cortex samples of a control, a mild cognitive impairment (MCI)- and a late stage AD patient. Sucrose gradient centrifugation fractions (fraction numbers are on the X-axis) were monitored for RNA content (UV 260 nm absorbance is depicted on the Y-axis using arbitrary units) [90]. In MCI, which often represents early stage AD, and late stage AD, the content of 60S large ribosomal subunit- and 80S ribosome/polysomes is decreased. Reproduced, with permission, from [90]. b, Slot blot analysis of the peak 40S, 60S and 80S fractions for the content of the marker of oxidized nucleic acids, 8-hydroxyguanosine using an anti-8-hydroxyguanosine-specific antibody [90]. Samples from two representative cases of each group (control-, MCI-, and late stage AD) are shown. RNA specificity of the signal was confirmed using RNase-pretreated samples (see reference [90] for more details). Reproduced, with permission, from [90]. c, In healthy neurons, proficient ribosomal biogenesis ensures appropriate renewal of the ribosome pools and normal protein synthesis that is required for synaptic maintenance and synaptic plasticity. d, In degenerating neurons (for example, in AD-affected brain regions), nucleolar transcription may be reduced by such factors as lowered energy resources, DNA damage and/or stress signaling activated by the excessive generation of the reactive oxygen species (ROS) [11, 19, 27, 91]. That would result in lower production of ribosomes and consequent inability to replace ribosomes whose rRNA is damaged by oxidation (marked in black) [88–90]. As a result, the number of active ribosomes would decrease and the declining protein synthesis would be insufficient to support neuronal maintenance including structural and functional integrity of neurites and synapse [88–90].
The causes of nucleolar malfunction in AD are yet to be identified. They may include such AD-associated phenomena as accumulation of oxidative DNA damage, activation of oxidative stress signaling pathways such as JNK, and reduced energy resources (reviewed in [93–95]) (Fig. 4C, D). Indeed, epigenetic silencing of rDNA has been shown to protect against energy depletion in proliferating cells, where ribosomal biogenesis is a major energy drain [11]. Although, in neurons electrical activity or actin dynamics is expected to consume more energy than ribosomal production, reduction of the latter could offer extra resources to survive under unfavorable conditions [96]. While a transient protection from cell death may be beneficial, impaired ribosomal biogenesis with time would be expected to decrease translation affecting processes like maintenance of synapses [97] and synaptic plasticity (reviewed in [98]). As a result, the neuroprotective reduction in nucleolar transcription could contribute to dementia by promoting synapse loss and/or synapse dysfunction.
Although the biological consequences of neurodegeneration-associated nucleolar deficiencies remain to be verified experimentally, questions arise whether the nucleolus may be a therapeutic target to reduce dysfunction of the degenerating nervous system. In support of this possibility, hypertrophic nucleoli have been observed in both cortical and hippocampal neurons from asymptomatic subjects with AD pathology [86, 99]. Therefore, stimulation of nucleolar transcription may offer a strategy to compensate for neurodegenerative damage to the brain.
Concluding remarks
Nearly 200 years after the first observations of neuronal nucleoli, recent work has provided new insights into the functional significance of nucleolar dynamics in neurons including its role in neuronal growth and stress response. Conversely, there is a new appreciation for how nucleolar insufficiency may contribute to several neurodevelopmental syndromes and neurodegenerative diseases. There are, however, several unresolved issues which are of particular significance for further progress on nucleolar research in the nervous system (Box 1). First, the mechanisms through which nucleoli regulate neuronal growth and stress responses remain to be identified. Second, whether partial nucleolar insufficiency plays a direct role in pathogenesis of such neurodegenerative diseases as AD is unclear. Third, experiments are warranted to determine whether the neuronal nucleolus can be a therapeutic target for neuroprotection and/or neurorepair. For example, pro-nucleolar interventions could be harnessed to correct neurodevelopmental disruptions or stimulate repair of the damaged nervous system. Conversely, transient inhibition of the nucleolar activity could support neuronal survival under such acute insults as stroke or neurotrauma. Taken together, studies of the neuronal nucleolus appear to have an exciting period ahead with a potential to change our mechanistic understanding of neuronal growth, maintenance and degeneration.
