Summary
The AAA ATPase p97 and its UBA-UBX cofactors are thought to extract ubiquitinated proteins from membranes or protein complexes as a prelude to their degradation. However, ubiquitinated targets have not yet been identified for many cofactors, leaving their biological function unclear. Previous analysis has linked the p97 pathway to Cullin-RING ubiquitin Ligases (CRLs); here we demonstrate that the p97 cofactor UBXD7 mediates the p97-CRL interaction through its conserved ubiquitin-interacting motif (UIM). UBXD7, and its yeast ortholog Ubx5, associate only with the active, NEDD8- or Rub1-modified form of cullins. Disruption of the Ubx5 UIM motif results in a loss of CRL binding and consequently impedes degradation of a Cul3 substrate. These results uncover an unexpected and conserved role for NEDD8 in linking CRL ubiquitin ligase function to the p97 pathway.
The abundant homohexameric AAA ATPase, p97/VCP (Cdc48 in yeast), participates in a wide range of cellular processes, including cell cycle regulation, endoplasmic reticulum (ER) associated degradation (ERAD), membrane fusion, and autophagy 1. In many of these processes, p97 is thought to recognize ubiquitinated substrates and separate them from tightly bound partner proteins. Substrate specificity is established through interactions with a plethora of p97 cofactors. In humans, the largest group of cofactors consists of at least 13 proteins that interact with the N-terminal region of p97 through an ubiquitin regulatory X (UBX) domain. Five of these proteins (p47, UBXD7, UBXD8, FAF1, and SAKS1) also have an ubiquitin-binding (UBA) domain, classifying them as UBA-UBX proteins. A recent proteomic analysis revealed that in addition to binding ubiquitin conjugates, UBA-UBX proteins interact with over two dozen ubiquitin ligases2, including several members of the Cullin–RING ubiquitin Ligase (CRL) family.
CRLs are multisubunit complexes comprising three core components – a RING finger protein, a cullin and, with the exception of CUL3 based CRLs, a cullin-specific adaptor protein 3. The latter binds interchangeable substrate specificity factors, which in turn recruit substrates for ubiquitination. For example the CUL1 adaptor SKP1 recruits over 42 different F-box proteins to CUL1 4,5, whereas Elongin C recruits 41 BC-box proteins to CUL2 and CUL5 6. CRL activity is stimulated following the covalent attachment of an ubiquitin-like molecule, NEDD8, to a conserved lysine residue in cullin 7,8, and continuous neddylation and deneddylation cycles are required for the proper regulation of CRL function9.
With up to 240 complexes in human cells, CRLs constitute the largest group of ubiquitin E3 ligases, accounting for >40% of all ubiquitin ligases and ~20% of protein degradation via the proteasome 10. For p97 this could mean an expansion in potential ubiquitylated substrates that require its function for their degradation. However it is currently unclear how p97 is recruited to CRLs, so we examined the interactions between UBA-UBX proteins and CRLs. We found that only UBXD7 specifically associated with the neddylated form of CRLs and this involved a direct interaction between its conserved UIM and the conjugated NEDD8 on CRLs. This UIM-NEDD8 interaction is conserved in yeast and contributes to CRL substrate degradation.
RESULTS
UBXD7 preferably binds CUL2 and CUL4
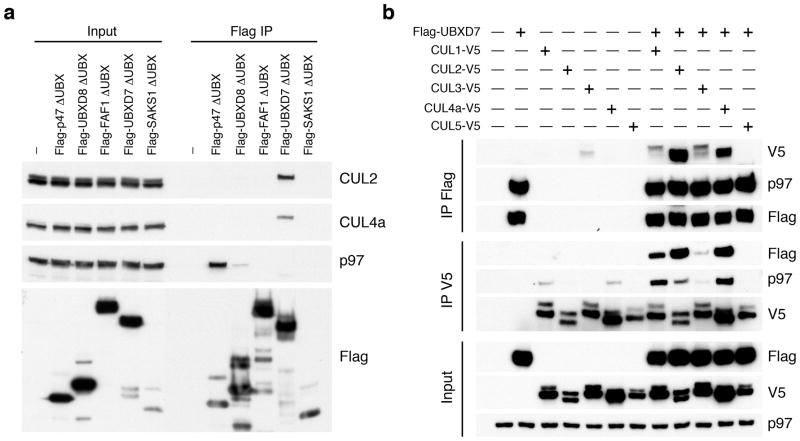
UBA-UBX adaptor interactions with CRLs may be mediated indirectly via p97. To understand how the p97 network is connected to CRLs, we examined whether CRL binding is specific for a certain UBA-UBX adaptor. We deleted the UBX domain from Flag-tagged versions of the five human UBA–UBX domain proteins (p47, UBXD8, FAF1, UBXD7 and SAKS1) to minimize cross-association with other p97-bound proteins. The expressed proteins were recovered by immunoprecipitation (IP) and evaluated by immunoblotting. As expected, only p47-ΔUBX, which has a second p97 contact site11, retained its ability to interact with p97 (Fig. 1a). Unexpectedly, only UBXD7-ΔUBX interacted with endogenous CUL2 and CUL4a.
Figure 1. UBXD7 associates with all cullins except CUL5.
(a) Flag-tagged UBA-UBX proteins (p47, UBXD8, FAF1, UBXD7 and SAKS1) lacking the UBX domain (ΔUBX) expressed in 293T cells were immunoprecipitated (IP) with anti-Flag antibodies and probed to detect endogenous binding partners CUL2, CUL4, and p97 as indicated. -, no transfection.
(b) Same as in a, except cells were transfected with (+) or without (−) vectors encoding Flag-UBXD7 and V5 epitope-tagged CUL 1 through 5. Immunoprecipitations were carried out with antibodies against the Flag or V5 epitope.
