Summary
To assess the genetic consequences of induced Pluripotent Stem Cell (iPSC) reprogramming, we sequenced the genomes of ten murine iPSC clones derived from three independent reprogramming experiments, and compared them to their parental cell genomes. We detected hundreds of single nucleotide variants (SNVs) in every clone, with an average of 11 in coding regions. In two experiments, all SNVs were unique for each clone and did not cluster in pathways, but in the third, all four iPSC clones contained 157 shared genetic variants, which could also be detected in rare cells (<1 in 500) within the parental MEF pool. This data suggests that most of the genetic variation in iPSC clones is not caused by reprogramming per se, but is rather a consequence of cloning individual cells, which “captures” their mutational history. These findings have implications for the development and therapeutic use of cells that are reprogrammed by any method.
Introduction
The discovery of methods to create induced pluripotent stem cells (iPSCs) in 2006 revolutionized the field of regenerative medicine. Yamanaka, et al. and Thomson, et al. showed that expression of a small set of transcription factors in mouse or human somatic cells can reprogram a small subset to a pluripotent state (Takahashi et al., 2007; Takahashi and Yamanaka, 2006; Yu et al., 2007). Subsequent modifications to the original reprogramming protocols have decreased safety concerns and increased efficiency. Reprogramming is now possible without the use of c-myc (Chang et al., 2009; Huangfu et al., 2008b; Wernig et al., 2008). It can be performed with single polycistronic lentiviruses to limit integration-site dependent mutagenesis (Carey et al., 2009; Chang et al., 2009; Shao et al., 2009), or by transiently expressing specific cDNAs, RNA molecules, or the reprogramming proteins themselves in somatic cells (Okita et al., 2008; Stadtfeld et al., 2008; Warren et al., 2010; Zhou et al., 2009). The use of hypomethylating agents and histone deacetylase inhibitors has also enhanced the efficiency of iPSC generation (Huangfu et al., 2008a; Huangfu et al., 2008b; Mikkelsen et al., 2008).
iPSC lines provide novel models for the study of human diseases, and gene-corrected iPSCs may provide novel therapeutic reagents (Hanna et al., 2007; Raya et al., 2009; Yamanaka, 2009). Jaenisch, Townes, and colleagues rescued a humanized sickle cell anemia mouse model by transplanting gene-corrected iPSCs that had been induced to undergo hematopoietic differentiation (Hanna et al., 2007). More recently, Cantz, et al., generated mice from tetraploid embryo aggregations of gene-corrected iPSCs, proving that genetic manipulation does not diminish the totipotent potential of iPSCs (Wu et al., 2011).
Although advancements have been made in the generation of iPSCs, the mechanism behind reprogramming is not yet completely understood (Yamanaka, 2009). Epigenetic mechanisms are clearly important; reprogramming is associated with new DNA methylation patterns that presumably reflect the activation of endogenous pluripotency genes, and repression of differentiation genes. Ecker and colleagues defined the methylomes of iPSCs at single base resolution; although they are globally similar to ES cells, iPSCs have unique patterns of DNA methylation (Lister et al., 2011). However, the reprogramming of fibroblasts deficient for Dnmt3A and Dnmt3B (the DNA methyltransferases responsible for de novo DNA methylation) revealed that neither of these genes is required for reprogramming (Pawlak and Jaenisch, 2011). This information, coupled with the fact that iPSCs have “memory” of the parental cells from which they were derived (Kim et al., 2010), suggests that there may be additional, currently unrecognized factors that are relevant for iPSC generation.
The role of genetic variation in reprogramming is less clear. Although iPSC lines generally have normal karyotypes (Park et al., 2008; Takahashi et al., 2007; Wernig et al., 2007; Yu et al., 2007), more recent analyses of iPSC genomes suggest that there may be more subtle genetic consequences of reprogramming. Hall and colleagues used whole genome sequencing data to detect structural variants (SVs) in three iPSC lines derived from a single reprogramming experiment; they found a very small number of new SVs in the iPSC lines, suggesting that reprogramming does not cause genomic instability (Quinlan et al., 2011); in contrast, array-based studies revealed a large number of copy number variants within iPS genomes, which evolved with passaging (Hussein et al., 2011; Laurent et al., 2011; Martins-Taylor et al., 2011). Since submission of this paper, a report by Ji, et al. confirmed this finding, using whole exome sequencing of human iPSC lines (Ji et al., 2011). Gore, et al. performed whole exome sequencing of 22 human iPSC lines generated with multiple reprogramming strategies, and a variety of donor cell types. They detected an average of 5 non-synonymous point mutations per exome, which were enriched in genes found in the COSMIC database of cancer-associated genes (Forbes et al., 2008; Gore et al., 2011); the authors concluded that iPSC genomes often contain mutations that may be related to cancer pathogenesis, raising a potentially important safety issue.
In this study, we performed three experiments utilizing whole genome sequencing to define all of the genetic variations associated with reprogramming. Our data suggest that most mutations in iPSCs are random and benign, occurring prior to reprogramming; these pre-existing mutations are “captured” by the cloning event itself. In one experiment, all tested iPSC clones were derived from the same pool of rare founding cells that contained a shared set of mutations. Although these mutations may simply mark “elite” cells with a reprogramming advantage (a model that was thought to be unlikely by Yamanaka (Yamanaka, 2009)), some could potentially contribute to reprogramming fitness by cooperating with reprogramming factors. Our data suggest that preexisting mutations in somatic cells will be captured by any reprogramming strategy that is used, since cell cloning is required for iPSC generation. By sequencing large numbers of iPSC clones, recurring mutations in pathways that affect reprogramming efficiency may be discovered; these mutations may reveal novel approaches to improve reprogramming efficiency and safety.
Results
Experimental system
To investigate the genetic events associated with transcription factor-mediated reprogramming, we generated murine iPSC lines using an established polycistronic lentivirus containing three transcription factors, OCT4, SOX2, and KLF4 (OSK) (Chang et al., 2009). Although it is now possible to reprogram somatic cells using non-integrating vectors, the use of an integrating reprogramming vector was necessary to provide a definitive and unique genetic mark for each iPSC clone, which was crucial for all subsequent steps of the analysis. We transduced mouse fibroblasts derived from three different mouse strains. The three donor mice had very different breeding histories: the embryo used to make mouse embryonic fibroblasts (MEFs) in experiment 1 was a WT littermate from an intercross between Ybx1 +/− founders (Lu et al., 2005). The donor for the WT tail tip fibroblasts (TTFs) used in experiment 2 was a WT littermate from an intercross between Casp9 +/− mice (Zheng et al., 2000). The MEFs used in experiment 3 were derived from a Gusb −/− embryo from an intercross between congenic Gusb +/− mice, a murine disease model for Mucopolysaccaridosis type VII (MPSVII) that has been maintained as an inbred strain at the Jackson Laboratory. Details of cell culture, transduction, and iPSC generation are provided in the Experimental Procedures, and are listed in Table 1.
