Abstract
Noise caused by stochastic fluctuations in genetic circuits (transcription and translation) is now appreciated as a central aspect of cell function and phenotypic behavior. Noise has also been detected in signaling networks, but the origin of this noise and how it shapes cellular outcomes remain poorly understood. Here, we argue that noise in signaling networks results from the intrinsic promiscuity of protein-protein interactions, and that this noise has shaped cellular signal transduction. Features promoted by the presence of this molecular signaling noise include multimerization and clustering of signaling components, pleiotropic effects of gross changes in protein concentration, and a probabilistic rather than linear view of signal propagation.
Keywords: interactome, SH2 domain, SH3 domain, probability in signaling, evolution, receptor tyrosine kinase, specificity
Noise in biological systems
Noise is increasingly appreciated as a force shaping biology. Recent research has revealed genetic circuits that are subject to stochastic fluctuations, or noise, at the [sta1] level of their components. By stochastically influencing state-switching systems, especially those based on positive feedback loops, genetic noise can cause substantial variations in expression of many genes. Thus, genetic noise can cause genetically identical cells to behave differently 1, 2. This noise-driven genetic regulation allows cell state choice to be probabilistic and can cause phenomena such as resistance to antibiotics or anticancer drugs 3, 4. Genetic noise has therefore emerged as a central factor in how biological systems function and evolve 3, 5.
The observation that noise in transcriptional regulation shapes biological systems raises the question of whether other physiological processes are also subject to the effects of noise. Recently it has been shown that the information transduced in cellular signaling pathways is significantly limited by noise 6, 7. The molecular basis of this noise and how it shapes cellular pathways is poorly understood 7. Based on the accumulated data on kinase-mediated signaling, we here propose that noise is generated by the interconnected and promiscuous nature of the protein-protein interactions (PPIs) that are required to transduce the signals. We further propose that this noise has fundamentally shaped kinase-mediated signaling and possibly cellular signal transduction in general.
Promiscuity in PPI networks
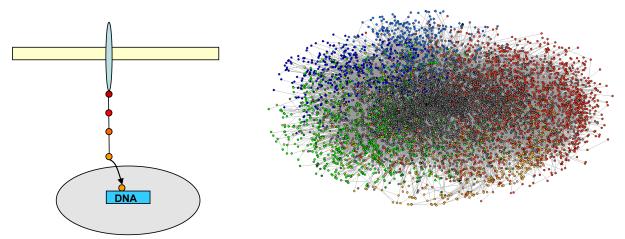
If signaling cascades are assumed to be linear relays of events, then each signal has to be transmitted through several mutually exclusive bimolecular PPIs from its origin (e.g., a plasma membrane-localized receptor tyrosine kinase; RTK) to an effector (e.g., a transcription regulator). The human interactome is believed to contain between 130,000 and 650,000 binary interactions, most of which are currently uncharacterized 8-10(Figure 1, Box 1). Each PPI involved in propagating a specific signal through the cell must therefore compete against the bulk of nonspecific competitors. This is particularly difficult when the specific signals and signal recognition domains are extremely similar to the nonspecific ones, as is the case in kinase-mediated signaling.
Figure 1.
Classical linear signaling versus the high-connectivity interactome. The high number and intricacy of protein-protein interactions revealed by the human interactome 8-10 (Figure taken from http://www.unihi.org/) (right panel) and kinome 12 is in contrast to the traditionally depicted linear signaling cascades, where each component of the pathway interacts with only a few others (left panel), from the membrane-localized receptor (elongated blue oval) to the nucleus (gray oval).
Box 1 Interactomes.