Box 1. Outstanding questions.
What are the regulatory mechanisms adjusting nucleolar transcription to neuronal needs? Which signaling pathways play a critical role in that process? Do epigenetic mechanisms contribute to such a regulation?
What are the mechanism(s) mediating the neurite growth effects of increased nucleolar transcription? Can such mechanism(s) include local delivery of de-novo made ribosomes to the growing neurites and support of local protein synthesis?
Does the nucleolus determine regenerative growth of neurons? If so, can the nucleolus be harnessed for the therapeutic neurorepair?
Does the neuronal nucleolar insufficiency contribute to neurodevelopmental disorders such as Bowen-Conradi-, ANE, Cockayne-, Prader-Willi- or Rett syndrome?
Does neuronal activity regulate nucleolar transcription? Is such a regulation required for synaptic plasticity?
What are the neuronal effector mechanisms of the nucleolar stress?
Is neuronal growth and/or neuronal maintenance affected by such non-classical functions of the nucleolus as non-rRNA processing, transcription factor regulation, extranuclear export of proteins and RNAs or chromatin maintenance?
Is the nucleolar insufficiency a cause or a result of neurodegeneration?
What are the biological consequences of partial nucleolar insufficiency in degenerating neurons?
Can the nucleolar modulation be a valid therapeutic approach to neurodevelopmental disorders and/or neurodegeneration?
What are the contributions of the glial nucleolus to neuronal growth and maintenance?
Acknowledgments
This work was supported by the National Institutes of Health (1R01NS073584 and 1P30RR031159), the National Science Foundation (IOS1021860), the Commonwealth of Kentucky Challenge for Excellence, and the Norton Healthcare. The authors wish to thank Drs. Steven R. Ellis, Scott R. Whittemore, Theo Hagg and all members of the Hetman laboratory for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Harris H. The cradle of histology. In: Harris H, editor. The birth of the cell. 1st. edn. Yale University Press; 2000. pp. 82–93. [Google Scholar]
- 2.Sirri V, et al. Nucleolus: the fascinating nuclear body. Histochem. Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drygin D, et al. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 4.Freed EF, et al. When ribosomes go bad: diseases of ribosome biogenesis. Mol. Biosyst. 2010;6:481–493. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kressler D, et al. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Olson MO. Sensing cellular stress: another new function for the nucleolus? Sci STKE. 2004;2004:pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 7.Boulon S, et al. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casafont I, et al. The giant fibrillar center: a nucleolar structure enriched in upstream binding factor (UBF) that appears in transcriptionally more active sensory ganglia neurons. J. Struct. Biol. 2007;159:451–461. doi: 10.1016/j.jsb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Kalita K, et al. Inhibition of nucleolar transcription as a trigger for neuronal apoptosis. J. Neurochem. 2008;105:2286–2299. doi: 10.1111/j.1471-4159.2008.05316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–629. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Murayama A, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J, et al. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell. 2003;11:405–413. doi: 10.1016/s1097-2765(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 13.Laferte A, et al. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 15.Hannan KM, et al. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James MJ, Zomerdijk JC. Phosphatidylinositol 3-kinase and mTOR signaling pathways regulate RNA polymerase I transcription in response to IGF-1 and nutrients. J. Biol. Chem. 2004;279:8911–8918. doi: 10.1074/jbc.M307735200. [DOI] [PubMed] [Google Scholar]