Table 1.
Pluripotency and genomic characterization of iPSC clones
| Experiment (Parental Cells) | Clone | Age of Mouse | Days in Culture for transduction | ES-like morphology | Alkaline Phosphatase staining positive | Flow Cytometric Staining | WGS Haploid Coverage | OSK Coverage | OSK Integration Positions | SNV Validation Platform | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pre | Post | SSEA1 + | Nanog + | Oct 3/4 + | |||||||||
| 1 (MEF) | e13.5 | 55.178 | |||||||||||
| 1 (MEF-derived iPSC) | 4 | e13.5 | 6 | 29 | Yes | Yes | Yes | Yes | Yes | 46.689 | 105.175 | 6:116062660 11:54261983 17:29179146 |
Exome Sequencing |
| 5 | e13.5 | 6 | 29 | Yes | Yes | Yes | Yes | Yes | 55.191 | 176.645 | 7:20201630 9:34109963 15:8307932 16:36841079 |
Exome Sequencing | |
| 9 | e13.5 | 6 | 29 | Yes | Yes | Yes | Yes | Yes | 57.668 | 104.485 | 12:56888952 X:82219170 |
Exome Sequencing | |
| 2 (TTF) | 65d | 22.727 | |||||||||||
| 2 (TTF-derived iPSC) | 1 | 65d | 20 | 16 | Yes | Yes | Yes | Yes | Yes | 26.56 | 148.404 | X:23294264 | Exome Sequencing |
| 2 | 65d | 20 | 16 | Yes | Yes | Yes | Yes | Yes | 24.377 | 66.531 | 7:102410141 9:41255493 |
Exome Sequencing | |
| 3 | 65d | 20 | 16 | Yes | Yes | Yes | Yes | Yes | 30.255 | 75.629 | 3:65184739 7:66559629 10:30896454 12:70688422 14:97803433 |
Exome Sequencing | |
| 3 (GusB−/− MEF) | e13.5 | 17.877 | |||||||||||
| 3 (GusB−/− MEF- derived iPSC)) | 1 | e13.5 | 17 | 24 | Yes | Yes | Yes | Yes | Yes | 28.896 | 20.387 | 5:16530376 9:72205724 |
454-Flx |
| 2 | e13.5 | 17 | 24 | Yes | Yes | Yes | Yes | Yes | 23.958 | 12.279 | 4:139018701 14:84892429 |
454-Flx | |
| 5 | e13.5 | 17 | 24 | Yes | Yes | Yes | Yes | Yes | 19.591 | 12.603 | 14:6229265 | 454-Flx | |
| 6 | e13.5 | 17 | 24 | Yes | Yes | Yes | Yes | Yes | 22.005 | 13.74 | 12:99479785 16:38231164 |
454-Flx | |
All clones were examined for morphology and alkaline phosphatase reactivity, as well as expression of the pluripotency markers SSEA-1, Nanog, and Oct4. All clones had characteristics of embryonic stem cells (Table 1). Since experiment 3 utilized MEFs derived from a disease model known to have growth and developmental defects (MPSVII is caused by a frameshift mutation in the Gusb gene that creates a null allele), we extensively characterized these iPSC lines (Meng et al., 2010; Sands and Birkenmeier, 1993). Affymetrix Mouse Exon 1.0ST arrays were used to compare expression patterns in MPSVII iPSC lines, and embryo-derived MPSVII ES cells (GEO Accession GSE36017). Unsupervised hierarchical clustering analysis showed that the iPSC clones and ES cell lines clustered randomly, suggesting that their global patterns of gene expression are highly similar (Figure S1a). The methylation status of the Oct4 and Nanog gene promoters was analyzed by bisulfite modification of genomic DNA from each of the four lines, along with ES cell and MEF controls. The promoter region of each gene was amplified after bisulfite treatment with bisulfite-specific primers, followed by deep digital sequencing of the amplicons on the Roche/454 FLX platform. Each CpG dinucleotide was covered by an average of 2,944 reads (range 107 to 6,129), and the percentage of methylated C residues was determined at each position. The Oct4 and Nanog promoters were extensively methylated in MEFs, but were relatively unmethylated in ES cells or iPSCs (Figure S1b). Lastly, NOG mice were injected with 1 million iPSCs from each of the four iPSC lines; each line formed cystic teratomas containing all 3 germ layers (Figure S1c).
Sequence analysis of the genomes of the parental fibroblasts (founder mice)
It was essential to sequence the genomes of the “parental” MEFs or TTFs from which the iPSC clones were derived as the appropriate comparator genome for each experiment. The founding animals used in experiments 1 and 2 were wild type littermates of mice containing targeted mutations made in 129/SvJ ES cells that were injected into C57Bl/6 blastocysts. After germline transmission of the mutant allele, the mice were backcrossed to C57Bl/6 mice. The embryo used in experiment 1 was backcrossed only twice, while the adult mouse used for experiment 2 was extensively backcrossed to C57Bl/6 mice (obtained from the Jackson Laboratory). To determine the fraction of 129/SvJ variants in the founder genomes, we compared all SNVs detected in these genomes to both the reference B6 genome, and the 129/SvJ genome, which we sequenced previously for the same purpose (Wartman et al., 2011). Our most recent alignment and filtering algorithms revealed that the 129/SvJ genome contains 4,434,171 SNVs compared to the B6 genome. We first compared each of the fibroblast genomes to the reference (C57Bl/6) genome. In the founding MEFs from experiment 1, a total of 497,691 SNVs were detected (237,319 were heterozygous, and the remainder homozygous). Of these variants, 476,002 (95.64%) were identical to 129 SNVs; the high number of 129 variants reveals the short backcrossing history of these mice. The TTFs from experiment 2 contained a total of 8,002 SNVs (5,702 heterozygous, and the remainder homozygous), of which 4,125 (51.55%) were identical to 129 SNVs; this data reveals the extensive backcrossing of these mice to B6. Finally, the MEFs used in experiment 3 contained a total of 7,278 SNVs (5,192 heterozygous, and the remaining homozygous), of which 4,317 were identical to 129 SNVs (59.32%). Despite the long history of intercrossing of these mice, a very small amount of residual 129 strain “contamination” is still apparently present in their genomes. The large number of private variants present in each mouse genome demonstrate the absolute necessity of sequencing the founder cell genomes as the comparator for derivative iPSC genomes.