To assess the cullin binding preference of UBXD7, V5-tagged cullin constructs (CUL1-5) were co-expressed with Flag-tagged UBXD7. Even though similar levels of p97 were found in association with UBXD7, strikingly different amounts of V5 tagged cullins were recovered (Fig. 1b). UBXD7 displayed the most efficient binding towards CUL2, CUL4a, and CUL4b (Fig. 1b and Supplementary Fig. 1) followed by weaker interaction with CUL1 and CUL3 and no interaction with CUL5. The UBXD7 binding preference for endogenous CUL2 and CUL4 was confirmed in a reciprocal pull-down. Interestingly, even though cullins were present in input lysates in both neddylated and unneddylated forms, UBXD7 appeared to associate with only a single species. Taken together, our data confirm UBXD7 as a CRL binding partner, consistent with previous studies6,12.
UBXD7 interacts exclusively with the active form of Cullins
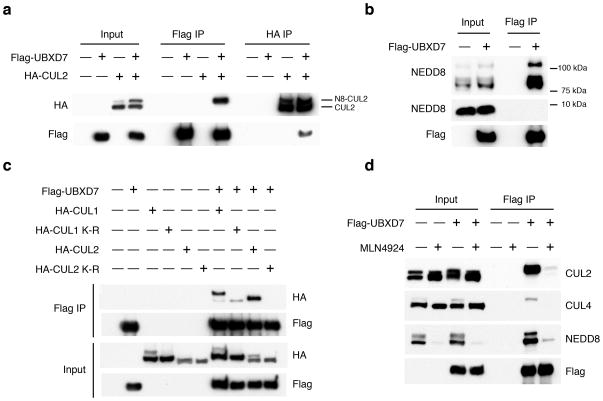
To determine whether UBXD7 associated with active or inactive CRLs, we co-expressed Flag-UBXD7 with HA-CUL2. Expression of UBXD7 resulted in a slight increase in neddylated HA-CUL2 (Fig. 2a). This may arise from the ability of UBXD7 to inhibit deneddylation of CUL1 by CSN in a purified system (R.J.D. and E. Emberley, unpublished data). However despite the presence of more unneddylated than neddylated HA-CUL2 in the lysate, UBXD7 exclusively bound the neddylated form. Because of the apparent selectivity for neddylated cullins, we also probed Flag-UBXD7 pull-downs with NEDD8 specific antibodies, and detected multiple endogenously neddylated species in the cullin size range, but no unconjugated NEDD8 (Fig. 2b).
Figure 2. The UBXD7-CRL interaction is neddylation dependent.
(a–b) 293T cells were transfected with (+) or without (−) the indicated expression constructs and immunoprecipitated (IP) with anti-Flag or anti-HA antibodies prior to being probed with the antibodies indicated. N8-CUL2, NEDD8-conjugated CUL2.
(c) Same as in a. HA-CUL1 (K/R) contains the K720R substitution and HA- CUL2 (K/R) contains the K689R substitution.
(d) Same as above, except 1 hour prior to cell lysis, cells were treated with the NEDD8 conjugation inhibitor, MLN4924.
We next examined whether cullin neddylation mediates the CRL-UBXD7 interaction. Neddylation-deficient CUL1 K720R and CUL2 K689R mutants exhibited minimal UBXD7 binding (Fig. 2c), and inhibition of NEDD8 conjugation via the NEDD8-E1 inhibitor MLN492410 resulted in accumulation of deneddylated CUL2 and CUL4a proteins that failed to bind UBXD7 (Fig. 2d). Altogether these results clearly identified UBXD7 as a neddylation-dependent CRL binding protein.
UBXD7 interacts with neddylated Cullins via its UIM
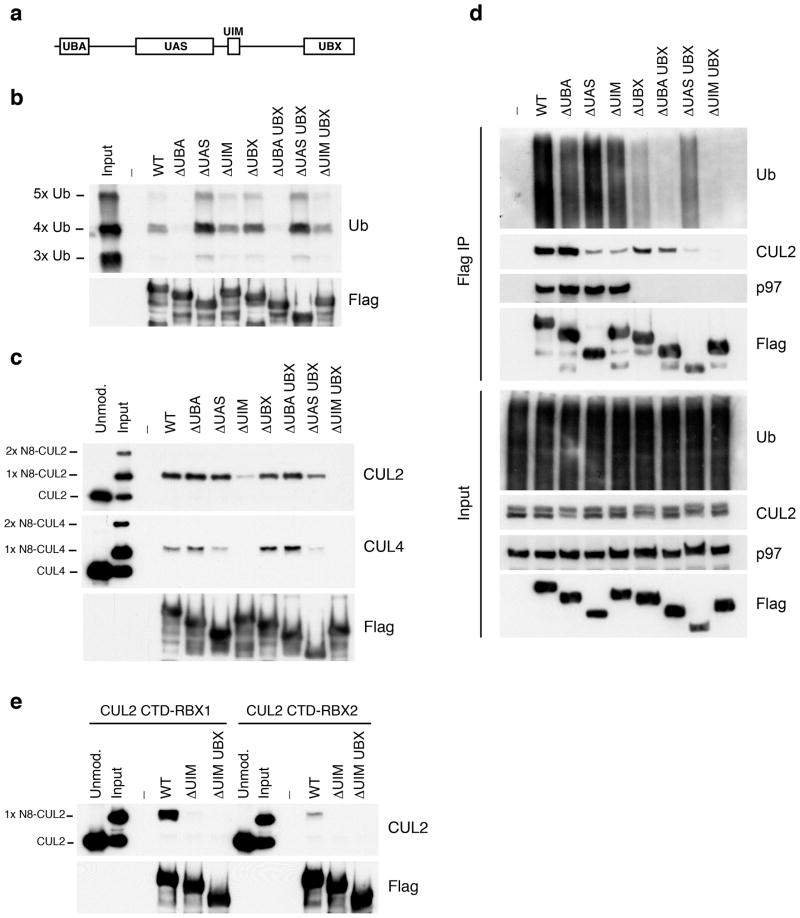
Because UBXD7 has ubiquitin-binding UBA domain and UIM (Fig. 3a), it could be attracted to neddylated CRLs via their bound, ubiquitinated substrates. Using pull-down assays with purified K48-linked ubiquitin chains and Flag-tagged UBXD7 mutant proteins, we found that the UBA domain and not the UIM contributed to polyubiquitin binding (Fig. 3b). Next, we looked at binding to purified CUL2 and CUL4a in the absence of polyubiquitin chains. When mixtures with equal amounts of neddylated and unneddylated cullins were incubated with recombinant Flag-UBXD7, only the neddylated forms were recovered after Flag pull-down (Fig. 3c). This interaction largely depended on the UIM; deletion of the UAS domain had a more modest effect on CUL4a but not CUL2 binding. By contrast, removal of the UBA or the UBX domains had no effect. The UIM dependence was also seen with recombinant CUL1-RBX1 (Supplementary Fig. 2a) and CUL3-RBX1 complexes (data not shown). Additional mapping experiments showed that a fragment containing just the UBXD7 UIM plus surrounding sequences bound neddylated CUL2 (Supplementary Fig. 2b).