Interactomes are a comprehensive list of protein-protein interactions (for detailed and critical reviews please see 7, 46, 47). Three different approaches have been used to establish interactome networks: (i) compilation and curation of interactions from published literature. These interactions are usually established from one or a few types of physical or biochemical interactions and are therefore not exhaustive. Also in considering data from peer-reviewed material there are no global standards for accuracy, reproducibility and sensitivity. (ii) Computational predictions. These methods are mainly based on using sequence similarities, protein structural information, gene-order conservation, co-presence and co-absence of genes in completely sequenced genomes to transfer the interactions from one organism to another via orthology mapping. These approaches are fast, but imperfect because of use of indirect ‘orthogonal’ information without experimental verification. (iii) Systematic high-throughput experimental mapping strategies applied at the scale of whole genomes or proteomes. Mapping of binary interactions is typically based on variations of the yeast two-hybrid system. Mapping of components of protein complexes is normally carried out by affinity purification followed by some type of mass spectrometry-based protein identification. Due to advances in technology, whole genome interactomes can now be established within a relative short time period. Implementation of statistical tests allows estimating the overall completeness, accuracy and sensitivity of a high-throughput mapping approach. Limitations of experimental large scale protein maps result from the fact that these approaches count all protein-protein interactions above a certain detection threshold (typically with Kd’s above 30-50 μM). Biologically relevant interactions with lower affinity will remain undetected, whereas nonrelevant interactions above the detection limit will be included. Currently, experimental interactome maps are established without taking into account the high degree of spatiotemporal organization within cells. A significant amount of the detected interactions may therefore not be physiologically relevant because the proposed binding partners would not meet in an intact cellular complex.
In kinase-mediated signaling, the initial event is often linked to phosphorylation of a substrate. Investigations of the kinase-phosphatase interaction network revealed a highly intricate and degenerate array of collaborative interactions that “creates[sta2] the conditions for indiscriminate chatter” 11 within and outside of the network 12. In other words, a central step in this type of signal transduction was shown to result in arbitrary nonspecific interactions and (de)phosphorylation events, in stark contrast to an orderly specific and linear signaling cascade. Phosphorylation events are typically recognized by small phosphoresidue binding domains, such as Src homology (SH) 2 or phosphotyrosine binding (PTB) domains 13-16. Signal propagation normally also involves other protein-protein recognition domains, such as SH3 domains. Mammalian cells can express more than 100 different types of proteins containing each of these domains. For each specific interaction, the biologically relevant domain must compete against all the homologous nonspecific domains for similar consensus binding motifs. Thus, from a simplistic point of view where these domain-containing proteins were expressed at equivalent concentrations, for a specific interaction to prevail against all the potential nonspecific competitors, the difference in affinity between the specific and nonspecific binding events would have to be significantly greater than 2 orders of magnitude (i.e. 1 specific interaction has to compete against >100 non-specific interactions)[sta3]. In some cases the differences in affinities would have to be even greater. For example, in the human cell, more than 300 SH3 domains compete not only with one another but also with WW, GYF, UEV, and EVH1 domains for similar proline-rich binding motifs (which are found in almost 25% of all human proteins) 15, 17. Thus, in the context of other competing interactions, likely differences in expression levels and fluctuations in local protein concentrations, to ensure that a specific interaction will occur, it is likely that the affinity of the interaction will have to be at least 3 orders of magnitude greater than those of the background interactions.[sta4] Such a large difference in affinity between the specific interaction and all nonspecific interactions is seldom observed; in fact, the interactions of binding domains with different ligands often show only a limited affinity range and in some cases show significant promiscuity 13-15. This promiscuity can in part be linked to the fact that in many cases the binding sites presented to ligands are highly similar (Figure 2). The limited range in observed binding affinities is also a consequence of the fact that PPIs must be short-lived to allow cells to cease signaling in a timely manner. For this, PPI off-rates need to be fairly short, and since on-rates are limited by diffusion, affinities are necessarily found within a limited range. In many cases, high affinities would result in a complex with too long a lifetime. For example, even an affinity in the half-micromolar range is sufficient to allow the HIV-1 Nef protein to activate the transforming potential of certain Src kinases by binding to their SH3 domains 18.
