Abstract
The role of interdomain linkers in modular polyketide synthases is poorly understood. Analysis of the 6-deoxyerythronolide B synthase (DEBS) has yielded a model in which chain elongation is governed by interactions between the acyl carrier protein domain and the ketosynthase domain plus an adjacent linker. Alanine scanning mutagenesis of the conserved residues of this linker in DEBS module 3 led to the identification of the R513A mutant with a markedly reduced rate of chain elongation. Limited proteolysis supported a structural role for this Arg residue. Our findings highlight the importance of domain-linker interactions in assembly line polyketide biosynthesis.
Multimodular polyketide synthases (PKSs) are modular enzymatic assembly line proteins that catalyze the biosynthesis of numerous polyketide antibiotics1. Each catalytic module minimally consists of a ketosynthase (KS), an acyltransferase (AT), and an acyl carrier protein (ACP) domain. The KS domain receives the growing polyketide chain from the ACP domain of the previous module, while the AT domain transfers an α-carboxyacyl extender unit onto the phosphopantetheine arm of the ACP domain. The KS then catalyzes decarboxylative condensation between the growing chain and the extender unit, leading to the formation of a β-ketoacyl thioester-ACP intermediate (chain elongation reaction). Following varying degrees of β-carbon modification by auxiliary enzymes in the module, this intermediate is eventually translocated to the KS domain of the next module. Alternatively, chain growth is terminated by a thioesterase (TE) domain. Understanding the mechanism for this orderly progress of the growing polyketide chain represents a fundamental challenge in assembly line enzymology.
The 6-deoxyerythronolide B synthase (DEBS) from Saccharopolyspora erythraea is perhaps the most well studied multimodular PKS2,3. Its ACP domains show marked specificity towards their cognate KS partners during chain elongation as well as intermodular chain translocation4,5. We have recently demonstrated that the specificity of these two catalytic reactions is controlled by distinct protein-protein interfaces6–8. Whereas the ACP recognition elements for both reactions were mapped in considerable detail, less is known about their precise sites of interaction with the KS-AT partner protein. For example, it was proposed that, during chain elongation, the ACP domain interacts with the KS domain as well as the linker connecting the KS and the AT domains (KS-AT linker)6. Here we have subjected our model to further interrogation, using DEBS module 3 harboring an appended TE domain (M3+TE) as a test case (Figure 1). The advantages of DEBS module 3 are two-fold. First, it lacks any apparent active auxiliary enzyme, and is therefore a bona fide “minimal” PKS module. Second, the majority of this 367 kD homodimer has been structurally characterized9.
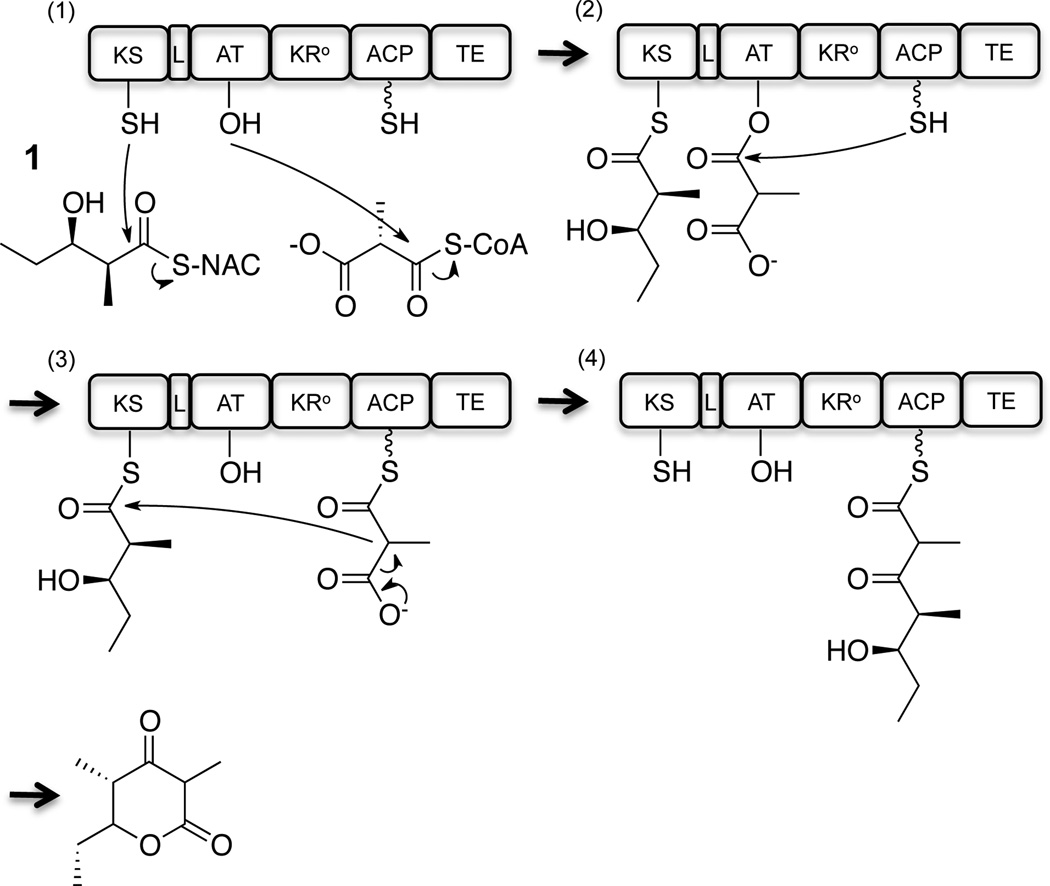
Figure 1.