Figure 3. UBXD7 directly interacts with neddylated CRLs via its UIM.
(a) Wild type UBXD7 protein, indicating its domains. UBA, ubiquitin-associated domain, UAS, UAS/Thioredoxin-like fold domain, UIM, ubiquitin-interacting motif, UBX, ubiquitin regulatory X domain.
(b) Recombinant wild-type Flag-UBXD7 or a deletion mutant was incubated with K48-linked polyubiquitin chains prior to immunoprecipitation and western blotting with indicated antibodies. Nx Ub refers to ubiquitin chains of increasing length.
(c) A mix (input) of unneddylated and neddylated recombinant full-length CUL2–RBX1 or CUL4a-RBX1 complex was incubated with recombinant WT Flag-tagged UBXD7 or deletion mutants. Following IP with anti-Flag antibodies, recovered proteins were detected by western blotting with indicated antibodies. Nx N8-CUL, cullin modified with 1 or 2 molecules of NEDD8.
(d) 293T cells were transfected with full length Flag-tagged UBXD7 or the indicated UBXD7 deletion mutants and treated with MG132. Lysates were immunoprecipitated with anti-Flag antibodies, and co-precipitated endogenous proteins were detected by western blotting with the indicated antibodies.
(e) Binding assays were performed as in (c) using a mix of unneddylated and neddylated recombinant CUL2 CTD–RBX1 or CUL2 CTD–RBX2 complex and recombinant WT Flag-tagged UBXD7 or deletion mutants.
To evaluate further the role of UBXD7’s four domains, the constructs employed above were expressed in 293 cells and their binding to endogenous CUL2, p97, and ubiquitin conjugates was determined (Fig. 3d). The results of this experiment supported our hypothesis that the UIM mediates assembly of UBXD7 with CUL2, in that the ΔUIM mutant exhibited a large reduction in binding to endogenous CUL2, a moderate reduction in binding to ubiquitin conjugates, and normal binding to p97. On the other hand, this experiment also yielded unexpected results: both the ΔUAS and ΔUBX mutants exhibited binding defects that were not seen with purified proteins (the former with CUL2 and the latter with ubiquitin conjugates). We do not understand the basis for these defects, which, based on the in vitro data, are likely to be indirect. Nevertheless, the ubiquitin-binding defect of the ΔUBX mutant points to a connection between p97 recruitment and ubiquitination of UBXD7 targets that merits future investigation.
In addition to neddylation, other features of the CRL complex can influence UBXD7 binding. Swapping the RING subunit RBX1 with the closely related RBX2 greatly diminished UIM-dependent binding of UBXD7 to neddylated CUL2 C-terminal domain (Fig. 3e). Moreover, mutation of a basic surface in CUL1 that mediates recruitment of the E2 enzyme CDC34 also reduced binding of UBXD7 (Supplementary Figure 2a). By contrast, deletion of CUL2’s N-terminal domain had little or no impact on UBXD7 recruitment (Supplementary Fig. 2c).