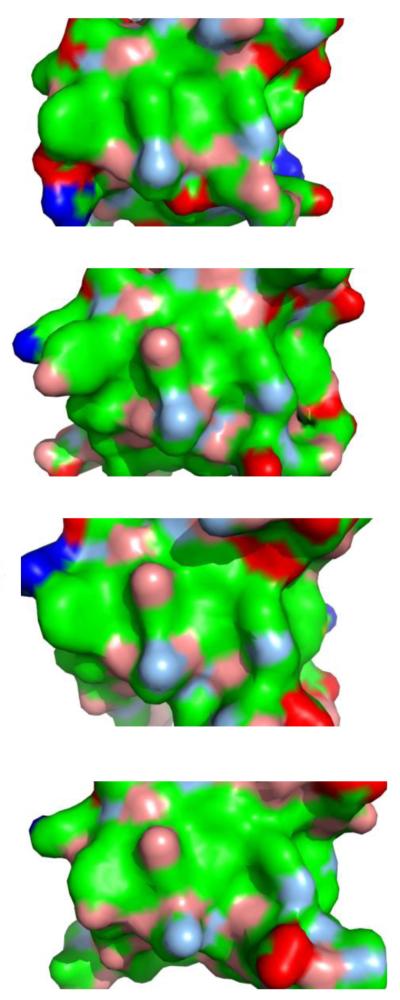
Figure 2.
Ligand binding surfaces of SH3 domains. Despite belonging to different proteins, and despite being implicated in different signaling pathways[sta12], the molecular characteristics of SH3 ligand binding surfaces are highly similar. From top to bottom:[sta13] CIN85 (PDB id 2BZ8), Fyn (1SHF), Grb2 C-terminal (2W0Z), Abl (1BBZ). Surfaces are colored according to blue, positively charged atoms; red, negatively charged atoms; green, hydrophobic atoms; salmon, polar oxygens; marine, polar nitrogens; yellow, sulfur.
The promiscuity in PPIs is further increased by another energetic component. Some proteins exhibit auto-inhibitory mechanisms, which help prevent these proteins from exposing promiscuous binding sites to the wrong partners in the wrong spatiotemporal context. However, the naturally occurring fluctuations between the inactive and active dynamic conformations, even in the inactivated protein 19, 20, are expected to produce opportunities for spurious nonspecific associations and basal enzymatic activity.
Promiscuity in PPIs is also observed in genome-wide interaction mapping.[sta5] For example, large-scale investigations in yeast and higher eukaryotes showed that although some SH3-ligand interactions have evolved a selectivity that allows strict exclusivity, many other SH3 interactions display significant cross-reactivity 21-24. Cellular signaling therefore appears to have evolved to incorporate a degree of promiscuity in the PPIs on which it is based. Such PPI promiscuity is also functional, because it allows the same protein to bind to different interaction partners and hence to participate in a different signaling event. Indeed, many signaling proteins form biologically relevant complexes with multiple distinct partners and need to tolerate some degeneracy in ligand recognition specificity 25.
Compartmentalization is without doubt important for limiting the number of erroneous protein-protein encounters within the cell 25. However, even if the 130,000 to 650,000 possible binary interactions were spread out over 100 different types of compartments, we would expect over 1000 possible interactions per microenvironment. Moreover compartmentalization can not be strict, because many signaling proteins partake in signaling events localized to different compartments, or need to migrate across large distances for function (for examples see 25, 26). Finally, diffusion between adjacent compartments is expected to result in local leaking of components from one microenvironment into the other. Therefore, while certainly required for functional fidelity in signaling, compartmentalization is unlikely to eliminate low-affinity non-specific protein-protein contacts.
