Abstract
Systemic delivery of siRNA to solid tumors remains challenging. In this study, we investigated the systemic delivery of an siRNA-nanoparticle targeting ribonucleotide reductase subunit M2 (RRM2), and evaluated its intratumoral kinetics, efficacy and mechanism of action. Knockdown of RRM2 by an RNAi mechanism strongly inhibited cell growth in head and neck squamous cell carcinoma (HNSCC) and non-small cell lung cancer (NSCLC) cell lines. In a mouse xenograft model of HNSCC, a single intravenous injection led to the accumulation of intact nanoparticles in the tumor that disassembled over a period of at least 3 days, leading to target gene knockdown lasting at least 10 days. A four-dose schedule of siRNA-nanoparticle delivering RRM2 siRNA targeted to HNSCC tumors significantly reduced tumor progression by suppressing cell proliferation and inducing apoptosis. These results show promise for the use of RRM2 siRNA-based therapy for HNSCC and possibly NSCLC.
Keywords: siRNA delivery, targeted nanoparticle, RNA interference, RRM2, TfR, tumor growth
1. Introduction
RNA interference (RNAi) has advanced to clinical trials with great promise following the Nobel-prize winning discovery in 1998 [1]. The pursuit of siRNA-based therapeutics designed to engage RNAi pathways has the potential to provide new, effective ways of imparting therapy to patients [2–4]. To treat most cancers and other disseminated diseases, it is necessary to administer the siRNA systemically. However, the systemic administration of siRNA in vivo faces a series of hurdles such as kidney filtration, uptake by phagocytes, aggregation with serum proteins, and enzymatic degradation by endogenous nucleases before reaching the cytoplasm of the target cell. As a class of new delivery vehicles, cell-specific targeted nanoparticles have the potential to provide increased efficacy and reduced toxicity relative to conventional therapeutics [5]. Several promising strategies have been adopted to improve the systemic delivery of siRNA in animal models [2, 3, 6–8], but their safe and effective delivery still remain challenging, particularly in the clinical setting.
Ribonucleotide reductase (RR) is the rate-limiting enzyme in the conversion of ribonucleotide 5′-diphosphates into 2′-deoxyribonucleotides, which are essential for DNA synthesis and replication. RR enzymatic activity is modulated by the level of its M2 subunit (RRM2), which is expressed only in the late G1/early S phase of the cell cycle when DNA replication occurs [9–13]. Overexpression of RRM2 plays an active role in tumor progression and in the cellular response to DNA damage [14, 15]. Elevated RR activity and RRM2 overexpression significantly increase the drug-resistant properties and the angiogenic and invasive potential of human cancer cells [12, 16–18]. RRM2 was identified as a diagnostic marker and an indicator of poor patient outcome of several cancers [19, 20], suggesting that RRM2 contributes to malignant progression and is a potential therapeutic target. In our study, we used a targeted nanoparticle for systemic delivery of RRM2 siRNA. The clinical version of this delivery system has been denoted CALAA-01, and is now in a Phase I clinical trial for systemic delivery of siRNA to treat patients with solid cancers [7, 21].
Human transferrin protein (Tf) ligand is displayed on the exterior of CALAA-01 as a targeting moiety to engage the Tf receptor (TfR), which has long been known to be upregulated in malignant cells [22] and is a common ligand used to target tumor cells [23–26]. It has been reported that CALAA-01 can deliver a higher fraction of siRNA within tumor cells by targeting TfR to achieve intracellular localization compared to nontargeted analogs [27]. This system recently provided the first example of dose-dependent accumulation of targeted nanoparticles in human tumors and also the first example of delivering an siRNA that was proven to mediate an RNAi mechanism of action [7].
Evaluation of CALAA-01 is further extended in the present work to demonstrate the intratumoral kinetics of the nanoparticle system and to observe its efficacy in tumor growth inhibition in head and neck cancer, which is one of the leading causes of cancer death worldwide [28]. Such studies are critical for understanding the effective dosing and prolonged knockdown of the targeted protein to achieve better therapeutic effect. Here, we explored the role of RRM2 by utilizing siRNA-mediated gene silencing that leads to growth suppression in vivo and in vitro. This could impact treatment of several other cancers. To our knowledge, this is the first animal study in head and neck cancer confirming that systemically delivered siRNA can produce specific gene inhibition in a mouse model and can contribute to inhibition of tumor progression by suppressing cell proliferation and by induction of apoptosis.
2. Materials and Methods
2.1. Cell lines
HNSCC cell lines Tu177 and Tu212 (provided by Dr. Gary L. Clayman, University of Texas M.D. Anderson Cancer Center), Tu686, 686LN and 886LN (provided by Dr. Peter G. Sacks, New York University College of Dentistry), SQCCY1 (provided by Dr. Shi-Yong Sun, Emory University) and M4e cells were derived from a metastasis xenograft mouse model, [29] and were cultured in DMEM/F12 (1:1) with 10% heat inactivated fetal bovine serum (FBS). A549, H292, H460 and H1299 lung cancer cell lines were kindly provided by Dr Shi-Yong Sun (Emory University, GA). These cell lines were maintained in RPMI 1640 with 10% FBS. BJ (normal human fibroblast) and BEAS-2B (bronchial epithelial cells) were obtained from Dr. George R. Stark (Cleveland Clinic Foundation, OH) and Dr. Xingming Deng (Emory Winship Cancer Institute, GA) respectively. All cells were maintained in a humidified incubator at 37°C, 5% CO2.
2.2. Western blotting
The primary antibodies were anti-RRM2, anti-GAPDH, from Santa Cruz Biotech, CA; anti-hTfR from Abcam, Cambridge, MA; and anti-β-actin from Sigma Aldrich. The secondary antibodies were from Santa Cruz Biotech. Western band quantification was performed using Image-Quant TL software (GE/Amersham Biosciences, Piscataway, NJ).
