Abstract
Alveolar rhabdomyosarcoma is an aggressive pediatric cancer of the myogenic lineage with frequent chromosomal translocations involving the PAX3 or PAX7 and FOXO1 genes. Based on previous studies indicating that the fusion genes are amplified in a subset of these cancers, we conducted a comprehensive molecular and clinical investigation of these amplification events. Using oligonucleotide arrays to localize amplicons, we found that the minimal 1p36 amplicon measured 0.13 Mb and only contained PAX7 whereas the minimal 13q14 amplicon measured 0.53 Mb and contained FOXO1 and the poorly characterized LOC646982 gene. Application of a fluorescence in situ hybridization assay to over 100 fusion-positive cases revealed that the fusion gene is amplified in 93% of PAX7-FOXO1-positive and 9% of PAX3-FOXO1-positive cases. While most cells in amplified PAX7-FOXO1-positive cases contained the amplicon, only a fraction of cells in the amplified PAX3-FOXO1-positive cases contained the amplicon. Expression studies demonstrated that the fusion transcripts were generally expressed at higher levels in amplified cases, and that the PAX7-FOXO1 fusion transcript was expressed at higher levels than the PAX3-FOXO1 fusion transcript. Finally, fusion gene amplification and PAX7-FOXO1 fusion status were each associated with significantly improved outcome; a multivariate analysis demonstrated that this predictive value was independent of other standard prognostic parameters. These findings therefore provide further evidence for a novel good prognosis subset of fusion-positive rhabdomyosarcoma.
INTRODUCTION
Rhabdomyosarcoma (RMS) is a family of soft tissue cancers that are related to the skeletal muscle lineage and frequently occur in children and young adults (Barr and Womer, 2009). RMS is composed of two major histologic subtypes, embryonal (ERMS) and alveolar (ARMS) (Parham and Ellison, 2006). As a category, ARMS tumors differ from ERMS tumors by frequent occurrence in the extremities and a worse prognosis, which is associated with a propensity for early metastasis, poor response to treatment and frequent relapse.
At the genetic level, chromosomal translocation events frequently occur in tumors of the ARMS subtype (Barr and Womer, 2009). The most frequent translocation is a t(2;13)(q35;q14) and there is also a less common variant t(1;13)(p36;q14) that occurs in a smaller subset of cases. These 2;13 and 1;13 translocations generate PAX3-FOXO1 and PAX7-FOXO1 fusion genes, respectively, which encode novel fusion transcription factors with oncogenic activity.
Although these chromosomal translocation events appear to be important events in RMS pathogenesis, genome-wide screens also indicate the frequent occurrence of gene amplification events in ARMS tumors (Pandita et al., 1999; Gordon et al., 2000; Bridge et al., 2002). As gene amplification provides an important mechanism for increasing oncogene copy number and expression, the associated overexpressed proteins are postulated to collaborate with the fusion oncoproteins generated by the translocation during oncogenesis. Our recent studies elucidated the molecular basis of the amplicons derived from the 2p24, 12q13-q14, and 13q31 chromosomal regions (Barr et al., 2009; Reichek et al., 2011). Each of these amplicons is associated with specific gene fusion subtypes and a differing clinical outcome. For example, the 12q13-q14 amplicon identified a poor prognostic subset of PAX3-FOXO1-positive tumors.
In earlier studies, we and others found amplification of PAX7-FOXO1 or less commonly PAX3-FOXO1 in ARMS tumors (Barr et al., 1996; Weber-Hall et al., 1996; Davis and Barr, 1997). These genetic events correspond to amplicons localized to 1p36, 2q34-qter, and 13q14 chromosomal regions in previous genome-wide screens. In this report, we use DNA-based arrays to localize the amplified region in RMS tumors and identify the consistently involved genes on genetic maps. The rearrangement and amplification status of numerous tumors are assessed with fluorescence in situ hybridization (FISH) assays. These findings are compared with quantitative RT-PCR assays, which analyze gene fusion status, subtype and expression. Finally, the clinical significance of gene fusion amplification and subtype is investigated by comparing these molecular findings with pathologic, molecular, and clinical data.