Noise arising from promiscuity
Taken together, the intricacy and multivalency of signal transduction, the lack of sufficient intrinsic specificity in many PPIs (both catalytic and noncatalytic) and the fluctuations in auto-inhibition appear to p[sta6]roduce a significant level of signaling promiscuity within the cell. This promiscuity in PPIs is expected to give rise to noise in the form of frequent spurious nonspecific binary associations. This noise is conceptually very different from the noise resulting from stochastic fluctuations of low-abundance genetic regulators. Whereas genetic noise typically causes stochastic bursts in protein production, with possible sudden dramatic changes in cell behavior, signaling noise causes a constant background. In kinase-mediated signaling, this background can consist of low levels of RTK activity through random transient interactions of receptors 20, phosphorylation of receptors and recruited proteins, binding of signaling proteins to receptors in nonfunctional complexes, and background phosphatase activity. A functional signal has to prevail above this background noise. Because fidelity in signaling is achieved despite this noise, the question arises of how this noise has shaped cellular signal transduction.
Multimerization to overcome the noise threshold
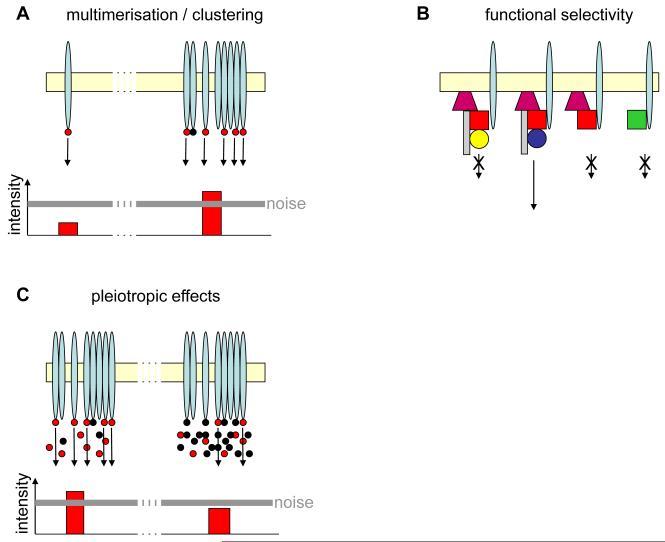
The noise resulting from transient nonspecific PPIs in interconnected signaling networks will generate a constant “chatter” that has to be surmounted[sta7] for signals to reach their destination. In terms of cellular signal transduction, signals can be amplified through oligomerization or clustering of the signaling components (Figure 3A). Indeed, dimerization of transcription factors has been shown to diminish noise in the genetic networks they control 27. Furthermore, it has been suggested that protein dimerization could dampen intrinsic and extrinsic noise 27. Also, eukaryotic signaling networks contain a significantly higher number of homologous interactions than expected from calculating the probability for their random occurrence 28, indicating that self-interaction has been evolutionarily favored. It has been suggested that ligand-induced oligomerization of cell surface receptors protects intracellular signaling against noise 29, 30. In the case of most RTKs, kinase signaling is initiated by extracellular ligand binding, which induces dimerization of RTKs. This cytokine-induced dimerization allows kinase domains to become juxtaposed, resulting in transphosphorylation of the RTKs 31. Receptor dimerization and clustering are very common, and the signal outcome is sensitive to changes in the status and lifetime of receptor multimers, with possible pathogenic consequences 31, 32. In addition, responses can be amplified through positive feedback loops and cooperative interactions between multiple partners 25.
Figure 3.
Examples of how signaling noise shapes cellular signal transduction. (A) Left: The signal (arrow) arising from one signaling receptor (elongated blue oval) at the membrane (beige) is not enough to overcome the threshold created by signaling noise. Right: Receptor multimerization is required to amplify the signal sufficiently to overcome the signaling threshold created by noise from promiscuous PPIs. Red dot: biologically relevant ligand; black dot: biologically nonrelevant ligand. (B) Functional selectivity through formation of multicomponent signaling complexes allows proofreading; a structurally similar but physiologically nonrelevant partner may bind to certain partially assembled components, but only the synergy of a correctly assembled complex (second from left) allows signal transduction to occur. First complex from left and first from right represent stalled complexes misassembled at different stages; second from right represents an incompletely assembled complex. Biologically nonrelevant homologues are indicated by the same shape and different color as the biologically relevant ligand in the correct signaling complex. (C) Grossly changing the concentration of one molecule may influence even signaling pathways for which this molecule is not a biologically relevant component. For example, before overexpression of the biologically nonrelevant ligand (black dot), a sufficient number of the biologically relevant ligand (red dot) could bind to its cognate receptor, initiating an above-threshold signal. After overexpression of the nonrelevant ligand, competition causes the number of biologically relevant ligands bound to the receptor to decrease. As a result, the ensuing signal remains below the noise threshold and cannot trigger a cellular response.