2.3. siRNAs duplexes
Unmodified RNA duplexes were gifts from Calando Pharmaceuticals (Pasadena, CA). Control siRNA was bioinformatically designed to minimize potential for targeting any human gene (Dharmacon, Lafayette, CO).
RRM2 siRNA (siR2):
5′-GCGAUUUAGCCAAGAAGUUCA-3′
Control siRNA (siC or siCON1):
5′-UAGCGACUAAACACAUCAAUU-3′
2.4. Lipofectamine transfection
siRNA was complexed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions and applied to each plate (cells 30~50% confluent). Transfection media was removed and replaced with new media after 5 hours.
2.5. 5′-RLM-RACE
5′-RLM-RACE (RNA Ligase Mediated Rapid Amplification of cDNA Ends) was performed according to the Invitrogen GeneRacer manual with modifications as described previously [7]. Briefly, RNA was ligated to GeneRacer RNA adaptor and the reverse-transcribed using SuperscriptIII (Invitrogen). RACE-PCR was performed using forward (GR5′) and reverse (R2Rev) primers in the GeneRacer adaptor and 3′ end of RRM2 mRNA, respectively, to span the predicted RRM2 cut site. PCR products were excised from 2% agarose gel and sequenced directly to confirm RACE band identities.
2.6. Cell growth assay
Sulforhodamine B (SRB) assay was used for cell growth determination [30]. siRNA-transfected cells were re-seeded (5×103 cells/well) in a 96-well plate 24 hrs after transfection. 72 hrs later, cells were fixed with 10% trichloroacetic acid. Plates were stained with 0.4% SRB for 10 min and bound SRB was dissolved in 10 mmol/L Tris base (pH 10.5). Cell growth was assessed by OD determination at 492 nm using a microplate reader. The percentage of survival was then calculated based on the absorbance values relative to the control siRNA-transfected samples.
2.7. Formulation of CALAA-01
The CALAA-01 formulation used in this study was described earlier [31]. The delivery components CDP, AD-PEG, AD-PEG-Tf were placed in one vial and the nucleic acid in a second vial. When the two solutions (1:1 volumes) were mixed, the nanoparticles self-assembled. siRNA and nanoparticle components were supplied by Calando Pharmaceuticals. The formulation gave a 70 nm diameter nanoparticle that contained ~10,000 CDP molecules, ~2000 siRNA molecules, ~4000 AD-PEG, and ~100 AD-PEG-Tf molecules [21].
2.8. Systemic delivery of CALAA-01 in a xenograft nude mouse model
Based on protocols approved by the Institutional Animal Care and Use Committee of Emory University, nude mice (athymic nu/nu) aged 4–6 weeks were purchased from Taconic. Each mouse was injected with 5×106 Tu212 cells subcutaneously in the right flank, and tumor volume was monitored (volume = 0.5×l×w2). Treatments commenced when the tumors had reached approximately 70 mm3. For detection of intratumoral kinetics of CALAA-01, we injected a single dose of nanoparticles (10mg/kg) via tail vein injection and collected tumors after 24h, 48h, 72h, 96h, 168h, and 240h (3 mice/group). Then we performed nanoparticle intratumoral kinetics assays by confocal microscopy, immunohistochemistry, and qRT-PCR.
In another animal model experiment (8 mice/group), we injected 5% dextrose in water (D5W), naked siRNA (siR2) or nanoparticle containing either siCON1 (siCON1) or siR2 (CALAA-01) via the tail vein in a four-dose schedule (Days 1, 3, 8 and 10) and sacrificed the mice on the 28th day after cancer cell transplantation.
2.9. Confocal microscopy
Snap-frozen tumor tissues were re-embedded in paraffin to generate tissue sections of 4 μm in thickness. Upon deparafinization with xylene and rehydration with a decreasing ethanol gradient, the sections were permeabilized in chilled acetone at −20 °C for 20 min. To detect intracellular localization of CDP/siRNA nanoparticles and/or expression levels of the RRM2 protein, PBS-rinsed sections were immersed in a PBS-based staining solution that contains adamantine-modified, PEGylated gold nanoparticles (Au-PEG-AD) at a final concentration of 1.5×1011 particles per mL and/or goat anti-human R2 polyclonal antibody (sc-10846; Santa Cruz Biotechnology), conjugated to Alexa Fluor 555 (Invitrogen), at a final concentration of 1 μg/mL. Staining proceeded in the dark at RT for 2 h. Brief PBS rinses removed any non-specifically bound gold particles and anti-R2 antibodies, before mounting the stained tumor sections with ProLong Gold antifade reagent with DAPI (Invitrogen). A Zeiss LSM 510 confocal scanning microscope was used to collect the images. The excitation wavelengths of DAPI, Au-PEG-AD, and Alexa Fluor 555 are 740 nm (two photon laser), 488 nm (argon laser), and 543 nm (HeNe laser), respectively. Their corresponding emission filters are 400–460 nm, 500–550 nm, and 560–610 nm, respectively.
2.10. qRT-PCR
Total tumor RNA was collected using the RNeasy mini kit according to the manufacturer’s instructions (Qiagen). Tumor RNA samples were reverse-transcribed using SuperScriptIII reverse transcriptase. 2 μl of prepared sample cDNA was used for triplicate Taqman real time-PCR as described elsewhere [32]. RRM2 levels were normalized to Tata Binding Protein (TBP) levels within the same sample.
2.11. Ribonucleotide reductase (RR) assay
RR activity was measured utilizing the CDP assay method as previously described [33]. Frozen tissue was cut into small pieces using a new razor blade. The tissue suspension was homogenized and tissue debris was removed by centrifugation at 16,000 g at 4° for 20 min. The supernatant was passed through a Sephadex G25 spin column, pre-equilibrated with buffer (50 mM of HEPES, pH 7.2, 2 mM of DTT) to remove endogenous nucleotides. The reaction mixture in a final volume of 50 μl contained the following: [3H]CDP (0.5 μCi;), HEPES (pH 7.2) (50 mM), DTT (6 mM), magnesium acetate (4 mM), ATP (2 mM), CDP (0.05 mM), and a specific amount of tissue extract. The incubation time for the reaction was 30 min at 37°C. [3H]dCDP was dephosphorylated by phosphodiesterase at 37°C for 2hrs. [3H]C and [3H]dC were separated with a C18 ion exchange column by HPLC and detected by β-RAM radioactive detector. The reaction was linear during the process.