MATERIALS AND METHODS
Tumor Cases
In this study, 201 frozen tissue samples were received from the Children’s Oncology Group (COG) Soft Tissue Sarcoma (STS) Tumor Bank. Subsets of this 201 sample collection were used in different parts of this study, and are summarized in Supplemental Table 1. All samples were reviewed at the time of original submission by pathologists affiliated with the COG STS Committee using existing criteria (Newton et al., 1995). Based on more recent changes, only cases with greater than 50% alveolar histology were included in this study. Although all these cases received a review diagnosis of ARMS, subsequent re-review of cases with available slides determined that a subset do not meet the current criteria for ARMS (Teot et al., manuscript submitted). Therefore, the above-described 201 cases will be considered a convenience sample of RMS cases that is enriched for ARMS cases. Our analysis will not specifically consider histologic subtype but will instead use fusion subtype as a basic system for categorizing these RMS cases. All 201 cases will be analyzed in this study, but since this study is specifically investigating fusion gene amplification status, we will focus predominantly on the fusion-positive RMS cases and will use the fusion-negative RMS cases as a comparison group.
Of the 201 RMS samples in this study, 166 samples were derived from patients enrolled on Intergroup Rhabdomyosarcoma Study (IRS) Group or COG STS clinical trials. These cases were distributed among the following trials: IRS IV pilot (n = 3), IRS IV (n = 47), IRS V pilot (n = 4), D9501 (n = 7), D9502 (n = 7), D9602 (n = 9), D9802 (n = 20), and D9803 (n = 69). The various trials from which these cases are drawn have similar chemotherapy control arms with comparable outcomes. In addition, the experimental arms did not show different outcomes when compared with the control arms. Therefore there is justification for combining the study populations of these 166 cases for outcome analysis in the present study. As for the remaining 35 of the original 201 samples, data from the corresponding cases were used to analyze various issues but not outcome in the present study,
This molecular correlative study was approved by the University of Pennsylvania Institutional Review Board.
DNA Preparation and Microarray Analysis
Genomic DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA), in accordance with the manufacturer’s instructions. Following quantification, DNA was analyzed on Affymetrix GeneChip Human Mapping 50K XbaI arrays of the 100K Mapping Panel (Affymetrix, Santa Clara, CA), following the manufacturer’s instructions. All analyses were performed in the University of Pennsylvania’s Microarray Core Facility. To further define the minimum region of amplification, selected samples were analyzed on Affymetrix GeneChip Human 250K StyI arrays of the 500K mapping panel. Data from the 50K and 250K oligonucleotide arrays were analyzed with Copy Number Analysis Tool (Affymetrix) and Copy Number Workflow (Partek Genomics Suite, St. Louis, MO), respectively, as previously described (Barr et al., 2009).
FISH
Interphase FISH was performed on touch imprints made from snap frozen or OCT-embedded tumor samples. To assay for 13q14 rearrangement and/or amplification, slides were hybridized with the Vysis LSI FOXO1 Dual Color Break Apart Probe (Abbott Molecular, Des Plaines, IL). This probe set consists of a SpectrumOrange-labeled probe lying 5′ of the FOXO1 gene and a SpectrumGreen-labeled probe lying 3′ of the FOXO1 gene. For each assay, cells were stained with a DAPI II counterstain (Abbott Molecular). The signals in a minimum of 100 tumor cells were counted. Based on a comparision of the FISH and RT-PCR data in this study, rearrangement is defined as a splitting of red and green signals in more than 17% of cells whereas splitting in less than 13% of cells is considered not rearranged. Cases with 13–17% splitting frequency are considered indeterminate for FOXO1 rearrangement. Amplification is defined as >10 green (3′) signals within a cell. Typically >20 signals were observed in amplified cells with this probe.
Quantitative RT-PCR
RNA was isolated from primary tumors using RNA STAT-60 (Tel-Test, Friendswood, TX) in accordance with manufacturer’s instructions. Fusion status was initially determined by RT-PCR analysis in all 201 cases, as previously described (Barr et al., 2009); the fusion status was indeterminate by RT-PCR analysis in two of these cases. In 144 of the initial 201 cases (of which 142 were derived from patients enrolled on IRS or COG STS clinical trials), the presence of the PAX3-FOXO1 and PAX7-FOXO1 fusion transcripts was detected and quantified with a modification of our previously published quantitative RT-PCR protocol (Table 1) (Barr et al., 2006). Following reverse transcription from a FOXO1-specific primer (FKHR-RT), the assay consists of PCR with consensus 5′ PAX3/PAX7 (PAX3/7-3) and 3′ FOXO1 (FKHR-R) primers and detection of PCR products with gene-specific PAX3 (3F3) and PAX7 (7F) probes. This arrangement thus determines both the presence and subtype of the fusion in a single assay. In a second reaction, expression of the 18S rRNA gene was quantified (using 18S primers and probe) to normalize the results and provide a final expression ratio. Consistent with its role as a reference gene, there is no significant difference in 18S rRNA expression between PAX3-FOXO1- and PAX7-FOXO1-positive tumors (data not shown). This 18S assay also assesses the quality of the RNA by comparing expression to a standard curve of a control ARMS cell line. To determine which negative gene fusion results were meaningful and to control for variability between runs, we set an 18S cut-off equivalent to the level in 0.5 ng of control RNA as evidence of a satisfactory sample. All assays were performed using an ABI Prism 7900 Sequence Detection System (Applied Biosystems, Carlsbad, CA).