Functional selectivity through integration of multiple signals
The promiscuity in PPIs reduces the fidelity of the signaling process and eliminates mutual exclusivity in the pathways. The cell therefore adopts mechanisms that allow nonspecific interactions but precludes these interactions from providing an appropriate signal. One such mechanism that is prevalent in the early signaling events accompanying cell surface receptor stimulation is the recruitment to a RTK of multiple proteins, which form large multiprotein signaling complexes (early signaling complexes, or signalosomes) 33. Each RTK molecule may adopt a large amount of different phosphorylation states that are associated with potentially different ligands and hence downstream responses 34. This process is constantly counteracted by phosphatase activity 34. Only when the correct complement of proteins in the appropriate stoichiometry are recruited to an adequately phosphorylated receptor is a functional signaling entity formed and a downstream response garnered (Figure 3B). Thus, interactions of nonspecific proteins occur, but nonfunctional signaling complexes will result. Although this functional selectivity ensures that only complexes of the correct composition can initiate downstream signals, it also results in a significant population of nonfunctional complexes. Whereas in a high-fidelity system only one complex (or, realistically, a few complexes) would be required to initiate signaling, in promiscuous systems the number of initial complexes required is much higher. The ephemeral, abortive signals of these complexes may add to the noise level.
Probabilistic rather than linear signaling
Background signaling noise therefore demands cell surface receptor multimerization in two different ways. First, the total number of receptors needs to be large to overcome the number of nonfunctional submembrane signaling complexes formed. Second, receptors need to multimerize so that the signal generated persists above the background noise threshold, even including leaking and erroneous diffusion of the signal during propagation. The cumulative potential for formation of non-functional or redundant complexes suggests RTK signaling is probabilistic. The traditional view of a tightly regulated and highly specific linear signaling cascade from cell surface receptors to cellular response (e.g., gene expression) therefore needs to be supplanted by a probabilistic model. In this model, the probability of a given RTK dimer triggering a cellular outcome is in fact extremely low because of the promiscuous nature of signaling, the low probability of achieving a functional complex, and the noise threshold (Figures 2,3). Signal transduction is therefore a ‘numbers game’, where enough cell surface receptors must be engaged initially to guarantee formation of a functional signaling complex against a background of redundant events. Indeed, cells express tens or even hundreds of thousands of receptors on their surfaces which are exposed to extracellular stimuli. Because the production of huge numbers of receptor molecules is energetically expensive, such numbers appear to be[sta8] required for successful signal transduction. This view is supported, for example, by the observation that the reduction of HLA molecules on the cell surface from 100,000 to about 10,000 by HIV-1 Nef is enough to protect HIV-infected cells against recognition and killing by cytotoxic T cells 35, 36. In other words, 10,000 HLA molecules are not enough to produce a signal strong enough to trigger a cellular response.
Does pre-activation and pre-clustering to keep reaction times short?
To overcome noise, the receptors, as well as subsequent signaling hubs, must first multimerize, then auto-activate, and then assemble large signaling complexes. This diffusion-limited process is slow and will delay cellular signaling responses. Reaction times can be reduced by pre-clustering signaling components and putting them into a “primed” state: a condition from which they can rapidly assemble into the fully active constellation. Then, rather than having to go from zero to above the noise level, signaling networks only have to transition from just below to just above the noise threshold. It is indeed often observed, although only poorly documented, that even in serum starved cells and in the absence of stimuli a significant percentage of many RTKs are already in a phosphorylated state, which is normally associated with an active signaling function 37-40. Ligand-based activation only tips the balance toward a higher percentage of fully active receptors 20. However, this small increase in the percentage of activated receptors appears to make all the difference in terms of signaling outcome 40. The primed states of signaling networks may be important in pushing cells into a state of criticality or supersensitivity, where a spike of activity above the noise level will be rapidly[sta9] perpetuated throughout the signaling cell 41, 42.