2.12. Immunohistochemistry
Upon deparaffinization and rehydration, tissue sections were permeabilized with 0.25% Triton-X-100/PBS for 5 min. Tissue sections were blocked with 2.5% horse serum for 30 minutes. To detect intracellular localization and expression levels of RRM2 and Ki67 proteins, we used goat anti-human R2 polyclonal antibody and mouse anti-human Ki-67 antibody (prediluted; Invitrogen, Carlsbad, CA), respectively, as primary antibodies, then conjugated to secondary antibody and counterstained cell nuclei using 4,6-diamidino-2-phenylindole (DAPI, Invitrogen, Carlsbad, CA). Mouse and rabbit IgG were used as negative controls. Terminal deoxynucleotidyl transferase dUTP nick and labeling (TUNEL) assay was performed according to the manufacturer’s protocol (Promega, Madison, WI).
2.13. Statistical analysis
All results represent the average of at least three separate experiments and are expressed as mean ±SD. Statistical analysis was performed using t-test. P<0.05 was considered statistically significant.
3. Results
3.1. RRM2 and TfR proteins are expressed in HNSCC and NSCLC cells and siR2 efficiently suppresses RRM2 protein
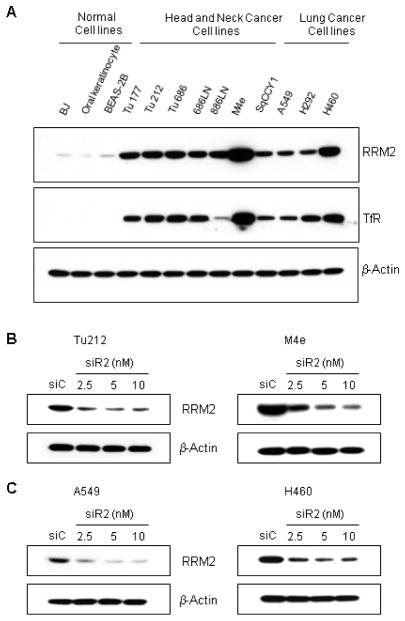
Several HNSCC and NSCLC cell lines were tested for RRM2 and TfR expression levels in order to identify a suitable model system to evaluate the efficacy of the CALAA-01 nanoparticle, which has Tf on the surface as a targeting ligand and encapsulates siRNA targeted to RRM2 [7]. HNSCC (Tu177, Tu212, Tu686, 686LN, 886LN, M4e and SqCCY1) and NSCLC (A549, H292 and H460) cells displayed various degrees (moderate to high level) of expression of TfR and RRM2 proteins compared to normal cells such as BJ (normal human fibroblasts), primary culture of BEAS-2B (bronchial epithelial cells) and oral keratinocytes (Figure 1A). These results suggest that HNSCC and NSCLC could be suitable models for in vivo studies with CALAA-01. The RRM2 sequence-specific siRNA (siR2) was designed previously to target RRM2 and has been characterized for potency, efficacy, and specificity [21, 34]. To explore the knockdown efficiency of siR2, HNSCC and NSCLC cell lines with moderate and high expression of RRM2, Tu212 and M4e, and A549 and H460, respectively, were examined. We transfected cells with siR2 or siC (non-targeting control siRNA) for 72h and observed >80% reduction in RRM2 protein levels following siR2 treatment when compared with siC (Figure 1B and C).
Fig. 1.
RRM2 and TfR proteins are expressed in HNSCC and NSCLC cells and siR2 efficiently suppresses RRM2 protein. (A) RRM2 and TfR expression in normal, HNSCC and NSCLC cell lines. Normal cell line, BJ (human fibroblast), primary culture of BEAS-2B (bronchial epithelial), normal oral keratinocytes, HNSCC (Tu177, Tu212, Tu686, 686LN, 886LN, M4e, SqCCY1) and lung cancer (A549, H292, H460) cell lines were cultured for 24 hours before cell lysis. Cell lysates were examined for expression of RRM2 (upper panel) and TfR (middle panel) by Western blotting. β-actin protein (lower panel) was immunoblotted to confirm equal loading amounts. One blot is pictured from three independent experiments. (B) HNSCC cell lines, Tu212 (left panel) and M4e (right panel), (C) NSCLC cell lines, A549 (left panel) and H460 (right panel), were transfected with 2.5, 5 and 10nM of RRM2-specific (siR2) or 10nM of control (siC) siRNA for 72h. Western blotting was performed for determination of RRM2 and β-actin expression.