The UIM of UBXD7 associates with conjugated NEDD8
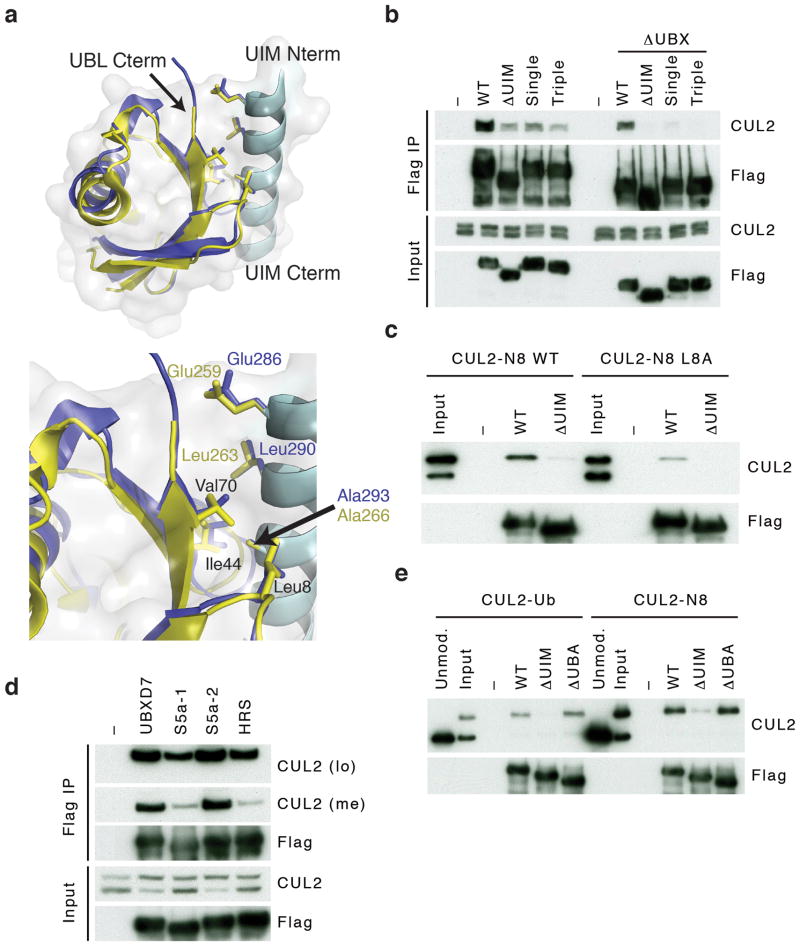
The UIM is a ~20 amino acid sequence motif 13 that forms a single α-helix and hydrophobic residues within the helix interact with the Leu8–Ile44–Val70 ‘hydrophobic patch’ of ubiquitin 14,15. Interestingly, these three residues are conserved in NEDD8 and form a hydrophobic surface identical to the one on ubiquitin 16, which could potentially be recognized by a UIM. Sequence alignment confirmed that UBXD7 contained the conserved residues characteristic for a UIM (Supplementary Fig. 3). Given the selectivity of UBXD7 for neddylated CRLs, we explored the possibility of a direct interaction between NEDD8 and the UIM of UBXD7. We used the crystal structure of the UIM of hepatocyte growth factor-regulated tyrosine kinase substrate (HRS) bound to ubiquitin as a template 17, and superimposed the structures of ubiquitin with NEDD8 and the HRS UIM with the UIM of UBXD7 (Fig. 4a). The resulting UBXD7 UIM-NEDD8 model was computationally refined using Rosetta Dock 18. The final low-energy model showed that residues in the UIM of HRS and the structurally equivalent residues in UBXD7 made similar contacts with ubiquitin and NEDD8 respectively.
Figure 4. The UIM of UBXD7 binds conjugated NEDD8 on CUL2.
(a) Modeling of the UBXD7 UIM - NEDD8 interaction using the HRS UIM -ubiquitin crystal structure as a template (PDB code 2D3G, yellow)17. The UBXD7 UIM in association with NEDD8 is shown in blue. This figure was made in PYMOL.
(b) Cells transfected with Flag-UBXD7 (wild type or ΔUBX) with a wild type, deleted (ΔUIM), or substitution mutant UIM were immunoprecipitated with anti-Flag beads and probed with the indicated antibodies.
(c) CUL2 was neddylated with wild type (WT) NEDD8 or L8A mutant NEDD8 and binding assays with Flag-UBXD7 were performed as described in Fig. 3c.
(d) Same as in (c), except transfections were carried out with wild type Flag-UBXD7 or Flag-UBXD7 in which the UIM was replaced with the UIM of HRS or the first (S5a-1) or second (S5a-2) UIM of S5a. Immunoprecipitations were carried out in either a low (CUL2 (lo)) or medium (CUL2 (me)) stringency binding buffer.
(e) Immunoprecipitation of recombinant wild type Flag-tagged UBXD7 or the indicated UBXD7 deletion mutants from a mix (input) containing recombinant full-length CUL2–RBX1 either unmodified or modified with monoubiquitin or NEDD8.
To validate this, we generated single (A293Q) and triple (E286R, L290E, A293Q) substitution mutants, in either full length UBXD7 or UBXD7-ΔUBX (Fig. 4b and Supplementary Fig. 3). Both mutants, but particularly UBXD7-ΔUBX, bound less endogenous neddylated CUL2 in a pull-down assay (Fig. 4b). Conversely, purified CUL2–RBX1 neddylated with a NEDD8 hydrophobic patch mutant protein (N8-L8A) showed a reduced binding affinity for purified UBXD7 (Fig. 4c). This decrease in association was not due to a change in the NEDD8-induced conformation because a mutant CUL1ΔWHB–RBX1 complex that spontaneously adopts the active conformation without neddylation 8,19 did not bind UBXD7 (Supplementary Fig. 4). Together these results support the idea that formation of a UBXD7–CRL complex is stabilized by a direct interaction between conjugated NEDD8 and the UIM of UBXD7.
Next we tested whether the UIM of UBXD7 is unique in its ability to recognize NEDD8 by replacing it with UIMs from the ubiquitin-binding proteins HRS or the proteasomal subunit S5a. In low stringency binding conditions (Fig. 4d, CUL2 (lo)) little difference was seen in the amount of recovered CUL2. However, when the stringency was increased (Fig. 4d, CUL2 (me)) both HRS UIM and the first UIM of S5a lost almost all their CUL2 binding ability. In contrast, the second UIM of S5a (S5a-2) was equivalent to UBXD7’s UIM.