Table 1.
Probes and Primers
| Name | Type | Sequence |
|---|---|---|
|
| ||
| FKHR-RT | RT Primer | CTG-GAT-TGA-GCA-TC |
| PAX3/7-3 | Forward Primer | CCT-CCA-ACC-MCA-TGA-ACC-C |
| FKHR-R | Reverse Primer | CCT-TCA-TTC-TGC-ACA-CGA-ATG-A |
| 3F3 | Probe | VIC-TGG-CAA-TGG-CCT-CTC-ACC-TCA-GAA-TT-TAMRA |
| 7F | Probe | 6FAM-AGC-AAC-GGC-CTG-TCT-CCT-CAG-AAT-TCA-TAMRA |
| FKHR-F1 | Forward Primer | CAA-TTC-GTC-ATA-ATC-TGT-CCC-TAC-A |
| FKHR-R1 | Reverse Primer | GCA-TCC-ACC-AAG-AAC-TTT-TTC-C |
| FKHR-P1 | Probe | 6FAM-AGC-AAG-TTC-ATT-CGT-GTG-CAG-AAT-GAA-GG-TAMRA |
| FKHR-F2 | Forward Primer | GCT-CAC-GCT-GTC-GCA-GAT-C |
| FKHR-R2 | Reverse Primer | TGC-TGT-CAC-CCT-TAT-CCT-TGA-A |
| FKHR-P2 | Probe | 6FAM-CGA-GTG-GAT-GGT-CAA-GAG-CGT-GCC-TAMRA |
| 18S-F | Forward Primer | CGG-CTA-CCA-CAT-CCA-AGG-AA |
| 18S-R | Reverse Primer | GCT-GGA-ATT-ACC-GCG-GCT |
| 18S-P | Probe | 6FAM-TGC-TGG-CAC-CAG-ACT-TGC-CCT-C-TAMRA |
| TTL2-F | Forward Primer | TCC-TGA-TTA-AGG-CAT-GCA-GTC-A |
| TTL2-R | Reverse Primer | CCC-TGG-CTC-CGT-CAG-TCT-T |
| TTL2-P | Probe | 6FAM-AAT-GGG-CCT-CAT-GGT-GGA-GCT-TAMRA |
We modified the above qRT-PCR assay to determine fusion transcript expression on a common scale for both PAX3-FOXO1 and PAX7-FOXO1 fusions (Table 1). To directly measure fusion transcript expression, the assay consists of reverse transcription with FKHR-RT, PCR with the PAX3/7-3 primer and a new further 3′ FOXO1 primer (FKHR-R1) followed by detection of PCR products with a 3′ FOXO1-specific probe (FKHR-P1). The second 18S control and normalization reaction is performed as described above.
Statistical Analyses
The unpaired Student’s t-test was used to compare the means of two groups with continuous measurements (e.g., gene expression). To compare means across more than two groups, a one-way ANOVA approach was used with Scheffe’s post-hoc test for pairwise comparisons (conducted in SPSS 12, SPSS Inc., Chicago, IL). To determine the effect of two factors - each with multiple levels - on the continuous measurement, a two-way ANOVA approach was used with Neuman-Keul’s post-hoc test for pairwise comparisons (conducted in SigmaStat Software 2.033, SPSS Inc.). The relationship between fusion status and amplification level was examined by Fisher’s exact test with Bonferroni’s post-hoc test for pairwise comparisons (conducted in SAS 9.2 software, SAS, Inc.). The Pearson correlation coefficient was calculated to evaluate the correlation between two continuous measurements. For all the above continuous responses, if the assessment of normal distributions failed, an appropriate transformation (e.g., logarithm) was implemented before the analyses.
For survival analyses, Kaplan-Meier curves were drawn for the variable with multiple groups, and the non-parametric log-rank statistic was used to assess the significance of outcome across groups. The Cox proportional hazards models were further fitted to evaluate the association of interesting variables with outcome in univariate (i.e., with only the variable of interest as the covariate) and multivariate analyses (i.e., with the variable of interest and other confounders).