3.2. siR2 silences RRM2 via an RNAi mechanism in HNSCC and NSCLC cell lines
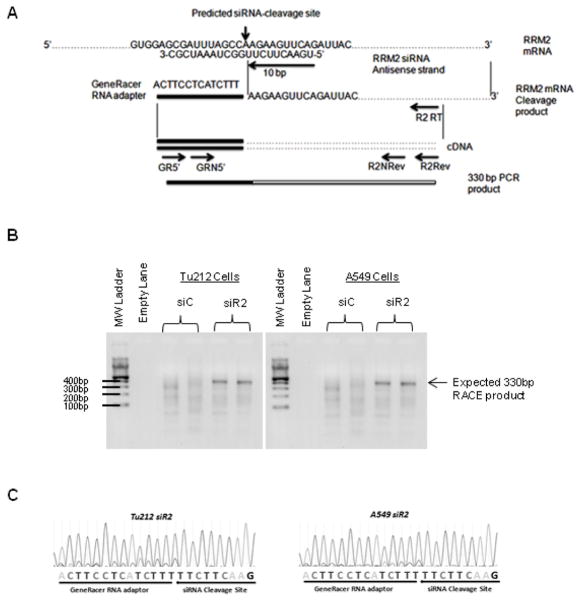
To confirm the RNAi-mediated mechanism of action of siR2, RNA from Tu212 (HNSCC) and A549 (NSCLC) cells transfected with siR2 or siC was subjected to 5′-RLM-RACE (Figure 2A–C). The schematic diagram in Figure 2A shows the predicted cleavage site of siR2 and the primers used for PCR amplification of the cleavage fragment. RRM2 mRNA fragments, whose 5′ ends matched the predicted siRNA-induced cleavage site (10 base pairs from the 5′ end of the antisense strand), and were of correct band size and sequence, were detected only in siR2-treated Tu212 and A549 cells (Figure 2B and C). These results suggest that siR2 can mediate an RNAi mechanism in both HNSCC and NSCLC cells.
Fig. 2.
siR2 silences RRM2 via an RNAi mechanism in HNSCC and NSCLC cell lines. (A) Schematic diagram showing the predicted cleavage site of RRM2 siRNA and primers used for PCR amplification of the cleavage fragment. GR5′: GeneRacer 5′ primer, GRN5′: GeneRacer 5′ nested primer, R2RT: RRM2 gene-specific RT primer, R2Rev: RRM2 gene-specific reverse primer, R2Nrev: RRM2 gene-specific nested primer. (B) 5′-RLM-RACE product detection of siRNA-induced RRM2 mRNA cleavage fragment in Tu212 (left panel) and A549 (right panel) cells. (C) Sequencing chromatographs obtained from positive RACE bands.
3.3. Depletion of RRM2 inhibits cellular growth in HNSCC and NSCLC
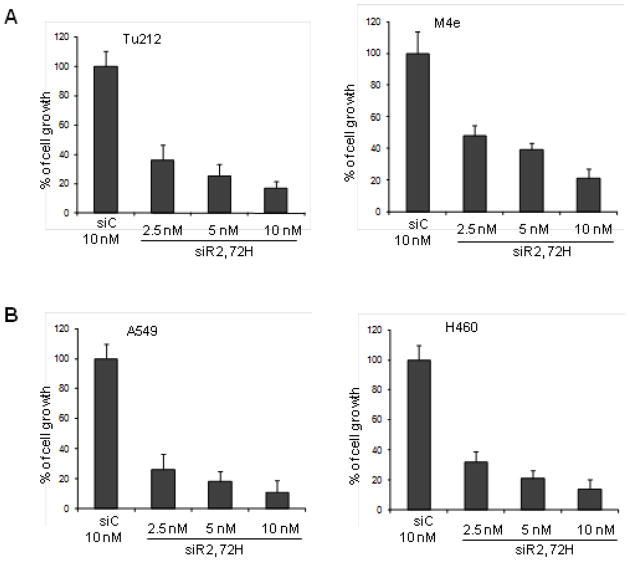
To explore the role of RRM2 in the regulation of cell proliferation, HNSCC and NSCLC cell lines with moderate and high expression of RRM2, Tu212 and M4e, and A549 and H460, respectively, were examined for cell growth inhibition by siR2 treatment. In both HNSCC and NSCLC cells, we found significant cell growth inhibition at nanomolar concentrations of siR2 compared to siC after 72h (Figure 3A and B). Treatment with 5 nM siR2 for 72h inhibited cell growth in both HNSCC (Tu212 ~75% and M4e ~60%) and NSCLC cells (A549 and H460 ~80%). Interestingly, we observed no significant difference in cell growth response between cell lines with moderate and high expression of RRM2. This finding demonstrated that once RRM2 was downregulated by siRNA, cellular growth was strongly inhibited regardless of RRM2 expression level. Our results provide evidence that RRM2 plays a vital role in cellular growth in HNSCC and NSCLC cells.
Fig. 3.
Depletion of RRM2 by siRNA inhibits cellular growth. (A) HNSCC cell lines, Tu212 (left panel) and M4e (right panel), were transfected with 2.5, 5 and 10nM of siR2 or 10nM of siC. After 72h, cell growth was measured by SRB assay (error bars are mean +SD from 3 independent experiments). (B) Cell growth assay in NSCLC cell lines, A549 (left panel) and H460 (right panel).