Positive effects of noise on fidelity, robustness, and evolvability of cellular signaling
An intriguing question is whether, counterintuitively, background noise could also improve fidelity in cellular signaling networks. The noise generated by the intricacy, multivalency, and promiscuity of PPIs introduces a threshold level for signal propagation. Because signals below this threshold will not be able to trigger a response, this threshold level of white noise makes the cell robust against erroneous endogenous signals, such as those originating from the very source of noise: the erroneous PPIs and basal enzymatic activity. By maintaining an appropriate noise level, cells would also protect themselves from erroneous exogenous signals such as, for example, ectopic growth factors or low doses of pathogenic activators.
Noise may also render cells more robust to inactivation of a signaling component (e.g., through a deleterious mutation). A certain type of noisy interaction may become significant if the biologically relevant binding partner is depleted. Thus, the PPI promiscuity that accounts for part of the endogenous noise may allow the closest homologous binding partner to ultimately illicit a significant cellular response if the biologically relevant partner is depleted. A homologous protein could take the lead to partially rescue a null mutant. This type of partial rescue has indeed been described (e.g., for Src kinases 43) and makes cells robust against protein deletions.
The evolution of cellular signaling networks is characterized by gene duplication followed by divergence and gain of new function 44. The signaling noise threshold may allow such duplicated genes to evolve into a new function without significantly perturbing existing communication networks.[sta10]
Influence of noise modulation on experimental results
If signaling has adjusted to, and relies on, a significant noise level, gross alterations in this noise level may, in turn, influence signaling in general. For example, strong overexpression or knockout/knockdown of abundant and promiscuous proteins may significantly alter the noise level inside the cell. As a result, gross changes in a particular protein concentration may pleiotropically affect even those cellular signaling pathways that do not use this protein, by altering the noise level (Figure 3C). According to the effects discussed here, this may lead to premature or delayed initiation of signaling cascades or to signal rerouting through significant involvement of proteins homologous to the one affected. More subtly, changes in the specificity of some signaling domains may affect not the total noise level but the “pressure” that noise exerts on particular interactions. Protein overexpression or knockout/knockdown are commonly used tools to assess the function of a particular protein. Potential pleiotropic effects of noise-level modulation would therefore be an important parameter to assess in experimental investigations to avoid false interpretations of the phenotype caused by gross changes in protein concentration.
Concluding remarks
Although the causes and effects of noise in genetic circuits have become well established, noise originating from promiscuity in PPIs has so far been widely neglected. Here, we argue that a certain level of PPI noise is unavoidable, because of the complex and multivalent nature of eukaryotic signaling networks, and that this sort of noise has profoundly shaped the way cells transmit signals. If cells have adapted to this noise level, then noise has become an integral part of the correct transmission of signals. If so, then many signaling cascades, especially those involving ubiquitous homology domains, may be indirectly linked through noise. This noise may be important in increasing the robustness of cellular signaling by dampening and aborting single erroneous events. Thus, erroneous endogenous signals, and possibly also a certain level of pathogenic intervention, may be dampened. In turn, the promiscuity giving rise to that noise may help cell function recovery through homologous pathways. Importantly, gross changes in protein expression, such as commonly achieved experimentally, may have pleiotropic effects on other cellular signaling networks. Here, we have discussed signaling noise principally from the perspective of kinase-mediated signaling. Similar noise may affect other signaling networks. For example, the recognition of histone modification has been shown to be influenced by noise 4, 45. Noise could therefore play a role in the fidelity of epigenetic memory. Because noise affects central regulatory switches of cell functions, alterations in noise level may play a role in human disease. Understanding how signaling noise affects cell function may therefore provide novel viewpoints on diseases such as cancer or on propagation of epigenetic memory.