The UIM replacement experiment suggested that the UIM of UBXD7 is not NEDD8 specific but rather that the recognition of NEDD8 is context dependent. This predicts that replacing conjugated NEDD8 on CUL2 with ubiquitin would not affect UBXD7 binding. The E2 enzyme UBCH5c can transfer ubiquitin onto the NEDD8 acceptor lysine of CUL1 and this mimics the activating effect of neddylation 7,8. Using conditions that favor this monoubiquitination reaction, we generated a mixture that contained both unmodified and monoubiquitinated CUL2 (Fig. 4e input). UBXD7 selectively bound monoubiquitinated CUL2 in a pull-down assay with an efficiency that was comparable to that seen for neddylated CUL2. Importantly, this interaction was dependent on the UIM and unaffected by deletion of the UBA domain.
The UIM of Ubx5 is required for the degradation of Rpb1
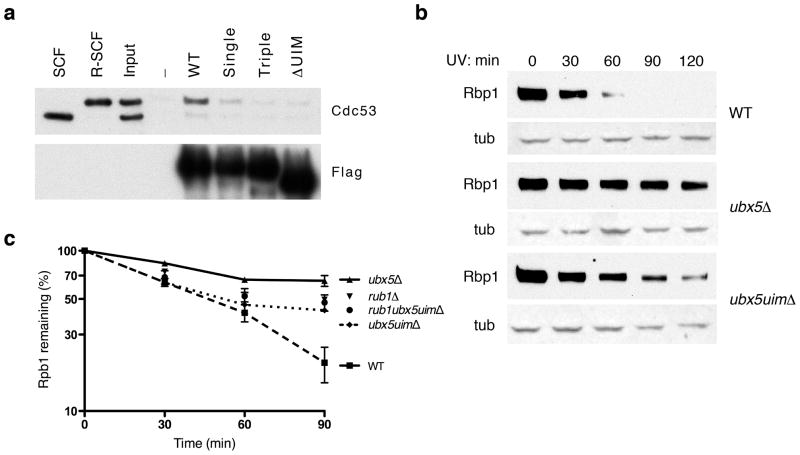
To address whether the association between UBXD7 UIM and NEDD8-conjugated cullins contributes to degradation of CRL substrates we turned to Saccharomyces cerevisiae where the first, and so far only, UBX-dependent CRL substrate has been described (other established CRL and p97-dependent substrates, including CDT1 (data not shown), are not dependent on UBXD7). We recently reported that UV induced, Cul3-dependent proteolysis of the large subunit of RNA polymerase II (Rpb1) depends on the Cdc48 cofactor Ubx5 20. Ubx5, like UBXD7, contains UBA, UAS, UBX, and UIM domains (Supplementary Fig. 5a and b), which is consistent with the suggestion that it is the yeast equivalent of mammalian UBXD7 21. Moreover, Ubx5 binds yeast Cul3 20, which associates with ElonginC and therefore is functionally most closely related to human CUL2/CUL5 22. To test directly whether Ubx5 binds yeast cullins in a manner dependent on Rub1 modification, we incubated purified Flag-Ubx5 protein with a 1:1 mixture of unmodified SCFCdc4 and SCFCdc4 modified with the yeast NEDD8 ortholog, Rub1. SCFCdc4 consists of yeast CUL1 (Cdc53) and Rbx1 (Hrt1), Skp1, and the F-box protein Cdc4. Analogous to UBXD7, Ubx5 only bound to rubylated Cdc53 and this interaction was disrupted by deletion or point mutation of the UIM domain (Fig. 5a).
Figure 5. The UIM in yeast Ubx5 promotes UV-dependent degradation of Rpb1.
(a) A mix (input) of unmodified SCF and rubylated recombinant SCF (R-SCF) was incubated with recombinant wild type Flag-tagged Ubx5 or the various Ubx5 mutants. Following immunoprecipitation with anti-Flag beads, the recovered proteins were probed with the indicated antibodies. Single and triple mutants are A368Q and E361R, M365E, A368Q respectively.
(b) Yeast cultures (wild type, ubx5Δ, ubx5uimΔ) were irradiated with UV and total cell lysates were prepared at the indicated times prior to detection by western blotting using antibodies against Rpb1 and tubulin (tub).
(c) Similar as in (b) except western blots were carried out with IR dye-linked secondary antibodies and quantified by LI-COR Odyssey following normalization with tubulin. Error bars show s.e.m., n=3 for each genotype.
To assess the role of Ubx5’s UIM domain we compared UV-induced degradation rates of Rpb1 in wild type, ubx5Δ, and a yeast strain, ubx5uimΔ, in which the UIM domain of endogenous UBX5 was eliminated by homologous recombination. Whereas Rpb1 was rapidly degraded in wild type cells, its degradation was delayed in ubx5uimΔ and further impaired in an ubx5Δ strain (Fig. 5b). Importantly, tagging the endogenous loci with a myc epitope confirmed that both wild type and Ubx5ΔUIM proteins were properly folded and expressed at identical levels (Supplementary Fig. 5c and d).
The intermediate effect on Rpb1 degradation in the ubx5uimΔ strain was also observed in a rub1Δ strain 23 suggesting that Cul3, Rub1, and the UIM domain of Ubx5 function in a common pathway. To address this directly, we generated an rub1Δ ubx5uimΔ strain and performed Rpb1 degradation studies. The single mutant rub1Δ behaved identical to the rub1Δ ubx5uimΔ strain, indicating an epistatic relationship between these mutations (Fig. 5c). These results are consistent with a functional, rubylation-dependent interaction between Ubx5 and cullins and demonstrate a role for the Ubx5 UIM domain in promoting degradation of Rbp1 in response to UV radiation.