RESULTS
Microarray Analysis of Amplicons in RMS Tumors
In our studies of amplification in RMS, we have been using a 50K oligonucleotide array dataset to analyze copy number changes in a panel of 57 RMS tumors selected from the larger 201 tumor set based on abundant frozen material, a representative distribution of fusion subtypes, and clinical trial eligibility (Supplemental Table 1). The fusion status of this panel is 31 PAX3-FOXO1-positive, 9 PAX7-FOXO1-positive, 16 fusion-negative, and 1 indeterminate fusion status (Barr et al., 2009; Reichek et al., 2011). In these 57 cases, we found a recurrent 13q14 amplification event in 9 cases including 1 PAX3-FOXO1-positive and 8 PAX7-FOXO1-positive cases. In addition, we also identified a recurrent 1p36 amplification event in 7 of the 8 PAX7-FOXO1 positive cases with the 13q14 amplicon. In the PAX3-FOXO1-positive case with a 13q14 amplicon, there was a 2q35 amplification event. Based on our previous studies of fusion gene amplification (Barr et al., 1996; Davis and Barr, 1997), we postulated that the 1p36, 2q35, and 13q14 amplicons involve the PAX3, PAX7, and FOXO1 genes.
To localize the minimum common region of 1p36 amplification, we identified the 1p36 amplicon endpoints in each amplified tumor. By comparing these endpoints, we initially situated the minimum common region of amplification to a 1.77 Mb segment (Figure 1A). To further delineate the minimal common amplified region, we analyzed four selected amplified cases on higher density 250K oligonucleotide arrays. We also analyzed the PAX7-FOXO1-positive case for which the 50K array detected a 13q14 amplicon but did not detect a 1p36 amplicon; a small 1p36 amplicon was detected on the 250K array in this latter case. The minimum overlapping 1p36 amplicon shared among these amplified cases was thereby reduced to 0.13 Mb. Comparison of this interval with the human genome map (NCBI Build 36.2) revealed that the minimal common amplified region contains only the PAX7 gene. Furthermore, the localization is sufficient to identify that the minimal region only contains the 5′ region of PAX7 and thus is consistent with amplification of the PAX7-FOXO1 fusion gene. Based on the genomic map, the 5′ PAX7 region is closer to the telomere and the 3′ PAX7 region is closer to the centromere.
Figure 1. Localization of the minimal common amplified regions in 1p36 (A) and 13q14 (B) chromosomal regions.
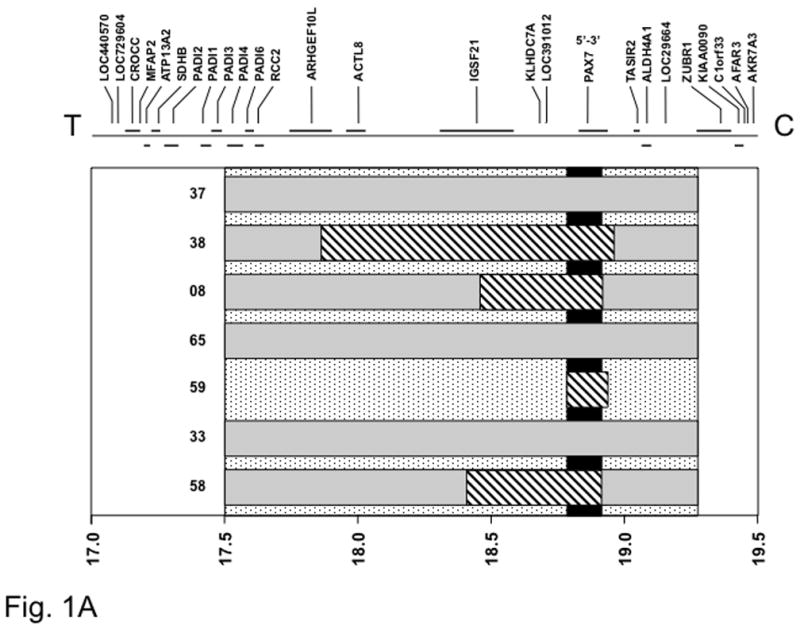
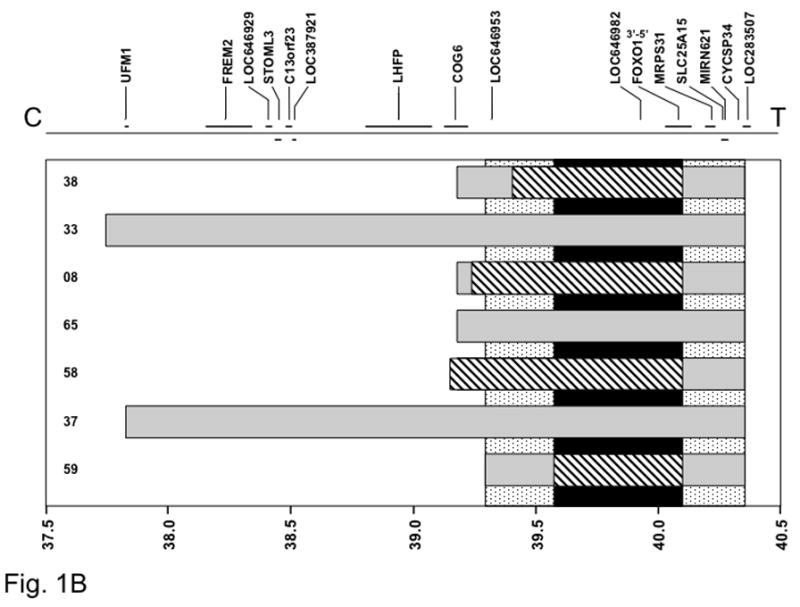