3.4. Intratumoral kinetics of CALAA-01 nanoparticles following a single intravenous injection
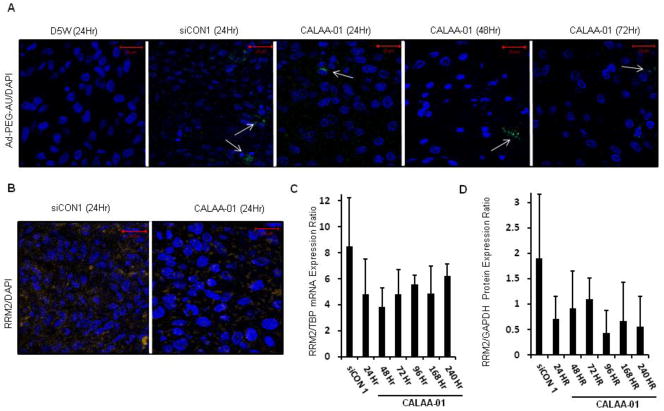
Since we confirmed that our model systems are suitable for in vivo studies, the systemic delivery of siRNA was extended to a Tu212 xenograft model by using CALAA-01 for cell-specific knockdown of RRM2. Results from the delivery of RRM2 siRNA via CALAA-01 in humans have suggested that gene inhibition can be achieved for lengthy time spans (ca. several weeks) [7]. Numerous factors may contribute to this observation: intrinsic intracellular siRNA stability, slow release of siRNA by the nanoparticles, and tumor growth dynamics. However, the contribution of each of these factors to the observed pharmacodynamics of the nanoparticle system has yet to be ascertained. In order to understand how long the intact nanoparticle protects and releases siRNA in the tumor environment, we sacrificed Tu212 tumor-bearing mice at several time points after single intravenous doses of the siRNA nanoparticle. We first examined tumors for the presence of nanoparticles using an adamantine-capped, PEGylated gold nanoparticle-based stain Au-PEG-AD, characterized elsewhere [7] that specifically binds to the cyclodextrin (CDP)/nanoparticle. Tumor sections from both control siRNA containing CDP/nanoparticle (siCON1)- and siR2-containing CDP/nanoparticles (CALAA-01) were positive for Au-PEG-AD staining, but not tumor sections from control animals treated with 5% dextrose (D5W) (Figure 4A). High Au-PEG-AD staining intensity was observed at 24 and 48 hours post dosing, and was markedly decreased at 72 hours. No further nanoparticle detection was achieved at later time points (data not shown), suggesting that no more intact nanoparticles remained within the tumor. Next, we examined the levels of RRM2 mRNA and protein. We observed a considerable decrease in RRM2 protein immunohistochemical staining in tumors collected from mice 24 hours after treatment with CALAA-01 compared to the siCON1-treated controls (Figure 4B). In order to quantify the amounts of RRM2 mRNA and protein expression, real time qRT-PCR and Western blotting analysis were employed (Figure 4,C and D). Both RRM2 mRNA and protein levels were decreased in tumors from animals treated with the CALAA-01 relative to the siCON1 nanoparticle at all time points tested – up to 10 days post treatment (Figure 4B–D). We have previously observed a similar level of mRNA knockdown in Ewing sarcoma tumors following delivery of siRNA targeting EWS-FLI1. We observed anti-tumor effects in that study also, suggesting that ~50% mRNA reduction is sufficient for anti-tumor effects in mice [35]. In patients we have observed up to 75% knockdown of RRM2 mRNA (although the patients received several doses of siRNA nanoparticles [7]).
Fig. 4.
Time course of nanoparticle intratumoral residency and RRM2 expression levels in Tu212 tumors of mice treated with single intravenous dose of 5% dextrose (D5W), control siRNA-containing CDP/nanoparticle (siCON1 10mg/kg) or siR2-containing CDP/nanoparticles (CALAA-01 10mg/kg). (A) Detection of CDP/siRNA nanoparticles with Au-PEG-AD stain (green) in tumors at 24, 48, and 72 hours after treatment (white arrows point to clusters of CDP/nanoparticles; nuclei stained with DAPI in blue). (B) Immunohistochemical analysis of RRM2 protein expression (orange) 24 hours after treatment with siCON1- or CALAA-01 nanoparticles counterstained with DAPI (blue). (C) Real time qRT-PCR analysis of tumor RRM2 mRNA expression over time (siCON1 24 hr, CALAA-01 24, 48, 72, 96, 168, 240 hr) (error bars are mean +SD, n=3). (D) Western blot analysis of tumor RRM2 protein expression over time (error bars are mean +SD from 3 independent blots).
A single intravenous injection of targeted nanoparticles can remain intact in a tumor for 3 days, leading to target gene knockdown for at least 10 days but for significant knockdown of RRM2 and tumor growth inhibition, multidosing of CALAA-01 might be required.
3.5. Efficacy of CALAA-01 in tumor growth reduction in a Tu212 xenograft model
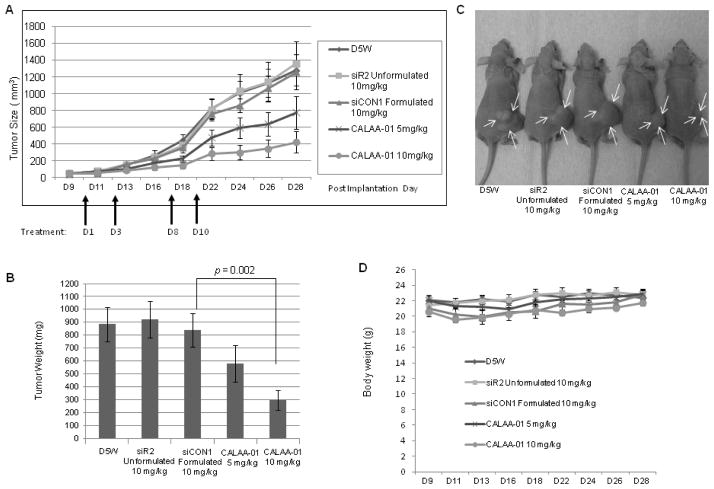
To investigate the anti-cancer therapeutic potential of CALAA-01, we established a Tu212 xenograft in vivo model. CALAA-01 (siR2 5mg/kg and 10mg/kg), nanoparticle siCON1, naked siR2 or D5W were injected via the tail vein in a four-dose schedule (Days 1, 3, 8 and 10) as previously described [7, 21].
We observed promising tumor growth inhibition in the CALAA-01 group. Over 28 days, tumor growth was reduced by an average 2.9-fold in the CALAA-01-treated group (10 mg/kg) compared to the nanoparticle-siCON1 group (10 mg/kg; p = 0.0004) (Figure 5A). Statistical analysis between groups is shown in the figure legend. Mice were sacrificed on day 28 and tumors were collected. Tumor weight was significantly reduced in the CALAA-01 (10 mg/kg) compared to siCON1 (10 mg/kg) group (p=0.002) (Figure 5B). Representative mice from each treatment group are shown in Figure 5C. No significant changes in body weight were observed throughout the study (Figure 5D). General health and behavior of the treated mice were also monitored and found to be suitable. We began with 8 mice per group but lost one mouse accidentally in the siCON1 group.
Fig. 5.