Genomic Architecture of iPSC Clones
Genomic DNA from each iPSC line and their parental fibroblasts were used to make libraries that were subjected to paired-end sequencing on the Illumina GAIIX or HiSeq 2000 platform. The three iPSC genomes from the first experiment were sequenced to an average haploid coverage depth of 55x; we learned that this was far greater than what was required for mutation discovery from inbred mouse genomes [29]; the average haploid coverage of the genomes for the second experiment was therefore ~23x, and for the third experiment, ~18x (Table 1); the variants for all genomes were deposited in the Short Read Archive (SRP011044). We first mapped the genomic integration sites of the polycistronic OSK lentivirus in every clone to establish that each clone was genetically unique. Each iPSC line had 1–5 distinct lentiviral insertion sites, which established the unique genetic identity of each clone (Table 1 and Figure 1A). For experiment 3, we used expression array data from each clone to demonstrate that the lentiviral insertions did not detectably alter expression of surrounding genes (two megabases upstream and downstream from each insertion site) in any clone (data not shown).
Figure 1. OSK lentivirus insertion sites for the three reprogramming experiments.
Insertion sites of the OSK lentivirus for each iPSC clone are indicated on a representation of the 20 mouse chromosomes. Sites of integration were determined by whole genome sequencing. Each iPSC line had 1–5 lentiviral insertion sites. The first number next to each insertion site refers to the experiment, and the second refers to the clone. Experiment 1 insertion sites are indicated in green, experiment 2 in red, and experiment 3 in blue.
The genome of each iPSC clone was compared to its own parental fibroblast genome to define all acquired variants in the iPSC clone. Compared to its own parental line, each iPSC clone contained a range of 190–773 SNVs (Figure 2A); an average of 11 SNVs (range 3–19) were located within coding regions (Figure 2B, Table S1). To assess the fidelity of our calling algorithms, we determined the number of 129 SNVs among the total SNVs detected in each iPS genome; if the algorithms were accurate, very few of the sequence variants should have been identical to 129 SNVs; this was indeed the case. For experiment 1, only 2/907 (0.22%) SNVs were from the 129 strain, for experiment 2, 8/2083 SNVs (0.38%) were 129 derived, and for experiment 3, 42/2130 SNVs (1.97%) were identical to 129 SNVs. Plotting the location of each SNV within the genome reveals that the variants are widely distributed throughout the genome (Figure S2). All four iPSC lines (and the starting MEF cell pool) derived from the Gusb −/− mice contained the expected homozygous single base deletion (del C) in the 10th exon of the GusB gene that creates the frameshift mutation that inactivates the gene (Sands and Birkenmeier, 1993).
Figure 2. Numbers of SNVs identified in each iPSC clone.
Panel A. Total number of SNVs (including a small fraction of loss of heterozygosity sites) detected in each iPSC clone compared to its parental fibroblasts, related to Figure S2.
Panel B. Total number of SNVs detected in the coding regions of each iPSC clone compared to its parental fibroblasts, listed in Table S1. Pathway analysis of these SNVs is shown in Table S5.
The digital readcounts provided by whole genome sequencing allowed us to calculate the variant allele frequencies of all SNVs. Variant allele frequency plots revealed that the variant frequency is distributed around a mean of ~50%, with a normal distribution (Figures 3A and 5). This suggests that the vast majority of variants are heterozygous, and present in nearly all cells within the iPSC sample.
Figure 3. SNVs in iPSC clones from experiments 1 and 2.
Panel A. Variant allele frequency plots of all SNVs for each of the iPSC clones from experiments 1 and 2. Two plots are shown for each clone: kernel density (top) and iPSC variant allele frequencies by sequence coverage (bottom). The traditional kernel density estimation (KDE, Experimental Procedures) reveals that each iPSC sample is comprised of a single dominant clone, with no apparent subclones.
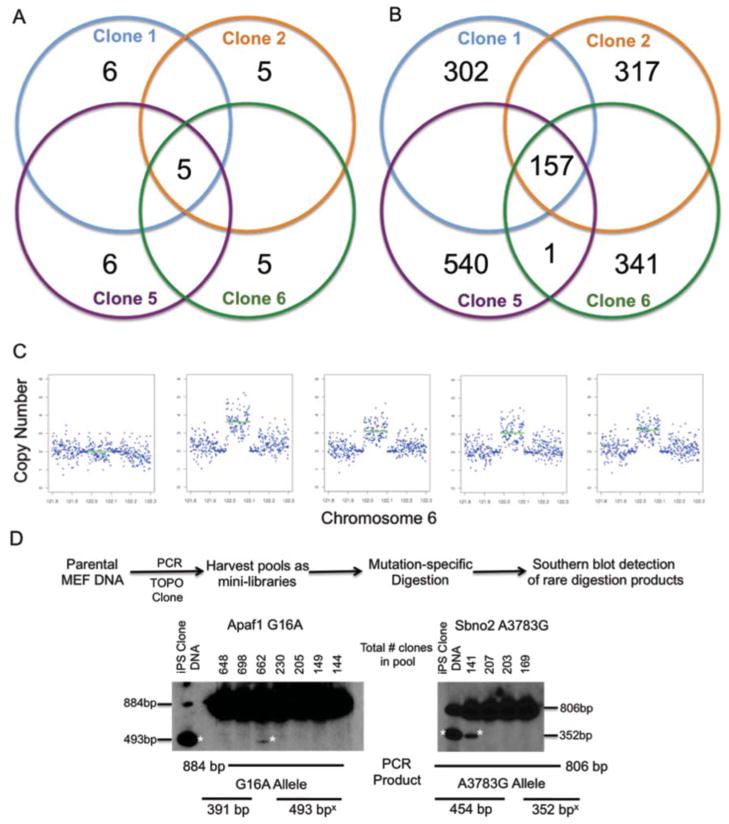
Panel B. Relationship of the total SNVs detected within each iPSC clone. No shared SNVs were detected in any clone in either experiment. Overlapping regions of the Venn diagram with no numbers contain no SNVs.
Figure 5. Comparison of shared vs. private variant allele frequencies for experiment 3.
Left and Center panels: Variant allele frequencies for the shared and private SNVs from the iPSC clones of experiment 3 were plotted. Two plots are shown for each clone: kernel density (top) and iPSC variant allele frequencies by sequence coverage (bottom). The shared SNVs have a single dominant clone with a peak variant allele frequency of ~50% (left panels). The private SNVs in each clone also have a major peak of variant allele frequency of ~50%, but the suggestion of second peak at ~25% is suggested in clones 1, 2 and 6. The second peak was confirmed only for clone 1 (red arrowhead).
Right panels: Validation of variant allele frequencies in iPSC clones. Using whole genome sequencing data, we selected ten variants with predicted VAFs of 50% vs. 25% for each of the iPSC clones, amplified all variants by PCR, and then performed deep sequencing on the 454-FLX platform. Variant allele frequencies (VAFs) were plotted for each clone for whole genome sequencing (X axis, VAF by WGS) vs deep sequencing (Y axis, VAF by deep 454). For clone 1, the variants with VAFs of ~25% were statistically confirmed by deep sequencing (p=0.000045). For the other three clones, the variants with predicted 25% VAF were not confirmed (p>0.05 for all three).