- 17.Mayer C, et al. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefanovsky VY, et al. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell. 2001;8:1063–1073. doi: 10.1016/s1097-2765(01)00384-7. [DOI] [PubMed] [Google Scholar]
- 19.Mayer C, et al. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005;19:933–941. doi: 10.1101/gad.333205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoppe S, et al. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. USA. 2009;106:17781–17786. doi: 10.1073/pnas.0909873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelletier G, et al. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol. Cell. 2000;6:1059–1066. doi: 10.1016/s1097-2765(00)00104-0. [DOI] [PubMed] [Google Scholar]
- 22.Hirschler-Laszkiewicz I, et al. The role of acetylation in rDNA transcription. Nucleic Acids Res. 2001;29:4114–4124. doi: 10.1093/nar/29.20.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J, et al. Dysregulation of upstream binding factor-1 acetylation at K352 is linked to impaired ribosomal DNA transcription in Huntington's disease. Cell Death Differ. 2011;18:1726–1735. doi: 10.1038/cdd.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel SB, Bellini M. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 2008;36:6482–6493. doi: 10.1093/nar/gkn658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hopper AK, Shaheen HH. A decade of surprises for tRNA nuclear-cytoplasmic dynamics. Trends Cell Biol. 2008;18:98–104. doi: 10.1016/j.tcb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pietrzak M, et al. Nucleolar disruption and apoptosis are distinct neuronal responses to etoposide-induced DNA damage. J. Neurochem. 2011;117:1033–1046. doi: 10.1111/j.1471-4159.2011.07279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tembe V, Henderson BR. Protein trafficking in response to DNA damage. Cell. Signal. 2007;19:1113–1120. doi: 10.1016/j.cellsig.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Moore HM, et al. Quantitative Proteomics and Dynamic Imaging of the Nucleolus Reveal Distinct Responses to UV and Ionizing Radiation. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.009241. M111:009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 2009;184:771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boyd MT, et al. The nucleolus directly regulates p53 export and degradation. J. Cell Biol. 2011;194:689–703. doi: 10.1083/jcb.201105143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latonen L, et al. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene. 2011;30:790–805. doi: 10.1038/onc.2010.469. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi T. How does genome instability affect lifespan?: roles of rDNA and telomeres. Genes Cells. 2011;16:617–624. doi: 10.1111/j.1365-2443.2011.01519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parrish JZ, et al. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu. Rev. Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- 35.Dijkhuizen PA, Ghosh A. Regulation of dendritic growth by calcium and neurotrophin signaling. Prog. Brain Res. 2005;147:17–27. doi: 10.1016/S0079-6123(04)47002-2. [DOI] [PubMed] [Google Scholar]
- 36.Zhou FQ, Snider WD. Intracellular control of developmental and regenerative axon growth. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1575–1592. doi: 10.1098/rstb.2006.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafarga M, et al. Changes in nucleolar morphology and volume of the supraoptic nucleus neurons during postnatal development of the rat. Brain Res. 1985;354:310–313. doi: 10.1016/0165-3806(85)90185-3. [DOI] [PubMed] [Google Scholar]
- 38.Clark P, et al. Ultrastructural and morphometric analysis of nucleolar and nuclear changes during the early growth period in hamster facial neurons. J. Comp. Neurol. 1990;302:749–760. doi: 10.1002/cne.903020407. [DOI] [PubMed] [Google Scholar]
- 39.Kinderman NB, et al. Ribosomal RNA transcriptional activation and processing in hamster facial motoneurons: effects of axotomy with or without exposure to testosterone. J. Comp. Neurol. 1998;401:205–216. [PubMed] [Google Scholar]