Chain elongation cycle catalyzed by M3+TE. The module consists of a KS, a KS-AT linker (L), an AT, an inactive KR (KR°), and an ACP domain. In the absence of an upstream module, the KS can be primed by a synthetic mimic of an ACP-bound diketide, 1, while the AT is acylated with a methylmalonyl extender unit (1), which is then transferred to the ACP domain (2). Decarboxylative condensation occurs (3), whereafter the extended polyketide chain is cyclized by the TE domain to generate a triketide lactone (4). The phosphopantetheine prosthetic arm is shown as a wavy line.
The KS-AT linkers in individual modules of DEBS are approximately 100 residues long, and consist of a three-stranded β-sheet packed against two α-helices on one side, representing a unique fold in the Protein Data Bank10. Guided by the assumption that mechanistically important residues in these linkers were also evolutionarily conserved, we compared the sequences of the KS-AT linkers of DEBS with those of other multimodular PKSs (Supplemental Figure 1). Based on the resulting alignment, we selected the most conserved Pro-473, Val-475, Val-476, Ser-477, Arg-479, Leu-484, Gln-487, Ile-491, Leu-509, Arg-513, His-516, His-518, Arg-519, Leu-534, and Ile-537 (numbering based on PDB ID: 2QO3), and constructed the corresponding alanine mutants in DEBS module 3+TE. All of the mutants were expressed as soluble proteins, and purified at yields of 15–20 mg/L. The rate of triketide lactone formation by each mutant protein was measured at 23 °C and 30 °C (Table 1). Most mutants showed initial rates comparable to the wild-type enzyme (0.1 min−1). The R513A mutant, however, showed a 7-fold decrease in activity (0.014 min−1) at 23 °C. Its rate was even more severely attenuated (> 20 fold relative to the wild-type enzyme) at 30 °C. To rule out gross protein structural changes, we determined the rates of both KS- and AT-catalyzed self-acylation in the R513A mutant. Both rates were comparable to the corresponding reaction rates of the wild-type enzyme. Moreover, both acylation reactions (0.5 min−1 and 1 min−1, respectively) were substantially faster than the overall M3+TE turnover rate (Supplemental Figure 2). These results support a model in which chain elongation is the rate-limiting step in the net turnover rate of M3+TE and that interactions between the ACP and the KS-AT linker play a critical role in this rate-limiting step.
Table 1.
Relative rates of triketide lactone formation of M3+TE mutants
| Protein | Relative to M3+TE at 23 °C | Relative to M3+TE at 30 °C |
|---|---|---|
| M3+TE | 1.00 | 1.00a |
| P473A | 0.95 | 0.93 |
| V475A | 0.96 | 0.88 |
| V476A | 1.03 | 0.94 |
| S477A | 1.06 | 0.85 |
| R479A | 0.83 | 0.68 |
| L484A | 1.16 | 1.14 |
| Q487A | 1.33 | 1.16 |
| I491A | 1.03 | 0.68 |
| L509A | 0.93 | 0.67 |
| R513A | 0.15 | <0.05 |
| H516A | 0.87 | 0.80 |
| H518A | 0.90 | 0.98 |
| R519A | 0.82 | 1.01 |
| L534A | 0.75 | 0.82 |
| I537A | 0.79 | 1.02 |
For each mutant, the initial rates were measured and compared with that of the wild-type enzyme.