Systemically delivered CALAA-01 in a four-dose schedule (days 1, 3, 8 and 10) efficiently reduced tumor growth in a Tu212 xenograft model. (A) Tumor growth (tumor volume = 0.5×l×w2) progression in five groups: D5W, naked siR2 (10 mg/kg), siCON1 (formulated 10 mg/kg) and siR2-containing nanoparticle (CALAA-01 5 mg/kg and 10 mg/kg). First dose started on Day 10 after implantation and tumor growth was monitored until Day 28 (endpoint of tumor volume 1500 mm3) (error bars are mean ±SE, n=8). Statistical analysis (all days) between groups was as follows: D5W vs CALAA-01 10 mg/kg: P=0.0002; siR2 Unformulated vs CALAA-01 10 mg/kg: P=0.0007; siCON1 vs CALAA-01 10 mg/kg: P=0.0004; D5W vs CALAA-01 5 mg/kg: P=0.03; siR2 Unformulated vs CALAA-01 5 mg/kg: P=0.04; siCON1 vs CALAA-01 5 mg/kg: P=0.09. (B) Mice were sacrificed, tumors were collected from each treatment group on Day 28 and tumors were weighed (error bars are mean ±SD, n=8). Statistical analysis between groups was as follows: D5W vs CALAA-01 10 mg/kg: P=0.0019; siR2 Unformulated vs CALAA-01 10 mg/kg: P=0.0016; siCON1 vs CALAA-01 10 mg/kg: P=0.0024. (C) Representative mouse from each group of D5W, naked siR2, siCON1, CALAA-01 5 mg/kg and 10 mg/kg. Arrows indicated tumor area. (D) Body weights throughout study in each treated group (error bars are mean ± SD, n=8).
To ascertain whether the RRM2 protein level was reduced in the tumor tissues, we measured relative RR activity and protein expression. Relative RR activity in the tumor tissue of each mouse from every group is shown in Figure 6A. We found CALAA-01 (10mg/kg) significantly reduced RRM2 activity in xenograft tumors by an average 1.88-fold compared to the siCON1 group (p=0.03) (Figure 6A). The tumor completely disappeared in one mouse from the CALAA-01 (10mg/kg) group, which was considered to have zero RR activity. We observed low RR activity in couple of mice with large tumors in the control groups, especially in the naked siRNA and D5W groups. Tumor progression was very fast in these mice and the tumors reached a large size. Ulceration in the tumor and necrotic tissues were detected in these mice, which may have affected the RR activity. To detect intracellular localization and expression levels of RRM2, we analyzed mice tumor tissues by immunohistochemistry. Significant reduction in RRM2 protein expression was observed after treatment of CALAA-01 (10mg/kg) as evidenced by tumor tissue staining with specific antibody (Figure 6B, middle panel). The nanoparticle CALAA-01 (10mg/kg)-delivered RRM2 siRNA significantly suppressed cell proliferation and induced apoptosis as evidenced by xenograft tumor tissue staining for Ki67 (Figure 6B, lower panel) and terminal deoxyribonucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) (Figure 6C), respectively. Our results suggest that siRNA delivered by nanoparticles significantly reduced the expression and activity of RRM2 expression in tumors, which facilitated inhibition of cell proliferation and induction of apoptosis followed by suppression of tumor growth progression.
Fig. 6.
RRM2 expression and cell proliferation were reduced followed by induction of apoptosis upon four doses of CALAA-01 in the Tu212 xenograft model. We used xenograft tumor and tumor tissue sections which were collected on Day 28 of the study, meaning 10 days after the last injection and during the time of sacrifice. The dosing schedule is illustrated in Figure 5A. (A) Relative RR activity in tumor tissues of each mouse from each group was plotted graphically (error bars are mean ±SD, n=8, siCON1 vs CALAA-01 10 mg/kg: P=0.03 and siR2 unformulated vs CALAA-01 10 mg/kg: P=0.01). (B) RRM2 and Ki67 expression detected in xenograft tissue by IHC analysis. Representative images shown from siCON1 and CALAA-01 10 mg/kg groups (brown stain for RRM2, and Ki67 and nuclei were counterstained by hematoxylin, blue; magnification X200). (C) Apoptosis was detected by xenograft tumor tissue staining with TUNEL (green) and nuclei (blue), Magnification X200.
4. Discussion
We describe findings that are highly significant and relevant in the context of RRM2-mediated tumor progression and provide vital clues for the development of novel cancer therapies. The role of RRM2 in HNSCC and NSCLC was explored, which could have profound impact on both understanding and targeting these diseases. Our results suggest that RRM2 plays a critical role in cell growth inhibition in both HNSCC and NSCLC cells. We have demonstrated the therapeutic potential of nanoparticle-formulated siRNA targeting RRM2 in HNSCC. We determined that knockdown of RRM2 via an RNAi mechanism leads to cell growth inhibition in HNSCC and NSCLC cells.
CALAA-01 is the first nanoparticle to demonstrate dose-dependent accumulation in tumors after systemic injection into patients, and to deliver siRNAs that turn off a desired gene using the RNAi mechanism [7]. Targeted delivery has yet to be optimized for most cell types and tissues. We further extended the evaluation of CALAA-01 in the present work to study its intratumoral kinetics and mechanism of action in an HNSCC model. We and many others have demonstrated that the mouse transferrin receptor will recognize human transferrin ligand and can facilitate the internalization of nanoparticles into mouse cells [36]. Transferrin was originally chosen as a targeting ligand because of this cross-reactivity across species. Therefore, we do not expect to see favorable uptake by human tissue in a mouse xenograft model. We ascertained that the siRNA delivered by single dose systemic administration of CALAA-01 in an in vivo mouse model can enable knockdown of mRNA and protein that lasts at least 10 days. We observed a consistent but not statistically significant decrease in RRM2 protein and RNA levels compared to siCON1-treated samples at all time points. Consequently, we performed another in vivo experiment in which CALAA-01 (10mg/kg) significantly reduced RRM2 activity and expression in a four-dose schedule.