BOX 2 Outstanding questions.
-
*
What conceptual and mathematical model best describes signaling based on a noisy, probabilistic network of pleiomorphic 7 ensembles, rather than based on a linear signaling cascade?
-
*
what experimental setup could assess noise at the level of networks or of individual proteins?
-
*
How sensitive are cellular processes to changes in noise levels?
-
*
Can gross changes in concentration of a particular protein have pleiotropic effects on signaling networks?
-
*
If defined noise levels are important for correct cellular function, can gross changes in noise level result in disease states?
Acknowledgments
We thank Gabor Balazsi, Hiro Akari, the editor and reviewers for their comments and interesting suggestions, and K. Muller for editorial assistance. This research is supported in part by the National Institutes of Health through MD Anderson’s Cancer Center Support Grant (CA016672), and by the G. Harold and Leila Y. Mathers Charitable Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balazsi G, et al. Cellular decision making and biological noise: from microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Losick R, Desplan C. Stochasticity and cell fate. Science. 2008;320:65–68. doi: 10.1126/science.1147888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheong R, et al. Information transduction capacity of noisy biochemical signaling networks. Science. 2011;334:354–358. doi: 10.1126/science.1204553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy ED, et al. How perfect can protein interactomes be? Sci Signal. 2009;2 doi: 10.1126/scisignal.260pe11. pe11. [DOI] [PubMed] [Google Scholar]
- 8.Chaurasia G, et al. UniHI 4: new tools for query, analysis and visualization of the human protein-protein interactome. Nucleic Acids Res. 2009;37:D657–660. doi: 10.1093/nar/gkn841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatesan K, et al. An empirical framework for binary interactome mapping. Nat Methods. 2009;6:83–90. doi: 10.1038/nmeth.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stumpf MP, et al. Estimating the size of the human interactome. Proc Natl Acad Sci U S A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy ED, et al. Cell signaling. Signaling through cooperation. Science. 2010;328:983–984. doi: 10.1126/science.1190993. [DOI] [PubMed] [Google Scholar]
- 12.Breitkreutz A, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ladbury JE, Arold S. Searching for specificity in SH domains. Chem Biol. 2000;7:R3–8. doi: 10.1016/s1074-5521(00)00067-3. [DOI] [PubMed] [Google Scholar]
- 14.Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 15.Ladbury JE, Arold ST. Energetics of Src homology domain interactions in receptor tyrosine kinase-mediated signaling. Methods Enzymol. 2011;488:147–183. doi: 10.1016/B978-0-12-381268-1.00007-0. [DOI] [PubMed] [Google Scholar]
- 16.Uhlik MT, et al. Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J Mol Biol. 2005;345:1–20. doi: 10.1016/j.jmb.2004.10.038. [DOI] [PubMed] [Google Scholar]
- 17.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–653. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Briggs SD, et al. SH3-mediated Hck tyrosine kinase activation and fibroblast transformation by the Nef protein of HIV-1. J Biol Chem. 1997;272:17899–17902. doi: 10.1074/jbc.272.29.17899. [DOI] [PubMed] [Google Scholar]
- 19.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 20.Chung I, et al. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf C, et al. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2004;2:94–103. doi: 10.1371/journal.pbio.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castagnoli L, et al. Selectivity and promiscuity in the interaction network mediated by protein recognition modules. FEBS Lett. 2004;567:74–79. doi: 10.1016/j.febslet.2004.03.116. [DOI] [PubMed] [Google Scholar]