The gray rectangles indicate the extent of amplification measured in each tumor using the Affymetrix GeneChip 50K array. The stippled vertical bar indicates the minimal common area of amplification based on this initial mapping data using the 50K array. The diagonal-striped rectangles indicate further localization of the amplicons in a selected set of tumors using the Affymetrix GeneChip 250K array. The black vertical bar indicates final localization of the minimal common area of amplification based on this more detailed analysis. At the top, genes present in the 1p36 (A) and 13q14 (B) regions are shown, based on NCBI Build 36.2 of the human genetic map, and at the bottom, a scale of genomic position (in megabases) is shown. The positions of the chromosome 1 (A) or 13 (B) centromere and 1p (A) or 13q (B) telomere are designated as C and T, respectively. The 5′ to 3′ transcriptional orientation of PAX7 (A) and FOXO1 (B) are also shown above the corresponding genes.
For the 13q14 chromosomal region, the minimal common amplified region was initially mapped to a 1.06 Mb segment, based on 50K array mapping data (Figure 1B). The amplicons from five of the cases were further examined on 250K arrays and the minimal common amplified region was further refined to a 0.53 Mb segment. The genetic map indicates that this minimal region contains the LOC646982 and FOXO1 genes. As evidence that these amplicons specifically amplify the fusion genes, only the 3′ end of FOXO1 is contained within the minimal region. Based on the genomic map, the 3′ FOXO1 region is closer to the centromere and the 5′ FOXO1 region is closer to the telomere.
The localization of the 2p35 amplicon was hampered by the presence of only one 2q35-amplified case in this panel of cases. Using mapping data from the 50K array, this amplicon was 0.84 Mb in size, and mapping on the 250K array only refined this size to 0.76 Mb (data not shown). This interval contained the 5′ PAX3 region as well as multiple additional genes.
FISH Studies of 13q14 Rearrangement and Amplification in RMS Tumors
To extend these findings, we used a commercially available set of FISH probes flanking the FOXO1 locus (Supplemental Figure 1). This FISH assay detects FOXO1 gene rearrangement (as part of the PAX3-FOXO1 or PAX7-FOXO1 gene fusion) by finding break-apart of the two probes (Matsumura et al., 2008). In addition, amplification of either fusion is detected by the presence of multiple copies of the 3′ probe in a dispersed pattern consistent with double minute chromosomes. We first analyzed 56 of the 57 cases previously analyzed on the copy number arrays. One fusion-negative case was not available for FISH analysis. As a demonstration of high concordance with the microarray results, this FISH assay detected FOXO1 amplification in 9/9 RMS cases in which a 13q14 amplicon was detected on the microarray, and detected a non-amplified pattern in 46/47 RMS cases in which a 13q14 amplicon was not detected on the microarray. In the one discordance, FOXO1 amplification was detected by FISH in one PAX3-FOXO1-positive case in which a 13q14 amplicon was not found on the microarray. The lack of amplicon detection on the microarray can be explained by the fact that the FISH assay detected a FOXO1 rearrangement in most cells in this case, but only found amplicons in 8% of the cells.