To our knowledge, this is the first animal study in head and neck cancer showing effective systemic delivery of siRNA to produce specific gene inhibition. Previously, multiple doses of siRNAs delivered via CALAA-01 have been shown to achieve gene inhibition for lengthy time spans (ca. several weeks) in a human [7]. Intratumoral kinetics of CALAA-01 such as siRNA stability, functionality and tumor growth dynamics have been addressed in our study. The PK behavior of these siRNA nanoparticles has been described previously [27]. We have demonstrated co-localization of fluorescent siRNA and nanoparticle staining in tumors from mice [7]. In this study we ascertained how long siRNA nanoparticles would be present in a tumor following a single dose. We demonstrated that the nanoparticles could only be detected in the tumor sections up to 72 hours following a single dose. This is a new observation for this delivery system and supports prior observations that dosing every 3 days is sufficient for maximal therapeutic effect [37]. Although Tf-targeted nanoparticles can deliver functional siRNA to the subcutaneous Tu212 tumors, their heterogeneous intratumoral distribution may limit siRNA delivery to all the tumor cells. Therefore, multiple doses might be important to reach new cells that have either not internalized any siRNA or have not internalized sufficient siRNA required for the phenotypic effect, such as cell death. A four-dose schedule of CALAA-01 significantly reduced the expression and activity of RRM2, which facilitated inhibition of cell proliferation and induction of apoptosis followed by inhibition of tumor growth progression. Throughout our study, we did not observe any adverse effects or any body weight changes. siRNA nanoparticles with transferrin targeting ligands do not cause gene knockdown in the liver. A hepatocyte specific targeting ligand is required to achieve measureable knockdown in the liver. No physiologic signs of toxicity were observed in any healthy tissue when the siRNA delivery system was delivered to non-human primates [31]. These results emphasize key features of CALAA-01, including its tumor specificity for cytotoxic targets and its duration of therapeutic effect for siRNA-based treatment for cancer therapy.
5. Conclusions
Our study offers promise in siRNA-based therapy for HNSCC and possibly NSCLC. RRM2-specific siRNA silences RRM2 by an RNAi mechanism and strongly inhibits cell growth in HNSCC and NSCLC cell lines. We have demonstrated the therapeutic potential of CALAA-01 in HNSCC. In a mouse xenograft model, a single intravenous injection shows intact nanoparticles in the tumor sections up to 72 hours, leading to target gene knockdown lasting at least 10 days. A four-dose schedule of CALAA-01 delivering siRNA targeted to HNSCC tumors significantly reduced tumor progression by suppressing cell proliferation and by induction of apoptosis without any potential toxicity issues. These findings signify the efficacy of the CALAA-01 delivery system against HNSCC tumors in mice and highlight RRM2 as an excellent anti-cancer target. Further investigation of the applicability of the delivery system in other cancer types is highly encouraged.
Acknowledgments
This work was supported by NIH/NCI grants U01CA151802 and U54CA119347, P50 CA128613 (Head and Neck Cancer SPORE) and, P30 CA138292. We thank Dr. Anthea Hammond for her critical and editorial review of this article.
Footnotes
Dr. Davis has founders stock in Calando Pharmaceuticals and there is no financial conflict of interest to other authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeshita F, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689–696. doi: 10.1111/j.1349-7006.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis ME, Chen ZG, Shin DM. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat Rev Drug Discov. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett DW, Davis ME. Physicochemical and biological characterization of targeted, nucleic acid-containing nanoparticles. Bioconjug Chem. 2007;18:456–468. doi: 10.1021/bc0603539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engstrom Y, Eriksson S, Jildevik I, Skog S, Thelander L, Tribukait B. Cell cycle-dependent expression of mammalian ribonucleotide reductase. Differential regulation of the two subunits. J Biol Chem. 1985;260:9114–9116. [PubMed] [Google Scholar]
- 10.Eriksson S, Martin DW., Jr Ribonucleotide reductase in cultured mouse lymphoma cells. Cell cycle-dependent variation in the activity of subunit protein M2. J Biol Chem. 1981;256:9436–9440. [PubMed] [Google Scholar]
- 11.Cerqueira NM, Pereira S, Fernandes PA, Ramos MJ. Overview of ribonucleotide reductase inhibitors: an appealing target in anti-tumour therapy. Curr Med Chem. 2005;12:1283–1294. doi: 10.2174/0929867054020981. [DOI] [PubMed] [Google Scholar]
- 12.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. RNA interference targeting the M2 subunit of ribonucleotide reductase enhances pancreatic adenocarcinoma chemosensitivity to gemcitabine. Oncogene. 2004;23:1539–1548. doi: 10.1038/sj.onc.1207272. [DOI] [PubMed] [Google Scholar]
- 13.Duxbury MS, Ito H, Benoit E, Zinner MJ, Ashley SW, Whang EE. Retrovirally mediated RNA interference targeting the M2 subunit of ribonucleotide reductase: A novel therapeutic strategy in pancreatic cancer. Surgery. 2004;136:261–269. doi: 10.1016/j.surg.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 14.Zhang YW, Jones TL, Martin SE, Caplen NJ, Pommier Y. Implication of checkpoint kinase-dependent up-regulation of ribonucleotide reductase R2 in DNA damage response. J Biol Chem. 2009;284:18085–18095. doi: 10.1074/jbc.M109.003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuta E, Okuda H, Kobayashi A, Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta. 2010;1805:141–152. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou BS, Tsai P, Ker R, Tsai J, Ho R, Yu J, Shih J, Yen Y. Overexpression of transfected human ribonucleotide reductase M2 subunit in human cancer cells enhances their invasive potential. Clin Exp Metastasis. 1998;16:43–49. doi: 10.1023/a:1006559901771. [DOI] [PubMed] [Google Scholar]