DISCUSSION
In our efforts to understand how the p97 pathway is linked to CRLs we discovered that the UBA-UBX protein UBXD7 selectively associated with neddylated cullins. UBXD7 is the only p97 adaptor with an UIM, and this motif enables UBXD7 and its yeast ortholog Ubx5 to bind neddylated cullins.
Several lines of evidence indicate that the UIM–NEDD8 interaction, though critical, is insufficient by itself to mediate the binding of UBXD7 to neddylated CRLs. This is not surprising as UIM–ubiquitin interactions are typically of low affinity (KD >100 μM)24. We propose that weak interactions between other sequences in UBXD7 and surfaces of the CRL that become exposed upon neddylation place the UIM in proper register to bind NEDD8. In this manner, the UIM–NEDD8 interface stabilizes a multidentate interaction between UBXD7 and active, neddylated CRLs. In support of this hypothesis, UBXD7’s UIM can be swapped for a canonical ubiquitin-binding UIM or NEDD8 can be replaced by ubiquitin, with little or no effect on UBXD7–CRL association. The exact nature of the rest of the UBXD7 binding surface remains unknown, but we note two things: first, it is likely to reside adjacent to the UIM–NEDD8 interface because the UIM plus flanking sequences are sufficient to bind neddylated CUL2 (Supplementary Fig. 2b), and second, UBXD7 is acidic (pI ~5), which could facilitate interaction with the ‘basic canyon’ in cullins25. Mutating the basic canyon impairs UBXD7 binding (Supplementary Figure 2a) while retaining ubiquitin ligase activity25. Full resolution of the details of UBXD7–CRL interaction awaits a crystal structure.
The NEDD8-dependent recruitment of UBXD7 biases the p97 pathway to engage CRLs that are active and potentially engaged in substrate ubiquitination. However this raises the question as to how UBXD7–p97 targets are selected. Our data point to some selectivity with respect to the cullin, with UBXD7 preferentially interacting with CUL2 and CUL4. The reason for this preference is unclear but could be related to differences in sequence or subcellular localization, possibly regulated by post-translational modification. For instance, two proteomics studies identified UBXD7 as a target of the ATM/ATR pathway 26,27 which fits well with the known function of mammalian CUL4 in DNA replication and DNA damage signaling and repair 28. However, UBXD7 associated with polyubiquitin conjugates in the absence of radiation, suggesting that not all targets are DNA damage pathway specific (Fig. 3d).
Additional control over recruitment could come from the substrate itself. CRL substrates with tightly folded domains, substrates that are part of multisubunit assemblages, or substrates associated with subcellular structures (eg chromatin) might require p97 unfoldase activity for efficient proteasomal degradation. We posit that when the proteasome encounters a difficult-to-resolve structure, the rate of degradation slows. According to this model, a temporarily stalled neddylated CRL–polyubiquitinated substrate–proteasome complex might comprise a signal that attracts UBXD7, and the lifetime of such a stalled complex would determine the statistical likelihood that the UBXD7-p97 pathway is engaged. For cullin complexes whose substrates don’t require p97 for degradation, the cycle of neddylation, substrate ubiquitination/degradation, and deneddylation might occur very quickly, providing limited opportunity for UBXD7 to bind.
Our data point to a positive role for the UBXD7 ortholog Ubx5 in the degradation of polyubiquitinated RNA polymerase II stalled at UV-induced lesions. However, we wish to note that all three determinants of the CRL complex (neddylation, Rbx1, and the basic canyon) that are important for UBXD7-CRL interaction also contribute to recruitment of CDC34, raising the possibility that UBXD7 might antagonize CRL activity. Interestingly, UBXD7 modestly inhibited SCFβ-TrCP/CDC34-dependent ubiquitination of a β-catenin peptide in vitro (G. K., unpublished data). If UBXD7 can function as a CRL antagonist in some contexts, it could explain our prior observation that levels of the CRL2VHL substrate HIF1alpha are reduced in UBXD7-depleted cells 2. Studies on the CRL regulators COP9 Signalosome (CSN) and CAND1 have revealed that these factors, which inhibit CRL activity in vitro, paradoxically behave as positive regulators in cells 29. UBXD7 may possess a similar dual nature that manifests itself depending upon the substrate and the manner in which it is assayed.
Our study revealed an unexpected role for NEDD8. First as an activating signal, NEDD8 conjugation causes a conformational change in the cullin, eliminating the CAND1 binding site and locking the enzyme in an active state 7,8. We propose that following this conformational change, NEDD8 and the newly exposed surfaces on cullin and RBX1 recruit UBXD7/Ubx5 which in turn links p97/Cdc48 to the CRL. For substrates whose dislocation from other factors and subsequent proteolysis are strongly dependent on p97/Cdc48 activity, recruitment of UBXD7/Ubx5 via NEDD8 promotes degradation. Selective recruitment of UBXD7 to nedddylated CRLs may also be employed in some circumstances to restrain their activity. Uncovering additional CRL substrates that engage the p97 pathway will be required to gain deeper insight into the full range of biological functions regulated by UBXD7.
METHODS
Cell culture conditions
Human kidney 293T cells were maintained in DMEM (BioWhittaker) supplemented with 10% (v/v) fetal bovine serum (Atlanta Biologicals), 4 mM L-glutamine, and 100 units of each penicillin and streptomycin (Invitrogen) in a 5% CO2-humidified incubator. Where it is indicated, cells were treated with 1 μM MLN4924 (Millennium Pharmaceuticals) for 1 hour or with 20 μM MG132 (EMD Biosciences) for 3 hours.
Expression Constructs
Constructs were generated using standard techniques and verified by DNA sequencing. Cloning details and primer sequences are provided in the supplementary methods. Constructs for the expression of V5 tagged Cullin 1–5 were provided by Thilo Hagen 30. All constructs used in this study are listed in the Supplementary Table 1 with their corresponding Deshaies laboratory database (RDB) number.