- 40.Storer PD, Jones KJ. Ribosomal RNA transcriptional activation and processing in hamster rubrospinal motoneurons: effects of axotomy and testosterone treatment. J. Comp. Neurol. 2003;458:326–333. doi: 10.1002/cne.10623. [DOI] [PubMed] [Google Scholar]
- 41.Gomes C, et al. RNA polymerase 1-driven transcription as a mediator of BDNF-induced neurite outgrowth. J. Biol. Chem. 2011;286:4357–4363. doi: 10.1074/jbc.M110.170134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaworski J, et al. Control of dendritic arborization by the phosphoinositide-3'-kinase-Akt-mammalian target of rapamycin pathway. J. Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar V, et al. Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J. Neurosci. 2005;25:11288–11299. doi: 10.1523/JNEUROSCI.2284-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dotti CG, et al. The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marmigere F, et al. The Runx1/AML1 transcription factor selectively regulates development and survival of TrkA nociceptive sensory neurons. Nat. Neurosci. 2006;9:180–187. doi: 10.1038/nn1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stegmuller J, et al. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1-APC target SnoN. Neuron. 2006;50:389–400. doi: 10.1016/j.neuron.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 47.Stoykova AS, et al. Different rates of synthesis and turnover of ribosomal RNA in rat brain and liver. J. Neurochem. 1983;41:942–949. doi: 10.1111/j.1471-4159.1983.tb09038.x. [DOI] [PubMed] [Google Scholar]
- 48.Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu. Rev. Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- 49.Twiss JL, Fainzilber M. Ribosomes in axons--scrounging from the neighbors? Trends Cell Biol. 2009;19:236–243. doi: 10.1016/j.tcb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 50.Jan YN, Jan LY. Branching out: mechanisms of dendritic arborization. Nat. Rev. Neurosci. 2010;11:316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W, et al. RNA transport and localized protein synthesis in neurological disorders and neural repair. Dev. Neurobiol. 2007;67:1166–1182. doi: 10.1002/dneu.20511. [DOI] [PubMed] [Google Scholar]
- 52.Koenig E, Martin R. Cortical plaque-like structures identify ribosome-containing domains in the Mauthner cell axon. J. Neurosci. 1996;16:1400–1411. doi: 10.1523/JNEUROSCI.16-04-01400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dinman JD. The eukaryotic ribosome: current status and challenges. J. Biol. Chem. 2009;284:11761–11765. doi: 10.1074/jbc.R800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S, Coulombe PA. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat. Rev. Mol. Cell Biol. 2010;11:75–81. doi: 10.1038/nrm2818. [DOI] [PubMed] [Google Scholar]
- 55.Zinzalla V, et al. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 56.Court FA, et al. Morphological evidence for a transport of ribosomes from Schwann cells to regenerating axons. Glia. 2011;59:1529–1539. doi: 10.1002/glia.21196. [DOI] [PubMed] [Google Scholar]
- 57.Court FA, et al. Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J. Neurosci. 2008;28:11024–11029. doi: 10.1523/JNEUROSCI.2429-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Armistead J, et al. Mutation of a gene essential for ribosome biogenesis, EMG1, causes Bowen-Conradi syndrome. Am. J. Hum. Genet. 2009;84:728–739. doi: 10.1016/j.ajhg.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nousbeck J, et al. Alopecia, neurological defects, and endocrinopathy syndrome caused by decreased expression of RBM28, a nucleolar protein associated with ribosome biogenesis. Am. J. Hum. Genet. 2008;82:1114–1121. doi: 10.1016/j.ajhg.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weidenheim KM, et al. Neuropathology of Cockayne syndrome: Evidence for impaired development, premature aging, and neurodegeneration. Mech. Ageing Dev. 2009;130:619–636. doi: 10.1016/j.mad.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 61.Assfalg R, et al. TFIIH is an elongation factor of RNA polymerase I. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sahoo T, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Smith AJ, et al. A deletion of the HBII-85 class of small nucleolar RNAs (snoRNAs) is associated with hyperphagia, obesity and hypogonadism. Hum. Mol. Genet. 2009;18:3257–3265. doi: 10.1093/hmg/ddp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cavaille J, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bazeley PS, et al. snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene. 2008;408:172–179. doi: 10.1016/j.gene.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leung KN, et al. Imprinting regulates mammalian snoRNA-encoding chromatin decondensation and neuronal nucleolar size. Hum. Molec. Genet. 2009;18:4227–4238. doi: 10.1093/hmg/ddp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Singleton MK, et al. MeCP2 is required for global heterochromatic and nucleolar changes during activity-dependent neuronal maturation. Neurobiol. Dis. 2011;43:190–200. doi: 10.1016/j.nbd.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tomiwa K, et al. The effects of cisplatin on rat spinal ganglia: a study by light and electron microscopy and by morphometry. Acta Neuropathol. (Berl) 1986;69:295–308. doi: 10.1007/BF00688308. [DOI] [PubMed] [Google Scholar]
- 70.Bigotte L, Olsson Y. Degeneration of trigeminal ganglion neurons caused by retrograde axonal transport of doxorubicin. Neurology. 1987;37:985–992. doi: 10.1212/wnl.37.6.985. [DOI] [PubMed] [Google Scholar]
- 71.McKeage MJ, et al. Nucleolar damage correlates with neurotoxicity induced by different platinum drugs. Br. J. Cancer. 2001;85:1219–1225. doi: 10.1054/bjoc.2001.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jamieson SM, et al. Oxaliplatin causes selective atrophy of a subpopulation of dorsal root ganglion neurons without inducing cell loss. Cancer Chemother. Pharmacol. 2005;56:391–399. doi: 10.1007/s00280-004-0953-4. [DOI] [PubMed] [Google Scholar]
- 73.Casafont I, et al. Effect of ionizing radiation in sensory ganglion neurons: organization and dynamics of nuclear compartments of DNA damage/repair and their relationship with transcription and cell cycle. Acta neuropathol. 2011;122:481–493. doi: 10.1007/s00401-011-0869-0. [DOI] [PubMed] [Google Scholar]
- 74.Mosgoeller W, et al. Brain RNA polymerase and nucleolar structure in perinatal asphyxia of the rat. Exp. Neurol. 2000;161:174–182. doi: 10.1006/exnr.1999.7232. [DOI] [PubMed] [Google Scholar]
- 75.Rieker C, et al. Nucleolar Disruption in Dopaminergic Neurons Leads to Oxidative Damage and Parkinsonism through Repression of Mammalian Target of Rapamycin Signaling. J. Neurosci. 2011;31:453–460. doi: 10.1523/JNEUROSCI.0590-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nagai T, et al. Cognition impairment in the genetic model of aging klotho gene mutant mice: a role of oxidative stress. Faseb J. 2003;17:50–52. doi: 10.1096/fj.02-0448fje. [DOI] [PubMed] [Google Scholar]
- 77.Anamizu Y, et al. Klotho insufficiency causes decrease of ribosomal RNA gene transcription activity, cytoplasmic RNA and rough ER in the spinal anterior horn cells. Acta Neuropathol. (Berl) 2005;109:457–466. doi: 10.1007/s00401-004-0971-7. [DOI] [PubMed] [Google Scholar]
- 78.Baltanas FC, et al. Nucleolar Disruption and Cajal Body Disassembly are Nuclear Hallmarks of DNA Damage-Induced Neurodegeneration in Purkinje Cells. Brain Pathol. 2011;21:374–388. doi: 10.1111/j.1750-3639.2010.00461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hetman M, et al. Neurotoxic mechanisms of DNA damage: focus on transcriptional inhibition. J. Neurochem. 2010;114:1537–1549. doi: 10.1111/j.1471-4159.2010.06859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kerr LE, et al. Nucleophosmin is a novel Bax chaperone that regulates apoptotic cell death. Oncogene. 2007;26:2554–2562. doi: 10.1038/sj.onc.1210044. [DOI] [PubMed] [Google Scholar]
- 81.Covey JM, et al. Protein-linked DNA strand breaks induced in mammalian cells by camptothecin, an inhibitor of topoisomerase I. Cancer Res. 1989;49:5016–5022. [PubMed] [Google Scholar]