- 23.Tonikian R, et al. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karkkainen S, et al. Identification of preferred protein interactions by phage-display of the human Src homology-3 proteome. EMBO Rep. 2006;7:186–191. doi: 10.1038/sj.embor.7400596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson TJ. Cell regulation: determined to signal discrete cooperation. Trends Biochem Sci. 2009;34:471–482. doi: 10.1016/j.tibs.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 26.Arold ST. How focal adhesion kinase achieves regulation by linking ligand binding, localization and action. Curr Opin Struct Biol. 2011;21:808–813. doi: 10.1016/j.sbi.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghim CM, Almaas E. Genetic noise control via protein oligomerization. BMC Syst Biol. 2008;2:94. doi: 10.1186/1752-0509-2-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ispolatov I, et al. Binding properties and evolution of homodimers in protein-protein interaction networks. Nucleic Acids Res. 2005;33:3629–3635. doi: 10.1093/nar/gki678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alarcon T, Page KM. Stochastic models of receptor oligomerization by bivalent ligand. J R Soc Interface. 2006;3:545–559. doi: 10.1098/rsif.2006.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macnamara S, et al. Stochastic analysis of the VEGF receptor response curve. In: Pham TD, Zhou X, editors. Computational models for Life Sciences-CMLS’07. American Institute of Physics; 2007. [Google Scholar]
- 31.Lemmon MA, chlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alarcon T, Page KM. Mathematical models of the VEGF receptor and its role in cancer therapy. J R Soc Interface. 2007;4:283–304. doi: 10.1098/rsif.2006.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Rourke L, Ladbury JE. Specificity is complex and time consuming: mutual exclusivity in tyrosine kinase-mediated signaling. Acc Chem Res. 2003;36:410–416. doi: 10.1021/ar020167s. [DOI] [PubMed] [Google Scholar]
- 34.Mayer BJ, et al. Molecular machines or pleiomorphic ensembles: signaling complexes revisited. J Biol. 2009;8:81. doi: 10.1186/jbiol185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins KL, et al. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 36.Akari H, et al. Nef-induced major histocompatibility complex class I down-regulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 down-regulation. J Virol. 2000;74:2907–2912. doi: 10.1128/jvi.74.6.2907-2912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunii K, et al. FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res. 2008;68:2340–2348. doi: 10.1158/0008-5472.CAN-07-5229. [DOI] [PubMed] [Google Scholar]
- 38.Takeda M, et al. AZD2171 shows potent antitumor activity against gastric cancer over-expressing fibroblast growth factor receptor 2/keratinocyte growth factor receptor. Clin Cancer Res. 2007;13:3051–3057. doi: 10.1158/1078-0432.CCR-06-2743. [DOI] [PubMed] [Google Scholar]
- 39.Bryant MR, et al. Phosphorylation and lipid raft association of fibroblast growth factor receptor-2 in oligodendrocytes. Glia. 2009;57:935–946. doi: 10.1002/glia.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed Z, et al. Direct binding of Grb2 SH3 domain to FGFR2 regulates SHP2 function. Cell Signal. 2010;22:23–33. doi: 10.1016/j.cellsig.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 41.Huang CY, Ferrell JE., Jr. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nykter M, et al. Gene expression dynamics in the macrophage exhibit criticality. Proc Natl Acad Sci U S A. 2008;105:1897–1900. doi: 10.1073/pnas.0711525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roche S, et al. Requirement for Src family protein tyrosine kinases in G2 for fibroblast cell division. Science. 1995;269:1567–1569. doi: 10.1126/science.7545311. [DOI] [PubMed] [Google Scholar]
- 44.Copley RR, et al. Eukaryotic domain evolution inferred from genome comparisons. Curr Opin Genet Dev. 2003;13:623–628. doi: 10.1016/j.gde.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 45.Choi JK, et al. Stochastic and regulatory role of chromatin silencing in genomic response to environmental changes. PLoS One. 2008;3:e3002. doi: 10.1371/journal.pone.0003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vidal M, et al. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seebacher J, Gavin AC. SnapShot: Protein-protein interaction networks. Cell. 2011;144:1000. doi: 10.1016/j.cell.2011.02.025. 1000 e1001. [DOI] [PubMed] [Google Scholar]