Wild-type M3+TE was 3-fold less active at 30 °C than at 23 °C.
Inverse temperature dependence is a widely recognized property of systems in which hydrophobic interactions are dominant, and is thought to be associated with a diminished amount of structured water around a nonpolar surface with increasing temperature, along with a vibrational contribution that increases in magnitude with increasing temperature11. Formation of triketide lactone by the wild-type M3+TE has a turnover number of 0.1 min−1 at 23 °C versus 0.033 min−1 at 30 °C (Table 1 and Supplemental Figure 3). This observation suggests that hydrophobic interactions between the KS-AT didomain and the ACP may dominate during chain elongation.
To investigate the effect of the R513A mutation on intermodular chain translocation, we employed a back-transfer assay in which the KS domain was first completely acylated with 14C-labeled 1, whereafter the rate of back-transfer of the acyl chain to a stand-alone ACP2 protein was monitored12. In contrast to the rate of intramodular chain elongation, the R513A mutant displayed an intermodular back-transfer rate comparable to that of wild type M3+TE (Supplemental Figure 4). None of the other mutants were perturbed in their ability to catalyze this reaction (Supplemental Figure 4). The marked difference between the effects of the R513A mutant on chain elongation versus chain translocation supports a model in which two ACP domains engage the same KS differently in the course of the two reactions6,8.
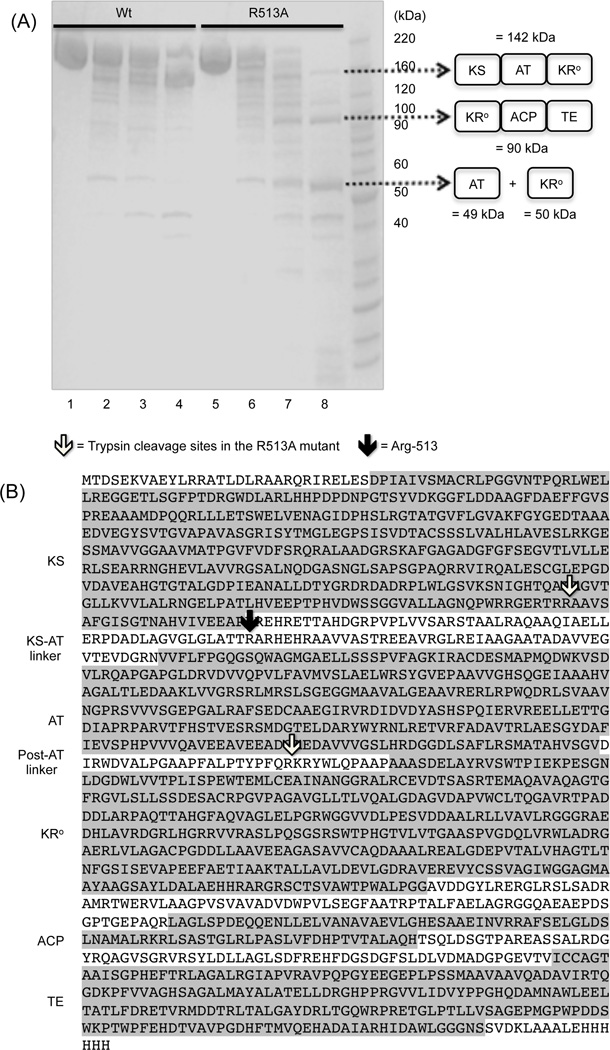
To further probe the role of the R513A mutation, wild-type and mutant M3+TE were subjected to limited proteolysis under identical conditions, and the reaction products were analyzed by SDS-PAGE. As seen in Figure 2A, the major product of prolonged proteolysis of the wild-type protein at given conditions was the 142 kDa KS-AT-KR fragment. In contrast, the corresponding fragment of the R513A mutant was susceptible to further proteolysis, suggesting that the mutant had a more flexible structure than the wild-type protein. N-terminal sequencing of the two most abundant 50–100 kDa proteolytic fragments revealed the presence of two new proteolytically susceptible sites in the R513A mutant that were absent in the wild-type protein13; these sites were mapped at near the C-terminal end of the KS domain and in the post-AT linker (see Figure 2B for proteolytic sites and definition of domain boundaries).