Immunoprecipitations and immunoblotting
Cells were lysed in either buffer A (50 mM HEPES pH 7.5, 5 mM Mg(OAc)2, 70 mM KOAc, 0.2% (v/v) Triton X-100, 10% (v/v) glycerol, and 0.2 mM EDTA) or EBC buffer (50 mM Tris at pH 8.0, 150 mM NaCl, 0.1 mM EDTA and 0.5% (v/v) NP-40) containing protease inhibitors (Protease Inhibitor Cocktail, Sigma) and phosphatase inhibitors (10 mM β-glycerolphosphate, 1 mM NaF, and 0.1 mM NaVO4). For Figure 4d, buffer A and EBC buffer are the low and medium stringency buffers respectively. Cleared lysates were immunoprecipitated with antibodies directed against the following epitopes, Flag (M2-agarose, Sigma), HA (3F10-agarose, Roche Applied Science), or V5 (V5-agarose, Sigma) and after 1–2 hours the resin was washed 3 times with lysis buffer. Recovered proteins were eluted from the beads by boiling in 2x Laemmli sample buffer, separated by SDS-PAGE and transferred to PVDF membrane (Immobilon-P, Millipore). Proteins were detected using antibodies to Flag (M2-HRP, Sigma), HA (3F10-HRP, Roche), V5 (V5-HRP, Sigma), p97 (H-120, Santa Cruz), CUL1 (71-8700, Zymed/Invitrogen), CUL2 (51-1800, Zymed/Invitrogen), CUL4a (2527-1, Epitomics), NEDD8 (Millenium Pharmaceuticals)10, Ubiquitin (SPA-200, Stressgene) and Cdc53 (y-300, Santa Cruz).
Protein expression and purification
Recombinant proteins for CUL2-RBX1 complex, Flag-UBXD7 and Flag-Ubx5 (WT, domain deletion, single and triple mutants) were expressed in bacteria and purified by standard methods. Detailed protocols are in supplementary methods. Procedures for purification of NEDD8 31, NEDD8-E2 (UBCH12) 31, NEDD8-E1 (APPBP1/UBA3) 31, Split-n-Co-express CUL1 32, UBCH5c 7, human ubiquitin E1 7, UBA1 (Yeast ub-E1) 33, yeast Cdc34 25, and SCFCdc4 34 were described previously. The purified proteins: CUL2CTD-RBX1, CUL2CTD-RBX2, Split-n-Coexpress CUL4a, CUL1ΔWHB, and NCE2 were generously provided by David Duda and Brenda Schulman (St. Jude Children’s Research Hospital).
Neddylation conditions and in vitro binding assay
Cullins were neddylated by incubating Cullin–RBX1 complexes (300 nM) with NEDD8-E1 (250 nM), UBCH12 (2 μM), and NEDD8 (10 μM) in 30 mM Tris-Cl pH 8.0 containing 2 mM ATP, 1 mM DTT and 5 mM MgCl2. For neddylating CUL2-RBX2 complexes, UBCH12 was substituted by NCE2/UBE2F. These neddylation reactions were complete in 5 minutes at 25°C and resulted in a 1:1 mixture of unneddylated:neddylated Cullin–RBX complex. Monoubiquitination of the NEDD8 acceptor lysine in CUL2 was carried out by mixing Ub-E1 (250 nM), UBCH5c (1 μM), CUL2-RBX1 (500 nM), and ubiquitin (20 μM) in 30 mM Tris-Cl pH 8.0 containing 2 mM ATP, 2 mM DTT and 5 mM MgCl2. Reactions were incubated at 25 °C for 1 hour and stopped by the addition of 100-fold excess of binding buffer. These reaction conditions resulted in a 1:1 mix of unmodified: mono-Ub CUL2.
Recombinant Flag-UBXD7 protein (1μM) was mixed with 10 nM Cullin–RBX (directly taken from neddylation reaction) in binding buffer A (50 mM HEPES pH 7.5, 5 mM Mg(OAc)2, 70 mM KOAc, 0.2% (v/v) Triton X-100, 10% (v/v) glycerol, and 0.2 mM EDTA). Binding reactions were carried out at 4 °C for 1 hour followed by 1 hour in the presence of anti Flag-beads (M2-agarose, Sigma). Beads were collected by centrifugation at 2,000 g for 1 min, washed 3 times with binding buffer and boiled in 2x Laemmli sample buffer. Samples were separated by SDS-PAGE and transferred to PVDF membrane (Immobilon-P, Millipore). Proteins were detected using antibodies against CUL1 (71-8700, Zymed/Invitrogen), CUL2 (51-1800, Zymed/Invitrogen), CUL4a (2527-1, Epitomics) and Flag (M2-HRP, Sigma).
Synthesis of K48-linked ubiquitin chains and in vitro binding assay
K48-linked ubiquitin chains were synthesized as previously described 35 using 0.8 μM yeast ubiquitin E1 (Uba1), 10 μM yeast Cdc34, and 0.7 mM ubiquitin (Sigma) in Tris buffer (30 mM Tris, 5 mM MgCl2, 200 mM NaCl, 2 mM DTT, 40 μM ATP) and incubated at 42°C for 16 hours. The mixture was bound and eluted from MonoQ resin using a linear salt gradient with a final concentration of 800 mM NaCl. Peak fractions containing tetra ubiquitin chains were pooled and dialyzed into Tris and stored at −80°C.