- 82.Parlato R, et al. Activation of an endogenous suicide response after perturbation of rRNA synthesis leads to neurodegeneration in mice. J. Neurosci. 2008;28:12759–12764. doi: 10.1523/JNEUROSCI.2439-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang H, et al. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc. Natl. Acad. Sci. USA. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johnson R, Strehler BL. Loss of genes coding for ribosomal RNA in ageing brain cells. Nature. 1972;240:412–414. doi: 10.1038/240412a0. [DOI] [PubMed] [Google Scholar]
- 85.Mann DM, et al. The progression of the pathological changes of Alzheimer's disease in frontal and temporal neocortex examined both at biopsy and at autopsy. Neuropathol. Appl. Neurobiol. 1988;14:177–195. doi: 10.1111/j.1365-2990.1988.tb00880.x. [DOI] [PubMed] [Google Scholar]
- 86.Iacono D, et al. Neuronal hypertrophy in asymptomatic Alzheimer disease. J. Neuropathol. Exp. Neurol. 2008;67:578–589. doi: 10.1097/NEN.0b013e3181772794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tagliavini F, Pilleri G. Basal nucleus of Meynert. A neuropathological study in Alzheimer's disease, simple senile dementia, Pick's disease and Huntington's chorea. J. Neurol. Sci. 1983;62:243–260. doi: 10.1016/0022-510x(83)90203-4. [DOI] [PubMed] [Google Scholar]
- 88.Honda K, et al. Ribosomal RNA in Alzheimer disease is oxidized by bound redox-active iron. J. Biol. Chem. 2005;280:20978–20986. doi: 10.1074/jbc.M500526200. [DOI] [PubMed] [Google Scholar]
- 89.Ding Q, et al. Ribosome dysfunction is an early event in Alzheimer's disease. J. Neurosci. 2005;25:9171–9175. doi: 10.1523/JNEUROSCI.3040-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ding Q, et al. Decreased RNA, Increased RNA Oxidation, in Ribosomes from Early Alzheimer's Disease. Neurochem. Res. 2006;31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- 91.Pietrzak M, et al. Epigenetic Silencing of Nucleolar rRNA Genes in Alzheimer's Disease. PLoS One. 2011;6:e22585. doi: 10.1371/journal.pone.0022585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmitz KM, et al. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol. Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 93.Markesbery WR, Lovell MA. DNA oxidation in Alzheimer's disease. Antioxid. Redox Signal. 2006;8:2039–2045. doi: 10.1089/ars.2006.8.2039. [DOI] [PubMed] [Google Scholar]
- 94.Zhu X, et al. Differential activation of neuronal ERK, JNK/SAPK and p38 in Alzheimer disease: the 'two hit' hypothesis. Mech. Ageing Dev. 2001;123:39–46. doi: 10.1016/s0047-6374(01)00342-6. [DOI] [PubMed] [Google Scholar]
- 95.Kapogiannis D, Mattson MP. Disrupted energy metabolism and neuronal circuit dysfunction in cognitive impairment and Alzheimer's disease. Lancet Neurol. 2011;10:187–198. doi: 10.1016/S1474-4422(10)70277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bernstein BW, Bamburg JR. Actin-ATP hydrolysis is a major energy drain for neurons. J. Neurosci. 2003;23:1–6. doi: 10.1523/JNEUROSCI.23-01-00002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCann CM, et al. Rapid synapse elimination after postsynaptic protein synthesis inhibition in vivo. J. Neurosci. 2007;27:6064–6067. doi: 10.1523/JNEUROSCI.0627-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klann E, Sweatt JD. Altered protein synthesis is a trigger for long-term memory formation. Neurobiol. Learn. Mem. 2008;89:247–259. doi: 10.1016/j.nlm.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iacono D, et al. The Nun study: clinically silent AD, neuronal hypertrophy, and linguistic skills in early life. Neurology. 2009;73:665–673. doi: 10.1212/WNL.0b013e3181b01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mann DM, Yates PO. Pathogenesis of Parkinson's disease. Arch. Neurol. 1982;39:545–549. doi: 10.1001/archneur.1982.00510210015004. [DOI] [PubMed] [Google Scholar]
- 101.McGowan PO, et al. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS ONE. 2008;3:e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]