Figure 2.
M3+TE and the R513A mutant were partially digested with 1 µg/ml trypsin at 30°C. (A) Time course of limited-trypsin digestion was monitored by SDS-PAGE. Lane 1–4: M3+TE (no digestion, 4 min, 16 min, 64 min) lane 5–8: R513A mutant (no digestion, 4 min, 16 min, 64 min). Domain composition of the proteolytic fragments was inferred from the observed molecular mass and N-terminal sequencing. (B) Sequence of M3+TE annotated with domain and linker boundaries. Proteolyzed fragments between 50–60 kDa and 90–100 kDa in the R513A mutant, which are indicated by black dashed arrows in Figure 2A, were extracted, and their N-termini were identified by Edman degradation. Trypsin cleavage sites are indicated by white arrows. Black arrow indicates the Arg-513 residue.
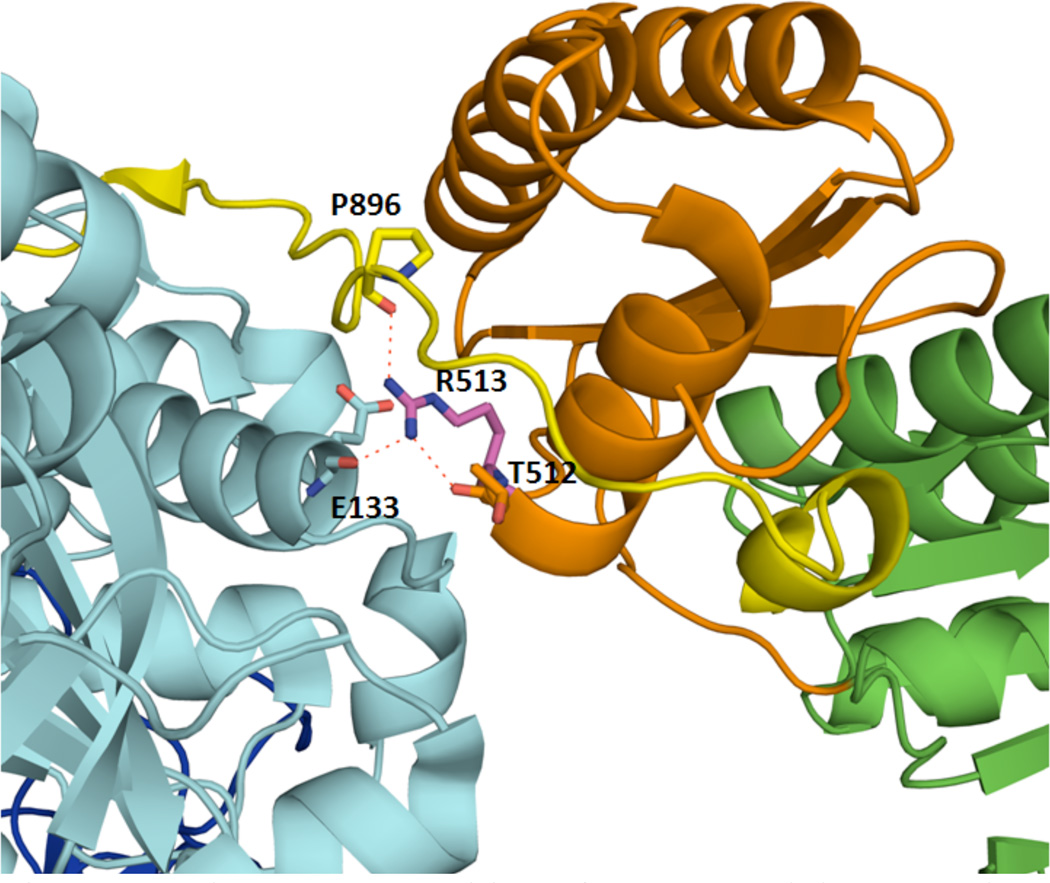
Analysis of the KS-AT structure suggests a possible explanation for these unexpected findings from limited proteolysis. As shown in Figure 3, the side chain of Arg-513 in the KS-AT linker of DEBS module 3 forms hydrogen bonds with the backbone amide groups of Glu-133 in the KS domain, Thr-512 in the KS-AT linker, and Pro-896 located in the post AT linker9. Thus, the reduced turnover rate of the R513A mutant is not due to a direct role of the Arg-513 in interaction with the ACP domain, but instead is most likely due to the important role of this residue in stabilizing the demonstrated compact quaternary structure of the KS-AT didomain, which in turn shapes the ACP docking site during chain elongation. Pro-896 in particular is located immediately upstream of the YPFQRKRYW sequence (residues 898–906 in the post-AT linker), which is highly conserved in multimodular PKSs14,15. This sequence interacts with the side-chains of Glu-126, Phe-106, Phe107, and Trp129 in the KS domain9. Interestingly, the YPFQRKRYW site underwent internal proteolysis in the R513A mutant, whereas trypsin cleaved the wild-type enzyme only after the sequence13. This suggests that R513 stabilizes the compact structure of this sequence. We also note that the post-AT linker has been shown to be required for triketide lactone synthesis by a combination of stand-alone KS3 protein and a stand-alone AT3 protein16. Together, these findings lend further support to our model for ACP/KS-AT interaction during chain elongation.