Indels and Structural Variants (SVs)
We tested for somatically acquired SVs in all three experiments by comparing sequencing read depths across the genomes of the iPSC clones vs. parental fibroblasts. We found no high confidence SVs in experiment 1, and two SVs in experiment 2. In clone 1, a 740 kb, single copy amplification was found on chromosome 11 between positions 87,990,000 and 88,730,000. In clone 2, a single copy amplification of 850 kbp was found on chromosome 14 between positions 11,530,000 and 12,380,000. In experiment 3, we found one single copy number amplification that was common to all four iPSC clones, which is described in detail below.
A large number of small insertions and deletions (indels) were predicted in all iPSC clones from all three experiments. However, we did not secondarily validate these variants because the frequency of false positives is very high with our current calling algorithms (see Experimental Procedures). However, for experiment 3, we detected a set of 32 identical indels (out of several hundred predicted in each clone) that were common to all four iPSC clones (Table S3); because these indels were identical in all four clones, we are confident that they are real.
Mutational consequences of SNVs
The calling algorithms used for SNV detection, and the sensitivity and specificity of the calling algorithms, are detailed in the Experimental Procedures section. All of the SNVs reported within coding regions were secondarily validated by exome sequencing for experiments 1 and 2, or by Roche/454-FLX sequencing of specific PCR amplified regions for experiment 3 (Table S1). Each iPSC clone contained an average of 11 validated coding region SNVs per genome (including missense, nonsense, splice site, and silent mutations). None of these were identical to 129/SvJ SNVs. In addition to the coding mutations, each clone contained hundreds of predicted SNVs in non-coding regions of the genome (Figure 2A). For the three iPSC clones in experiments 1 and 2, all of the SNVs were unique to each iPSC genome, and there was no overlap among the variants detected in each clone or experiment (Figure 3B).
However, among the 459–698 total SNVs detected in each iPSC genome in experiment 3, we detected 157 SNVs (5 within coding regions) that were common to all four iPSC lines (Figures 4A and 4B, Table S2). We tested for the 5 shared coding region SNVs in two additional iPSC lines generated from the same experiment, and both were heterozygous for all 5 variants (data not shown). Each of the iPSC clones in this experiment had unique lentiviral insertion sites, and each had a set of private mutations that did not overlap among the clones; each clone was therefore genetically distinct. We tested for two of the shared mutations (Apaf1 G16A and Sbno2 A3783G) in a second reprogramming experiment using TTFs derived from a different Gusb −/− mouse, and detected neither SNV in any of 10 iPSC clones tested (data not shown). Using expression array data from each of the Gusb −/− iPSC clones, we found that none of the shared SNVs altered the expression of any “nearest neighbor” genes (data not shown).
Figure 4. Shared genomic variants identified in all four iPSC clones from experiment 3.
Panel A. Relationship of the SNVs detected within the coding regions of each iPSC clone. Private and shared mutations were found in all four clones. Overlapping regions of the Venn diagram with no numbers contain no SNVs. Pathway analysis of these SNVs is shown in Table S5.
Panel B. Relationship of the total SNVs (including a small fraction of loss of heterozygosity sites) for each iPSC clone. Overlapping regions of the Venn diagram with no numbers contain no SNVs. Shared SNVs are listed in Table S2 and shared indels are listed in Table S3.
Panel C. An identical structural variant was identified in all four iPSC clones from experiment 3, spanning ~130,000 bp on chromosome 6.
Panel D. Detection of shared variants in rare cells from the parental MEF pool. Regions containing the SNVs within the Apaf1 and Sbno2 genes were amplified from the Gusb −/− MEF starting pool from experiment 3, and these amplicons were directly ligated into the pCR2.1 vector. The ligation products were transformed into E. coli; ampicillin resistant colonies were counted on each 10 cm plate, and pooled. Plasmid DNA was prepared from each pool, and digested with StuI and EcoRI to detect the novel StuI sites generated by the Apaf1 G16A and Sbno2 A3783G variants. Digestion products were separated on 5% polyacrylamide gels, transferred to nitrocellulose, and analyzed by Southern blotting. Expected fragment lengths are indicated below the Southern blots. * indicates the variant-specific digestion products. Quantification data from Illumina sequencing validation is shown in Table S4.
To further confirm that the four iPSC clones from experiment 3 had a shared set of genetic variants, we also searched for shared indels and structural variants, as noted above. Somatic indel prediction analysis identified 32 identical indels that were present in all four iPSC clones (Table S3); none had a translational effect. We also identified a shared amplified region of ~130 kilobases on chromosome 6 that was present in all four iPSC clones, but not in the parental MEFs (Figure 4C). Two genes are located within this region, Mug2 (a protease inhibitor) and Gm10319 (a predicted gene); Gm10319 was not significantly overexpressed in the iPSC clones compared to Gusb −/− ES cells; the mouse Exon1.0 array did not contain probesets for Mug2 so we could not evaluate its expression (data not shown).
The identification of a large set of shared genetic variants strongly suggests that all four of these iPSC lines arose from rare cells within the MEF pool that contained a set of identical variants. However, none of the shared SNVs were reliably detected in the parental MEFs even with deep readcounts obtained when the mutations were validated (average 2,308x, range 776x to 4,312x coverage for each SNV, data not shown). Importantly, rare variant detection on the 454-FLX platform is limited by an error rate of ~1% (Gilles et al., 2011), suggesting that the shared variants were present in less than 1% of the total cells in the MEF pool. In an attempt to detect these rare cells among the parental MEFs, we decided that we must limit our analysis to several hundred cells at a time. We devised a strategy that took advantage of novel StuI restriction sites created by two of the shared coding region mutations (i.e. Apaf1 G16A and Sbno2 A3783G). We amplified regions containing these variants with PCR, and then cloned these amplicons into the pCR2.1 vector (Invitrogen, Carlsbad, CA). Plasmid DNA preparations derived from mini-libraries containing hundreds of cloned amplicons were then digested with EcoRI (to release the entire insert from the plasmid backbone) and StuI (to detect clones containing the variant alleles), and the digestion products were resolved using Southern blot analysis. Figure 4D shows representative blots of the Apaf1 and Sbno2 analyses. DNA from a pool of clones from one of the iPSC lines reveals that StuI cuts ~50% of the starting DNA. However, most pools of clones derived from the MEFs revealed no digestion products; only occasional pools contain very small amounts DNA cleaved by StuI, suggesting that no more than a few clones within the pool contain the variant allele. To estimate the frequency of the clones containing the variant allele in these positive pools, the same pools were sequenced on an Illumina HiSeq 2000, with >50,000x coverage of the variant position. The variant allele was detected in 0.153% of the Apaf1 reads in a pool containing 662 colonies; suggesting that only one clone contained the mutation. In the other experiment, the variant allele was detected in 1.016% of the Sbno2 reads in a pool containing 141 colonies, suggesting that 1–2 clones contained the mutation (Table S4). The total variant frequency was calculated to be 0.037% and 0.199% for Apaf1 and Sbno2, respectively, by taking into account the total number of clones from all negative pools within the same experiment (indicated in Figure 4D). This analysis suggests that less than 1 cell in 500 from the starting pool contained the shared variants detected in all of the iPSC clones.