After assaying an additional 118 RMS cases to bring the total to 174 cases assayed by FISH, we detected FOXO1 amplification in 35 of 174 cases (20%). Considering fusion subtypes, this amplification is present in 28/30 PAX7-FOXO1-positive (93%), 7/79 PAX3-FOXO1-positive (9%) and 0/63 fusion-negative (0%) cases. In two cases without FOXO1 amplification, the fusion status was indeterminate by RT-PCR analysis. Using the Fisher’s exact test, we found a significant association between fusion status and 13q14 amplification (p<0.0001). Moreover, the proportion of amplified cases was significantly higher in the PAX7-FOXO1-positive than in the PAX3-FOXO1-positive (p<0.0001) and fusion-negative (p<0.0001) groups. In addition, a significant difference was found between the PAX3-FOXO1 and fusion-negative groups (p=0.05). All pairwise comparisons were adjusted by using Bonferroni’s method.
In tumors with fusion gene amplification, we noted a significant difference in the frequency of amplicon-containing cells between PAX7-FOXO1- and PAX3-FOXO1-positive tumors (p<0.0001) (Figure 2A). Not only were most PAX7-FOXO1-positive tumors positive for fusion gene amplification, but also the far majority of cells within each tumor contained the fusion gene amplicon (mean, 72%; SD, 27%). In contrast, in the rare cases of PAX3-FOXO1-positive tumors with fusion gene amplification, these tumors generally contained few cells with the amplicon (mean, 20%; SD, 16%). It should be noted that the non-amplicon containing cells in these “amplified” PAX3-FOXO1 tumors generally had FOXO1 rearrangements, thus indicating that the low frequency was not because of normal cell contamination.
Figure 2. Comparison of molecular parameters among fusion subtypes.
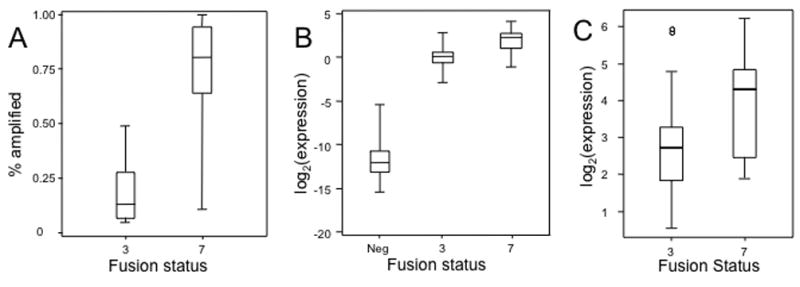
A. FISH methodology was used to quantify the fraction of amplified cells in each case with fusion gene amplification. The distribution for each fusion subtype is represented graphically by a Box and Whisker plot. The horizontal line dividing the box into two parts represents the median. The lower and upper edges of the boxes indicate the 25th and 75th percentiles, and line segments illustrate the lower and upper extremes, except for outliers (indicated in subsequent figures by circles). B. Expression of a PAX3-FOXO1 or PAX7-FOXO1 fusion transcript was quantified by qRT-PCR in RNA isolated from RMS samples. Use of a FOXO1 probe permits quantification of the two fusions on a common scale. Values were normalized to corresponding 18S RNA expression levels and represent the ratio of the means (+/−SD) of triplicate measurements. C. LOC646982 expression was measured by qRT-PCR and normalized for 18S RNA expression as described above. Abbreviations: 3, PAX3-FOXO1; 7, PAX7-FOXO1; Neg, Fusion-negative.
In a final set of comparisons, we analyzed the concordance between the FISH and RT-PCR findings in these cases. In 174 cases with FISH data, 109 cases were determined to be fusion-positive by RT-PCR. The FISH assay found a FOXO1 rearrangement with or without amplification in 101 of 109 cases (93%). In four additional cases that were fusion-positive by RT-PCR, the far majority of the cells (62%–97%) demonstrated selective loss of the 5′ FOXO1 signal (Supplemental Figure 1C). This clonal loss of the 5′ signal (associated with the nonfunctional reciprocal FOXO1-PAX3 fusion gene) and maintenance of the 3′ signal (associated with the functional PAX3-FOXO1 fusion gene) was only found in cases that were fusion-positive by RT-PCR and was interpreted as a variant FOXO1 rearrangement pattern, thus increasing the concordance frequency to 96% (105/109). In the final four fusion-positive cases, the FISH assay detected 14–17% cells with breakapart signals, and the FISH result in these cases was considered as indeterminate based on our criteria for the indeterminate range (3/109, 3%). In the remaining cases with FISH data, 63 cases were fusion-negative by RT-PCR. In the 63 “fusion-negative” cases, 59 cases were negative for FOXO1 rearrangement by FISH (94%). In three cases (5%), we detected 13–16% of cells with breakapart signals and these cases were considered indeterminate for the FOXO1 rearrangement. Finally, one case that was negative by RT-PCR was positive for FOXO1 rearrangement by this FISH assay; these results were confirmed by repeat assays. The reason for this one rare instance of discordance between RT-PCR and FISH is not known.