- 17.Zhang K, Hu S, Wu J, Chen L, Lu J, Wang X, Liu X, Zhou B, Yen Y. Overexpression of RRM2 decreases thrombspondin-1 and increases VEGF production in human cancer cells in vitro and in vivo: implication of RRM2 in angiogenesis. Mol Cancer. 2009;8:11. doi: 10.1186/1476-4598-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duxbury MS, Whang EE. RRM2 induces NF-kappaB-dependent MMP-9 activation and enhances cellular invasiveness. Biochem Biophys Res Commun. 2007;354:190–196. doi: 10.1016/j.bbrc.2006.12.177. [DOI] [PubMed] [Google Scholar]
- 19.Souglakos J, Boukovinas I, Taron M, Mendez P, Mavroudis D, Tripaki M, Hatzidaki D, Koutsopoulos A, Stathopoulos E, Georgoulias V, Rosell R. Ribonucleotide reductase subunits M1 and M2 mRNA expression levels and clinical outcome of lung adenocarcinoma patients treated with docetaxel/gemcitabine. Br J Cancer. 2008;98:1710–1715. doi: 10.1038/sj.bjc.6604344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morikawa T, Maeda D, Kume H, Homma Y, Fukayama M. Ribonucleotide reductase M2 subunit is a novel diagnostic marker and a potential therapeutic target in bladder cancer. Histopathology. 2010;57:885–892. doi: 10.1111/j.1365-2559.2010.03725.x. [DOI] [PubMed] [Google Scholar]
- 21.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–668. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 22.Gatter KC, Brown G, Trowbridge IS, Woolston RE, Mason DY. Transferrin receptors in human tissues: their distribution and possible clinical relevance. J Clin Pathol. 1983;36:539–545. doi: 10.1136/jcp.36.5.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao HQ, Roy K, Troung-Le VL, Janes KA, Lin KY, Wang Y, August JT, Leong KW. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release. 2001;70:399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- 24.Kircheis R, Blessing T, Brunner S, Wightman L, Wagner E. Tumor targeting with surface-shielded ligand--polycation DNA complexes. J Control Release. 2001;72:165–170. doi: 10.1016/s0168-3659(01)00272-3. [DOI] [PubMed] [Google Scholar]
- 25.Dash PR, Read ML, Fisher KD, Howard KA, Wolfert M, Oupicky D, Subr V, Strohalm J, Ulbrich K, Seymour LW. Decreased binding to proteins and cells of polymeric gene delivery vectors surface modified with a multivalent hydrophilic polymer and retargeting through attachment of transferrin. J Biol Chem. 2000;275:3793–3802. doi: 10.1074/jbc.275.6.3793. [DOI] [PubMed] [Google Scholar]
- 26.Bellocq NC, Pun SH, Jensen GS, Davis ME. Transferrin-containing, cyclodextrin polymer-based particles for tumor-targeted gene delivery. Bioconjug Chem. 2003;14:1122–1132. doi: 10.1021/bc034125f. [DOI] [PubMed] [Google Scholar]
- 27.Bartlett DW, Su H, Hildebrandt IJ, Weber WA, Davis ME. Impact of tumor-specific targeting on the biodistribution and efficacy of siRNA nanoparticles measured by multimodality in vivo imaging. Proc Natl Acad Sci U S A. 2007;104:15549–15554. doi: 10.1073/pnas.0707461104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Su L, Pirani AA, Wu H, Zhang H, Shin DM, Gernert KM, Chen ZG. Understanding metastatic SCCHN cells from unique genotypes to phenotypes with the aid of an animal model and DNA microarray analysis. Clin Exp Metastasis. 2006;23:209–222. doi: 10.1007/s10585-006-9031-0. [DOI] [PubMed] [Google Scholar]
- 30.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 31.Heidel JD, Yu Z, Liu JY, Rele SM, Liang Y, Zeidan RK, Kornbrust DJ, Davis ME. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc Natl Acad Sci U S A. 2007;104:5715–5721. doi: 10.1073/pnas.0701458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Juhasz A, Frankel P, Cheng C, Rivera H, Vishwanath R, Chiu A, Margolin K, Yen Y, Newman EM, Synold T, Wilczynski S, Lenz HJ, Gandara D, Albain KS, Longmate J, Doroshow JH. Quantification of chemotherapeutic target gene mRNA expression in human breast cancer biopsies: comparison of real-time reverse transcription-PCR vs. relative quantification reverse transcription-PCR utilizing DNA sequencer analysis of PCR products. J Clin Lab Anal. 2003;17:184–194. doi: 10.1002/jcla.10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou BS, Ker R, Ho R, Yu J, Zhao YR, Shih J, Yen Y. Determination of deoxyribonucleoside triphosphate pool sizes in ribonucleotide reductase cDNA transfected human KB cells. Biochem Pharmacol. 1998;55:1657–1665. doi: 10.1016/s0006-2952(98)00042-2. [DOI] [PubMed] [Google Scholar]
- 34.Heidel JD, Liu JY, Yen Y, Zhou B, Heale BS, Rossi JJ, Bartlett DW, Davis ME. Potent siRNA inhibitors of ribonucleotide reductase subunit RRM2 reduce cell proliferation in vitro and in vivo. Clin Cancer Res. 2007;13:2207–2215. doi: 10.1158/1078-0432.CCR-06-2218. [DOI] [PubMed] [Google Scholar]
- 35.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing’s sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 36.Choi CH, Alabi CA, Webster P, Davis ME. Mechanism of active targeting in solid tumors with transferrin-containing gold nanoparticles. Proc Natl Acad Sci U S A. 2010;107:1235–1240. doi: 10.1073/pnas.0914140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartlett DW, Davis ME. Impact of tumor-specific targeting and dosing schedule on tumor growth inhibition after intravenous administration of siRNA-containing nanoparticles. Biotechnol Bioeng. 2008;99:975–985. doi: 10.1002/bit.21668. [DOI] [PubMed] [Google Scholar]