In addition to the shared variants in the iPSC clones of experiment 3, there were also 302–540 unique (“private”) SNVs in each of the clones (Figure 4B). We compared the variant allele frequencies of the shared SNVs vs. the private SNVs. As expected, the shared variants all had variant allele frequency distributions with a single peak at about 50%, since these mutations must have occurred prior to reprogramming (Figure 5, left panels). While the majority of the private SNVs are also present in ~50% of reads, clones 1, 2, and 6 contained a small shoulder in the distribution at ~25% variant allele frequency, suggesting that a subclone may be present within these samples (Figure 5, middle panels). The presence of a true subclone in clone 1 was validated by examining the deep readcounts of 10 randomly selected mutations with a variant allele frequency of ~25%, vs. 10 with a ~50% variant allele frequency (Figure 5, right panels). In clones 2 and 6, the presence of a subclone was not confirmed.
Other than the shared variants detected in experiment 3, there was no overlap in the genes containing SNVs among the three experiments. We also compared all of the genes with coding region SNVs in our dataset to that of Gore et al., who sequenced the exomes of 22 human iPSC lines. A single gene, ATM/Atm, contained an SNV in both datasets: in our study, clone 5 from experiment 1 contained a splice site mutation at the 5′ end of intron 23; one of the 22 human iPSC clones contained a nonsynonymous SNV in ATM (L752P) (Gore et al., 2011).
Gore, et al. also reported that their human iPSC clones displayed a significant enrichment of mutations in genes found in the COSMIC database (Gore et al., 2011). We therefore examined the 89 unique genes with coding region SNVs in our 10 iPSC clones, and found that 36 (40.4%) were in the COSMIC database (Table S1). We also searched the COSMIC database for the 247 genes with homozygous variants in the three parental mice used in this study (compared to the B6 reference genome); 36.1% of these genes are likewise in COSMIC, a value that is not significantly different from that of the iPSC clones (p=0.5733).
We performed a pathway analysis using the MuSiC suite (N. Dees, et al., submitted) to test for significantly mutated genes (SMGs), and common pathways that might be affected by mutations among the 10 clones. The only SMGs were the 4 that contained shared missense variants identified in experiment 3. A total of 50 pathways contained genes that were mutated (Table S5). Only 13 of these had a P-value of less than 0.05; all included one of the shared variants from experiment 3. There were no pathways with mutations common to all 10 clones.
Discussion
In this study, we defined the genetic landscapes of 10 independent murine iPSC clones by whole genome sequencing. In the first two experiments, we identified several hundred SNVs in each individual clone (with an average of 11 in coding sequences), but found no overlap among the mutations in any of the clones. In the third experiment, however, we identified a subset of shared variants in all four iPSC clones, as well as a set of “private” variants in each. Using genomic DNA from the parental MEFs used to create these clones, we were able to detect two of the shared mutations at a very low frequency, demonstrating that rare cells within the total MEF population contained these mutations. Our data suggest that many SNVs may occur at or before the time of reprogramming, but some may occur during expansion of the iPSC clones in culture. Regardless, all of the iPSC lines tested contained hundreds of mutations. Although most are probably functionally irrelevant, some could potentially contribute to reprogramming fitness.
The use of whole genome sequencing, with assessment of the variant allele frequencies of hundreds of mutations, is a very powerful tool that has allowed us to begin to assess whether the mutations detected in iPSC clones are caused by reprogramming, or whether they simply represent the genetic “history” of the cell that was reprogrammed. In the first two experiments, this issue could not be resolved. All SNVs in all six clones were unique, with variant allele frequencies of ~50%, suggesting that most of the SNVs were heterozygous and present in nearly all of the iPSCs in the clone. Three scenarios could explain these results: 1) the SNVs preexisted in the cell that was reprogrammed, and reflect “fixation” of the background mutations that were present in that cell by cloning, or 2) the variants concurrently arose in a “burst” of mutational activity when the lentivirus integrated, but they were not relevant for outgrowth of the clone, or 3) the SNVs all arose in a burst of mutational activity after reprogramming; one or more mutations was important for selection, and the entire group of mutations in that cell was genetically fixed by cloning. Based on the background rate of mutations in somatic cells (Warren et al., 1981), and the large numbers of mutations detected in each clone, we suspect that the latter two scenarios are unlikely, although they cannot be definitively ruled out with the data from the first two experiments.
In the third experiment, however, we detected a large set of shared mutations in all four iPSC clones that were sequenced, including SNVs, indels, and a structural variant; we detected 5/5 coding SNVs tested in two other iPSC clones generated in the same experiment. Importantly, these shared variants were not detected in 10 iPSC clones derived from another reprogramming experiment using fibroblasts from an independent Gusb deficient mouse, i.e. the shared mutations were not related to the mouse strain itself. Using techniques that could detect point mutations in rare cells, we were able to detect two of the shared mutations at a very low frequency (<1 in 500 cells) in the starting MEF pool; it is clear that this small subset of MEFs was more likely to undergo reprogramming than the ‘average’ MEF cell in the pool. Although the MEFs used in experiment 3 had a slightly higher overall reprogramming efficiency than those from experiment 1 (1/7,500 cells vs. 1/14,000 cells, see Experimental Procedures for details), many more clones would have to be sequenced to define the actual reprogramming advantage of the cells with the shared mutations. However, a relatively small improvement in efficiency (e.g. 5–10 fold) could probably have yielded the observed results, since we only sampled 6 clones. Although we do not know whether any of the shared mutations contributed to reprogramming fitness, sequencing the genomes of large numbers of iPSC clones from independent pools of starting cells may allow for the identification of recurrently mutated genes (or pathways) in iPSC clones; these data could potentially help to elucidate the genetic barriers to reprogramming, and provide novel approaches for improving the efficiency of the process.