Expression Studies in RMS Tumors
We next focused on fusion gene expression at the RNA level. In our initial fusion transcript qRT-PCR assay, we used distinct PAX3 and PAX7 probes to detect PAX3-FOXO1 and PAX7-FOXO1 and thus the expression of the two fusion transcripts could not be directly compared. To compare expression of both fusion transcripts on a common scale, we developed a variation of our qRT-PCR assay in which we used a 3′ FOXO1 probe that will detect either fusion transcript. We assayed expression in 144 of the 201 initial RMS cases with this “combined fusion” qRT-PCR assay. Comparison of these combined expression findings with the PAX3-FOXO1 or PAX7-FOXO1-specific expression findings showed excellent correlation (PAX3-FOXO1, Pearson correlation coefficient [r] = 0.95; PAX7-FOXO1, r = 0.99).
Using these combined expression findings, we found a significant difference in expression among the three fusion subtypes (p<0.0001) (Figure 2B). In a pairwise comparison, PAX7-FOXO1-positive tumors had significantly higher fusion expression than PAX3-FOXO1-positive tumors with a mean expression level that is 3.7 times higher (p<0.0001). This result was corroborated using a second set of qRT-PCR assays from the wild-type FOXO1 regions 5′ and 3′ of the fusion gene breakpoint (Supplemental Figure 2). As expected, the fusion-negative category did not express detectable fusion transcript, and had a level that was significantly less than both fusion-positive categories. In a second set of comparisons of amplification in only fusion-positive cases, fusion gene-amplified cases had significantly higher fusion gene expression than cases without amplification with a mean expression level that is 2.4 times higher (p=0.0006). However, when this comparison was restricted to PAX3-FOXO1-positive cases, there was no significantly increased fusion expression in amplified cases compared to non-amplified cases (p=0.3). There was an insufficient number of non-amplified PAX7-FOXO1-positive cases (n=2) to explore this issue in the PAX7-FOXO1-positive subset.
Finally, we evaluated LOC646982, the other gene consistently amplified in the 13q14 amplicon. The LOC646982 locus encodes a widely expressed gene (also known as TTL) with a poorly characterized protein product (Qiao et al., 2003). Analysis of LOC646982 expression showed significantly higher expression in PAX7-FOXO1- compared to PAX3-FOXO1-positive cases (p=0.013) (Figure 2C), and in 13q14-amplified compared to non-amplified cases (p=0.044). In each comparison, the mean expression level was only 1.4-fold higher, suggesting a less prominent expression increase in amplified LOC646982 compared to amplified 3′ FOXO1.
Association of 13q14 Amplification and Fusion Status with Clinical Outcome
To study an association with clinical characteristics, we focused on the fusion-positive subset of RMS cases, which includes all cases with the 13q14 amplification event. We compared 13q14 amplification data with pertinent clinical parameters in the 87 fusion-positive RMS cases with available amplification and clinical data (28 amplified and 59 non-amplified) (Table 2). The most notable associations for these 13q14-amplified tumors were younger age at diagnosis, extremity primaries, no nodal involvement and lower invasiveness. A similar comparison of PAX3-FOXO1- and PAX7-FOXO1-positive cases with clinical data (n=76 and n=30, respectively) demonstrated that the PAX7-FOXO1 fusion was also preferentially associated with these same four clinicopathologic features.
Table 2.