Figure 3.
The Arg-513 residue of DEBS module 3 (numbering based on PDB ID: 2QO3). Arg-513 is located at the boundary between the KS domain (light blue) and the KS-AT linker (orange). Its side-chain forms hydrogen bonds with the backbone amide groups of Glu-133 in the KS domain, Thr-512 in the KS-AT linker, and Pro-896 in the post-AT linker (yellow). The AT domain is shown in green.
In summary, the structural basis for protein-protein interactions is crucial to our understanding of the assembly line mechanism of multimodular PKSs and to the design of hybrid assembly lines. In this study, we have focused on the role of the relatively unconserved KS-AT linker. Our findings provide the first insights into the structural and the mechanistic roles of this linker in polyketide chain elongation.
Supplementary Material
ACKNOWLEDGMENT
This research was funded by grants from the NIH (GM 87934 to C.K. and GM 22172 to D.E.C.), by a Naito Foundation Fellowship to S.Y., and a Stanford Graduate Fellowship to S.K.
Footnotes
ASSOCIATED CONTENT
Supporting Information
This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Hertweck C. Angew. Chem. Int. Ed. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 2.Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Annu. Rev. Biochem. 2007;76:195–221. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 3.Cane DE. J. Biol. Chem. 2010;285:27517–27523. doi: 10.1074/jbc.R110.144618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu N, Cane DE, Khosla C. Biochemistry. 2002;41:5056–5066. doi: 10.1021/bi012086u. [DOI] [PubMed] [Google Scholar]
- 5.Chen AY, Schnarr NA, Kim CY, Cane DE, Khosla C. J. Am. Chem. Soc. 2006;128:3067–3074. doi: 10.1021/ja058093d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapur S, Chen AY, Cane DE, Khosla C. Proc. Natl. Acad. Sci. USA. 2010;107:22066–22071. doi: 10.1073/pnas.1014081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charkoudian LK, Liu CW, Capone S, Kapur S, Cane DE, Togni A, Seebach D, Khosla C. Protein Sci. 2011;20:1244–1255. doi: 10.1002/pro.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapur S, Lowry B, Yuzawa S, Kenthirapalan S, Chen AY, Cane DE, Khosla C. Proc. Natl. Acad. Sci. USA. 2012;109:4110–4115. doi: 10.1073/pnas.1118734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. Chem. Biol. 2007;14:931–943. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y, Kim CY, Mathews, Cane DE, Khosla C. Proc. Natl. Acad. Sci. USA. 2006;103:11124–11129. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sturtevant JM. Proc. Natl. Acad. Sci. USA. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu N, Tsuji SY, Cane DE, Khosla C. J. Am. Chem. Soc. 2001;123:6465–6474. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- 13.Kim CY, Alekseyev VY, Chen AY, Tang Y, Cane DE, Khosla C. Biochemistry. 2004;43:13892–13898. doi: 10.1021/bi048418n. [DOI] [PubMed] [Google Scholar]
- 14.Jenke-Kodama H, Borner T, Dittmann E. PLoS Comput. Biol. 2006;2:e132. doi: 10.1371/journal.pcbi.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridley CP, Lee HY, Khosla C. Proc. Natl. Acad. Sci. USA. 2008;105:4595–4600. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen AY, Cane DE, Khosla C. Chem. Biol. 2007;14:784–792. doi: 10.1016/j.chembiol.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.