Shared mutations in “sister” iPSC clones from the same reprogramming experiment have also recently been reported by Gore, et al. These authors performed whole exome sequencing of 22 human iPSC lines that were created with a variety of reprogramming approaches (including 4-factor retroviral and lentiviral vectors, 3-factor retroviral vectors, episomal plasmids, and messenger RNA). Among this set, seven pairs of iPSC lines were generated from the same parental cells; in three of the seven pairs, the authors detected shared SNVs (1–3 shared mutations were detected, along with a few private mutations in each of the members of the pair). None of these mutations was detected in other human iPSC clones, and none were detected in our set of shared mutations. Even though exome sequencing of iPSC clone pairs can detect only a small subset of the mutations found by whole genome sequencing, it is striking that 3 out of 7 sets of iPSC clones derived from the same parental cells had shared variants. When combined with our data, 4 of 10 sets of iPSC clones tested had shared variants; clearly this is not a rare event. Although the significance of the shared mutations is unknown, they do not appear to provide an overall selective advantage for cells before they are reprogrammed (since they are rare in the starting population). Although one or more of the shared mutations may cooperate with reprogramming factors to provide a selective advantage for the reprogramming, it is formally possible that none of the shared mutations are relevant for reprogramming fitness: they simply may be markers for rare cells that are fit for another reason (e.g. they may have stem-like properties (Yamanaka, 2009)). Many additional studies will be required to discern among these possibilities.
As noted above, we evaluated variant allele frequencies in the iPSC clones to help understand the timing of mutational events. The variant allele frequencies of most clones averaged 50% (Figures 3A and 5), suggesting that most variants are heterozygous, and present in nearly all of the cells in the sample. In experiment 3, we further examined the variant allele frequencies of the shared vs. private mutations in each clone (Figure 5). The shared variants had to be present prior to reprogramming, and had an average variant allele frequency of 50%, as expected. The private variants also had a major peak of variant allele frequencies at 50%, but one clone was confirmed to contain a minor, secondary peak at ~25% that represents a subclone. Clearly, this subclone must have arisen after reprogramming, since it is not present in all cells in the sample. Since we sampled clones at a single time in their existence, we do not know whether this late subclone was stable, rising, or falling in relation to the founding clone; serial sampling would be required to resolve this question. However, these results do suggest that iPSC clones may evolve with serial passaging, as previously reported (Laurent et al., 2011).
Based on this information, we propose a model of how mutations are acquired during iPSC cell development (Figure 6). Most of the time, individual iPSC clones arise from random, unique cells within the transduced parental cell population. Each cell has a unique set of pre-existing background mutations, which are fixed by the cloning of single cells (Figure 6A); it is formally possible that rare cells contain background mutations that provide improved fitness for reprogramming (which could help to explain why reprogramming is inherently inefficient), but we did not find evidence for a common pathway targeted by these mutations. However, there are some instances where pre-existing mutations are associated with a cell’s reprogramming fitness. These cells are more likely to be reprogrammed and detected as iPSC clones (Figure 6B). Between the time when the shared mutations and the reprogramming event occurs, additional private mutations are acquired (which may or may not further increase reprogramming efficiency). Finally, additional mutations can be acquired after reprogramming that can provide a selective advantage for the outgrowth of subclones.
Figure 6. Model for the acquisition of genetic variants during iPSC reprogramming.
Panel A. Model for the genetic variants detected in experiments 1 and 2. Each cell has a unique set of pre-existing background mutations, which are “captured” by the expansion and cloning of single cells with reprogramming. Not all cells transduced with the reprogramming vector yield iPSC clones, as suggested by the X’s. It is possible that the background mutations in some cells could contribute to reprogramming fitness.
Panel B. Model for the data obtained for experiment 3. Preexisting background mutations in rare cells mark their fitness for reprogramming. Private mutations must be acquired after the shared mutations, and could have arisen at different times: 1) between the time when the shared mutations were acquired and when reprogramming occurred, or 2) at the time of reprogramming, or 3) after reprogramming. Some mutations arising after reprogramming may provide a selective advantage for the outgrowth of subclones.
These data provide several important caveats for the field of somatic cell reprogramming. By cloning individual somatic cells, background genetic variants will invariably be fixed by the act of cloning, regardless of the strategy used to induce reprogramming. If fitness mutations are present in rare somatic cells, they could potentially cooperate with reprogramming factors, regardless of how the factors are delivered (stably integrated viruses, transiently expressed plasmids, mRNAs, or proteins). Although this may have important consequences for the use of iPSC clones in therapeutic settings, the sequencing of large numbers of iPSC clones may also provide new strategies for identifying genes that represent barriers for reprogramming, and suggest approaches to safely overcome them. Perhaps most importantly, these results strongly suggest that somatic cell reprogramming may not be a mutagenic event per se, and that knowledge gained from sequencing iPSC genomes may help to refine the process and make it safer for therapeutic purposes.
Experimental Procedures
Production of iPSC clones
We generated iPSC clones from fibroblasts obtained from three different mouse strains, fully described in the Results section. iPSC clones were generated from each line as previously described (Chang et al., 2009). Briefly, ~3×105 fibroblasts were seeded on 6 well plates and allowed to grow overnight. The next day, the cells were transduced with the OSK lentivirus at an MOI of 3–5:1. The cells were incubated with virus for 48 hours, trypsinized, and transferred to a 100-mm dish without a feeder MEF layer. Cells were grown for 2–3 weeks with daily media changes. After 17 days (Experiments 1 and 3) or 26 days (Experiment 2), individual colonies were picked and expanded on MEF feeder layers. For experiment 1, 48 iPSC colonies were identified from 675,000 transduced MEFs (1 in 14,000 cells). For experiment 2, 12 colonies were identified from 675,000 transduced TTFs (1 cell in 57,000). For experiment 3, 120 colonies were identified from 900,000 transduced MEFs (1 cell in 7,500).
Pluripotency characterization of iPSC clones
All ten iPSC clones were assessed for ES cell-like morphology and stained for alkaline phosphatase according to the manufacturer’s instructions (Millipore, Billerica, MA). Cells were analyzed by flow cytometry for the pluripotency markers SSEA-1, Nanog and Oct4. For intracellular Nanog and Oct4 detection, cells were fixed with 4% paraformaldehyde, and permeabilized with 1% saponin prior to incubation with antibodies.
llumina whole genome shotgun library construction
Libraries were prepared for whole genome sequencing essentially as described [35]. For exome sequencing, the sequencing libraries were hybridized with the SureSelectXT Mouse All Exon Kit (Agilent), which captures 49.6 Mb of the coding sequence from ~24,000 genes. KAPA qPCR was used to determine the quantity of library necessary to produce cluster counts appropriate for the Illumina HiSeq 2000.
Illumina sequencing
After diluting the libraries to a 10pM concentration, we utilized the paired-end flow cell and cluster generation kits to produce flow cells with an average cluster density ranging between 1.9 – 3 million clusters per tile. We employed the standard sequencing kits (Illumina HiSeq Sequencing Kit) and performed 2 × 100 cycles of nucleotide incorporation using a paired-end read approach. We used the Illumina sequencing pipeline, version 1.7 or 1.8, to analyze the data and produced files containing high quality (“passed filter”) reads with associated quality values.