For in vitro binding assay with ubiquitin chains, Flag-UBXD7 protein (1 μM) was incubated with 400 nM Ub chains in binding buffer A. Binding reactions were carried out at 4 °C for 1 hour followed by 1 hour in the presence of anti Flag-beads (M2-agarose, Sigma). Beads were collected by centrifugation at 2,000 g for 1 min, washed 3 times with binding buffer and boiled in 2x Laemmli sample buffer. Samples were separated by SDS-PAGE and transferred to PVDF membrane (Immobilon-P, Millipore). Proteins were detected using antibodies against ubiquitin (SPA-200, Stressgene) and Flag (M2-HRP, Sigma).
Yeast transformation and genomic integration
Yeast integration vector, pRS306-UBX5UIMΔ (RDB2610), was linearized with EcoRI and transformed into the W303 yeast strain. Transformants that grew on SD plates lacking uracil were analyzed by PCR to confirm correct integration at the UBX5 locus using the following forward primer (5′-tcccacggttaatgaacctcttcc) and reverse primer (5′-tcgtgttataagtgcttacatacc). Yeast strains with the correct integration were counter selected on 5-FOA and surviving clones were screened for uracil auxotrophy and genomic DNA was analyzed by PCR for UIM deletion using the following primers; forward (5′-tcccacggttaatgaacctcttcc), reverse (5′-gtacgcacagtgtcttccagagc).
The chromosomal UBX5 open reading frame was tagged with myc epitopes according to 36, using the following primers; forward (5′-gcttaaaaaatagttctttactacttgagaagcttgaccctgaaatagaacggatccccgggttaattaa), reverse (5′-ttacatacctaattacatctaggtacctgccaccatacaatttgtgaattcgagctcgtttaaac).
Rbp1 degradation assay
Rbp1 degradation assays were performed as described 20.
Supplementary Material
Acknowledgments
We thank Michael Rome for providing K48-linked polyubiquitin chains, Nathan Pierce for purified SCFCdc4, Thilo Hagen (National University of Singapore) and Ethan Emberley for cullin expression constructs, David Duda and Brenda Schulman (St. Jude Children’s Research Hospital) for providing purified recombinant CUL2, CUL3, CUL4a, CUL1ΔWHB and NCE2 proteins, Steven Lewis for help with the Rosetta Dock server, Millennium Pharmaceuticals Inc for MLN4924 and NEDD8 antibody, and the members of the Deshaies laboratory for helpful discussion during the course of this work. This work was supported by the Howard Hughes Medical Institute (HHMI) and a National Institute of Health Ruth Kirschstein Postdoctoral Fellowship (F32 GM088975; WdB). R.J.D. is an HHMI Investigator.
Footnotes
AUTHOR CONTRIBUTION
W.d.B. and R.J.D. conceived and designed the experiments. W.d.B. performed most of the experiments, except G.K. performed the structural modeling in Fig 3a and R.V. and R.O. made the yeast strains and carried out the Rbp1 turnover studies in Fig. 4b and Fig. 4c. W.d.B. and R.J.D. wrote the manuscript with editorial input from the other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Ye Y. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Alexandru G, et al. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 4.Jin J, et al. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JE, et al. The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahrour N, et al. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem. 2008;283:8005–8013. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- 7.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosu DR, Kipreos ET. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008;3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 11.Uchiyama K, et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitinligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann K, Falquet L. A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- 14.Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 15.Fisher RD, et al. Structure and ubiquitin binding of the ubiquitin-interacting motif. J Biol Chem. 2003;278:28976–28984. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- 16.Whitby FG, Xia G, Pickart CM, Hill CP. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J BiolChem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 17.Hirano S, et al. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat Struct Mol Biol. 2006;13:272–277. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- 18.Gray JJ, et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 19.Yamoah K, et al. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci U S A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol Cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribar B, Prakash L, Prakash S. ELA1 and CUL3 are required along with ELC1 for RNA polymerase II polyubiquitylation and degradation in DNA-damaged yeast cells. Mol Cell Biol. 2007;27:3211–3216. doi: 10.1128/MCB.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabut G, et al. The TFIIH Subunit Tfb3 Regulates Cullin Neddylation. Mol Cell. 2011;43:488–495. doi: 10.1016/j.molcel.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 25.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 27.Stokes MP, et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci U S A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higa LA, Zhang H. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2007;2:5. doi: 10.1186/1747-1028-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 30.Chew EH, Hagen T. Substrate-mediated regulation of cullin neddylation. J BiolChem. 2007;282:17032–17040. doi: 10.1074/jbc.M701153200. [DOI] [PubMed] [Google Scholar]
- 31.Huang DT, Schulman BA. Expression, purification, and characterization of the E1 for human NEDD8, the heterodimeric APPBP1-UBA3 complex. Methods Enzymol. 2005;398:9–20. doi: 10.1016/S0076-6879(05)98002-6. [DOI] [PubMed] [Google Scholar]
- 32.Li T, Pavletich NP, Schulman BA, Zheng N. High-level expression and purification of recombinant SCF ubiquitin ligases. Methods Enzymol. 2005;398:125–142. doi: 10.1016/S0076-6879(05)98012-9. [DOI] [PubMed] [Google Scholar]
- 33.Petroski MD, Deshaies RJ. In vitro reconstitution of SCF substrate ubiquitination with purified proteins. Methods Enzymol. 2005;398:143–158. doi: 10.1016/S0076-6879(05)98013-0. [DOI] [PubMed] [Google Scholar]
- 34.Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Pickart CM. A 25-kilodalton ubiquitin carrier protein (E2) catalyzes multi-ubiquitin chain synthesis via lysine 48 of ubiquitin. J Biol Chem. 1990;265:21835–21842. [PubMed] [Google Scholar]
- 36.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.