Clonality analysis
Clonality estimates were determined using the mutation allele frequencies from whole genome sequencing. To minimize the effect of coverage outliers from likely false positives, we pre-filtered each site to ensure that the coverage fell within ±2 median absolute deviations from the median coverage of all non-repetitive predictions within each clone. We drew a kernel density estimate (KDE) plot for variant allele frequencies using the density function in “R”. A custom R function evaluated each KDE plot to determine the number of significant peaks, which served as an estimation of the number and relative composition of different sub-clones present within each iPSC clone.
To confirm the presence of subclones and better estimate their frequency, we randomly selected 10 sites from each major subclone (between 45–55% variant allele frequency) and each minor subclone (between 25–35% variant allele frequency). These sites were amplified by PCR from the original sample, and then sequenced on the 454-FLX platform to a median depth of 6,322 reads. A Student’s t-test was used to assess the significance of the difference in frequencies between the major and minor clones.
Significantly mutated gene analysis and pathway analysis
Since the mutational process in clones is analogous to the mutational process of a tumor, we used components of the Mutational Significance in Cancer (MuSiC) package (N. Dees et al., submitted) to determine significantly mutated genes (SMG) and pathways. The SMG component of MuSiC assigns mutations to various categories, such as transition or transversion, and then uses methods including convolution, Fisher’s test, and a likelihood test to combine the category-specific binomials to obtain an overall P-value. The result is appreciably more accurate than if these attributes were disregarded. The pathway analysis component of MuSiC is an implementation of PathScan (Wendl et al., 2011).
Sequence Analysis
Reads were aligned using BWA 0.5.5 (Li and Durbin, 2009) with quality trimming set at 5 to the NCBI build 37 of Mus musculus reference sequence augmented with an additional contig representing the complete OSK sequence. The resulting alignments were de-duplicated using Picard (http://picard.sourceforge.net) and quality recalibrated using the GATK (DePristo et al., 2011) base quality recalibrator (v1.0.3471). All 13 genomes, along with a 129/SvJ reference genome, were submitted to the SRA database under accession number SRP011044.
We called potential somatic single nucleotide variants using a modified version of SomaticSniper (Larson et al., 2011) to account for perfectly pure samples. Variants with a somatic score greater than 10 were filtered on a number of sequence features to remove false positives as described elsewhere (Koboldt et al., 2012). Sites passing these filters were additionally filtered to remove variants where the difference between the clone and control variant allele frequency was 30% or less and the read depth was 10 reads or less.
The software and parameters used for identifying somatic variants were as follows:
Recalibration
GATK v1.0.3471(--max_reads_at_locus 20000) with covariates for ReadGroup, QualityScore, Cycle, and Dinucleotide context.
Deduplication
Picard1.25 (Experiments 2 and 3) or Picard1.36 (Experiment 1)
SNV calling
bam-somaticsniper (Unreleased version. v1.0.0 should be similar) -P -f -q 1 -Q 10 –s 0.000001
Indel Calling
GATK(v1.0.5336) IndelGenotyperV2 --somatic -window_size 300
SV calling
BreakDancer(v1.1_20100719) breakdancer_max -t -q 10 –d
SquareDancer(unreleased; v0.1r177) squaredancer -q 25 -r 2 -k 25 -n 1 -c 1 -m 3 -e 0.5
CNV Calling
CNVHMM was described in Wartman, et al. (Wartman et al., 2011). Software has not been released.
ReadCount bam-readcount 0.3 -b –q. This software has not been released.
SNV Filtering
Readcount data was generated as described above, and filtered identically to the parameters in Koboldt, et al. (Koboldt et al., 2012). Those are:
Max average mapping quality difference (variant-reference) of 30
Max difference (variant-reference reads) of the mean mismatch quality sum of 50
Max difference (variant-reference) of average clipped read length of 25
Minimum flanking homopolymer length 5
Minimum average absolute distance to the end of the read (1- (absolute value of the distance to the center of the clipped read/center of the clipped read) of 0.1
Minimum fraction plus strand of variant reads (number of plus strand reads/total reads) of 0.01
Maximum fraction plus strand of variant reads of 0.99
Minimum number of variant reads of 4
Minimum average distance of variant to the effective 3′ end of 0.2
Minimum variant base frequency of 0.05
Non-genic variants in repetitive regions (as identified from the UCSC Genome Browser’s RepeatMasker track for mouse build 37) were also excluded from further analysis (Karolchik et al., 2011); 43.5% of the genome was excluded from our variant analysis.
Variants passing all filters were annotated as previously described (Wartman et al., 2011).
For experiment 3, sites chosen for validation were manually reviewed and validated using 454-FLX sequencing as previously described (Ding et al., 2010). Read counts were calculated by excluding reads where the called base was below a phred quality of 15 for WGS data, with a mapping quality of 10 or a base quality less than 20 for the high depth Illumina data, and for a base quality less than 20 for the 454-FLX data.
Based on 454 validation experiment we performed for 84 sites in experiment 3, and applying a cutoff of >20% variant allele frequency in the clone and <10% in the MEF sample, the estimated true positive rate (specificity) is 78.6% for calls filtered for greater than 10X depth and variant allele frequency difference of greater than 30%. Applying these filters resulted in an estimated sensitivity of 85% based on the same 454 validation data of unfiltered sites. We have previously described the false negative rate (sensitivity) in Larson, et al. (Larson et al., 2011) and our simulations suggest a sensitivity of > 99.9% for variants with 50% variant allele frequency when we have 30X haploid coverage.
Indels were called using the GATK (DePristo et al., 2011). OSK insertion sites were predicted using BreakDancer (Chen et al., 2009) and SquareDancer, an in-house implementation of the CREST algorithm (Wang et al., 2011). Copy number predictions were generated as previously described (Wartman et al., 2011) and manually reviewed to determine their accuracy.
Detection of shared variants in parental Gusb −/− MEFs
Rare variants were detected in the parental MEFs by novel StuI sites are generated by the Apaf1 G16A and Sbno2 A3783G variants. Regions surrounding each variant were amplified (Apaf1: 884 bp amplicon, 5′-CCCAAACACTTTGATGAACGA-3′ and 5′-CTATAAGGACCTTGCTGCGC-3′; Sbno2: 806 bp amplicon, 5′-GCTGCAGACTGACACA- GGAG-3′ and 5′-AGCAGAGGCTCCCATGACTA-3′). PCR products were cloned into the pCR2.1 vector according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). The number of clones in individual transformations was counted on each plate, and then all colonies were pooled into a single sample (see Table S3 for pool sizes), creating mini-libraries of amplicons from the parental MEF cell DNA. These mini-libraries were then digested with EcoRI to cut the amplicon from the pCR2.1 vector, and StuI to detect the presence of the variant allele (Apaf1: 391 and 493 bp products; Sbno2: 454 and 352 bp products). Digestion products were electrophoresed on 5% acrylamide gels and transferred to nitrocellulose. Nitrocellulose blots were hybridized with 32P-labeled probes (Apaf1: the 493 bp product; Sbno2: the 352 bp product), washed stringently, and subjected to autoradiography.
Supplementary Material
Highlights.
iPSC clones contain hundreds of SNVs that are unique to each clone
Most iPSC genomes do not contain recurrently mutated genes or pathways
Reprogramming can select for rare cells with shared genetic variants
Most SNVs are probably preexisting mutations “captured” by cloning
Acknowledgments
The authors thank the Siteman Cancer Center Embryonic Stem Cell Core, the Molecular and Genomic Analysis Core, and the Flow Cytometry Core for their expert contributions to this work. We would like to acknowledge the Analysis Pipeline group at the Genome Institute for developing the automated sequence analysis pipelines and Joshua McMichael for his assistance in figure design. We thank Drs. Suellen Greco, Jeffery Klco, and Trenton Shoeb for carefully reviewing the teratoma pathology. This work was funded by grants to Richard K. Wilson from the National Human Genome Research Institute (NHGRI U54 HG003079), to Tim M. Townes from the National Heart Lung and Blood Institute (HL057619), and to Timothy J. Ley from the National Institutes of Health (CA0101937 and DK38682) and the Barnes-Jewish Hospital Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Carey BW, Markoulaki S, Hanna J, Saha K, Gao Q, Mitalipova M, Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Lai YS, Pawlik KM, Liu K, Sun CW, Li C, Schoeb TR, Townes TM. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008;Chapter 10(Unit 10):11. doi: 10.1002/0471142905.hg1011s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles A, Meglecz E, Pech N, Ferreira S, Malausa T, Martin JF. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics. 2011;12:245. doi: 10.1186/1471-2164-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008a;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008b;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Narva E, Ng S, Sourour M, Hamalainen R, Olsson C, et al. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Ji J, Ng SH, Sharma V, Neculai D, Hussein S, Sam M, Trinh Q, Church GM, McPherson JD, Nagy A, et al. Elevated Coding Mutation Rate During the Reprogramming of Human Somatic Cells into Induced Pluripotent Stem Cells. Stem Cells. 2011 doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Kent WJ. The UCSC Genome Browser. Curr Protoc Hum Genet. 2011;Chapter 18(Unit18):16. doi: 10.1002/0471142905.hg1806s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012 doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, Ley TJ, Mardis ER, Wilson RK, Ding L. SomaticSniper: Identification of Somatic Point Mutations in Whole Genome Sequencing Data. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol Cell Biol. 2005;25:4625–4637. doi: 10.1128/MCB.25.11.4625-4637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins-Taylor K, Nisler BS, Taapken SM, Compton T, Crandall L, Montgomery KD, Lalande M, Xu RH. Recurrent copy number variations in human induced pluripotent stem cells. Nat Biotechnol. 2011;29:488–491. doi: 10.1038/nbt.1890. [DOI] [PubMed] [Google Scholar]
- Meng XL, Shen JS, Kawagoe S, Ohashi T, Brady RO, Eto Y. Induced pluripotent stem cells derived from mouse models of lysosomal storage disorders. Proc Natl Acad Sci U S A. 2010;107:7886–7891. doi: 10.1073/pnas.1002758107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Park IH, Lerou PH, Zhao R, Huo H, Daley GQ. Generation of human-induced pluripotent stem cells. Nat Protoc. 2008;3:1180–1186. doi: 10.1038/nprot.2008.92. [DOI] [PubMed] [Google Scholar]
- Pawlak M, Jaenisch R. De novo DNA methylation by Dnmt3a and Dnmt3b is dispensable for nuclear reprogramming of somatic cells to a pluripotent state. Genes Dev. 2011;25:1035–1040. doi: 10.1101/gad.2039011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Boland MJ, Leibowitz ML, Shumilina S, Pehrson SM, Baldwin KK, Hall IM. Genome Sequencing of Mouse Induced Pluripotent Stem Cells Reveals Retroelement Stability and Infrequent DNA Rearrangement during Reprogramming. Cell Stem Cell. 2011;9:366–373. doi: 10.1016/j.stem.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castella M, Rio P, Sleep E, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands MS, Birkenmeier EH. A single-base-pair deletion in the beta-glucuronidase gene accounts for the phenotype of murine mucopolysaccharidosis type VII. Proc Natl Acad Sci U S A. 1993;90:6567–6571. doi: 10.1073/pnas.90.14.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Feng W, Sun Y, Bai H, Liu J, Currie C, Kim J, Gama R, Wang Z, Qian Z, et al. Generation of iPS cells using defined factors linked via the self-cleaving 2A sequences in a single open reading frame. Cell Res. 2009;19:296–306. doi: 10.1038/cr.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Wang J, Mullighan CG, Easton J, Roberts S, Heatley SL, Ma J, Rusch MC, Chen K, Harris CC, Ding L, et al. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods. 2011;8:652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren ST, Schultz RA, Chang CC, Wade MH, Trosko JE. Elevated spontaneous mutation rate in Bloom syndrome fibroblasts. Proc Natl Acad Sci U S A. 1981;78:3133–3137. doi: 10.1073/pnas.78.5.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartman LD, Larson DE, Xiang Z, Ding L, Chen K, Lin L, Cahan P, Klco JM, Welch JS, Li C, et al. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J Clin Invest. 2011;121:1445–1455. doi: 10.1172/JCI45284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendl MC, Wallis JW, Lin L, Kandoth C, Mardis ER, Wilson RK, Ding L. PathScan: a tool for discerning mutational significance in groups of putative cancer genes. Bioinformatics. 2011;27:1595–1602. doi: 10.1093/bioinformatics/btr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wu G, Liu N, Rittelmeyer I, Sharma AD, Sgodda M, Zaehres H, Bleidissel M, Greber B, Gentile L, Han DW, et al. Generation of healthy mice from gene-corrected disease-specific induced pluripotent stem cells. PLoS Biol. 2011;9:e1001099. doi: 10.1371/journal.pbio.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zheng TS, Hunot S, Kuida K, Momoi T, Srinivasan A, Nicholson DW, Lazebnik Y, Flavell RA. Deficiency in caspase-9 or caspase-3 induces compensatory caspase activation. Nat Med. 2000;6:1241–1247. doi: 10.1038/81343. [DOI] [PubMed] [Google Scholar]
- Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.