Patient characteristics in fusion-positive cases compared by 13q14 amplification status or gene fusion status
| 13q14 amp status (n=87) | Gene fusion status (n=106) | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Amplified (n=28) | Not amp (n=59) | p-value | PAX3 (n=76) | PAX7 (n=30) | p-value | |
|
| ||||||
| Age | ||||||
| <1 | 3 (11%) | 1 (2%) | 0.0002 | 3 (4%) | 1 (3%) | <0.0001 |
| 1–9 | 20 (71%) | 22 (37%) | 28 (37%) | 25 (85%) | ||
| 10+ | 5 (18%) | 36 (61%) | 45 (59%) | 4 (13%) | ||
| Sex | ||||||
| male | 18 (64%) | 28 (47%) | 0.17 | 40 (53%) | 17 (57%) | 0.83 |
| female | 10 (36%) | 31 (53%) | 36 (47%) | 13 (43%) | ||
| Race | ||||||
| White | 17 (61%) | 35 (59%) | 0.32 | 47 (62%) | 19 (63%) | 0.20 |
| Hispanic | 4 (14%) | 12 (20%) | 12 (16%) | 4 (13%) | ||
| Black | 4 (14%) | 11 (19%) | 16 (21%) | 4 (13%) | ||
| Other | 3 (11%) | 1 (2%) | 1 (1%) | 3 (10%) | ||
| Primary Site | ||||||
| Other | 7 (28%) | 38 (64%) | 0.0037 | 45 (62%) | 8 (27%) | 0.021 |
| Extremity | 18 (72%) | 21 (36%) | 28 (38%) | 22 (73%) | ||
| Node status | ||||||
| N-0 | 18 (75%) | 29 (49%) | 0.020 | 34 (47%) | 24 (80%) | 0.0038 |
| N-1 | 6 (25%) | 30 (51%) | 38 (53%) | 6 (20%) | ||
| Size | ||||||
| <=5 cm | 12 (48%) | 20 (34%) | 0.23 | 23 (32%) | 14 (47%) | 0.18 |
| >5 cm | 13 (52%) | 39 (66%) | 50 (68%) | 16 (53%) | ||
| Tumor invasion | ||||||
| T-1 | 17 (68%) | 16 (27%) | 0.0006 | 20 (27%) | 21 (70%) | 0.0001 |
| T-2 | 8 (32%) | 43 (73%) | 53 (73%) | 9 (30%) | ||
| Stage | ||||||
| 1 | 2 (8%) | 9 (15%) | 0.069 | 8 (11%) | 3 (10%) | 0.083 |
| 2 | 10 (40%) | 8 (14%) | 11 (15%) | 11 (37%) | ||
| 3 | 8 (32%) | 22 (37%) | 29 (40%) | 11 (37%) | ||
| 4 | 5 (20%) | 20 (34%) | 25 (34%) | 5 (17%) | ||
| Group | ||||||
| I | 4 (16%) | 3 (5%) | 0.27 | 6 (8%) | 6 (20%) | 0.21 |
| II | 7 (28%) | 13 (22%) | 13 (18%) | 7 (23%) | ||
| III | 9 (36%) | 23 (39%) | 28 (38%) | 11 (37%) | ||
| IV | 5 (20%) | 20 (34%) | 26 (36%) | 6 (20%) | ||
| Risk category | ||||||
| Grp I/II, N0 | 9 (37%) | 12 (20%) | 0.35 | 14 (19%) | 12 (40%) | 0.13 |
| Grp III | 6 (25%) | 13 (22%) | 15 (21%) | 7 (23%) | ||
| N1 | 4 (17%) | 14 (24%) | 17 (24%) | 5 (17%) | ||
| Grp IV | 5 (21%) | 20 (34%) | 26 (36%) | 6 (20%) | ||
We next investigated the association of these molecular features with outcome data, using the Kaplan-Meier method to estimate survival distributions and the log-rank test to compare them by molecular status. In an analysis of 13q14 amplification status, overall outcome was significantly better for amplified cases relative to non-amplified cases (p=0.0011); the overall survival at five years for amplified and non-amplified cases was 80% and 40%, respectively (Figure 3A). The failure-free survival was also quantitatively better for amplified cases relative to non-amplified cases with a trend towards significance (p=0.053) (Figure 3B). A comparable analysis of fusion status demonstrated that a similar difference in outcome between PAX7-FOXO1- and PAX3-FOXO1-positive cases, which was statistically significant for both overall (p=0.0012) and failure-free survival (p=0.046) (Figure 3C, D). In further studies, the subset of 13q14-amplified PAX3-FOXO1-positive cases had a quantitatively improved overall outcome (66% five-year survival) compared to the non-amplified PAX3-FOXO1-positive cases (40% five-year survival), but this result was not significant (p=0.32) due to the small number of amplified cases (n=5). There were not enough non-amplified PAX7-FOXO1-positive cases to pursue a similar analysis in the PAX7-FOXO1-positive subset.
Figure 3. Relationship of outcome to 13q14 amplification and fusion status in fusion-positive RMS patients.
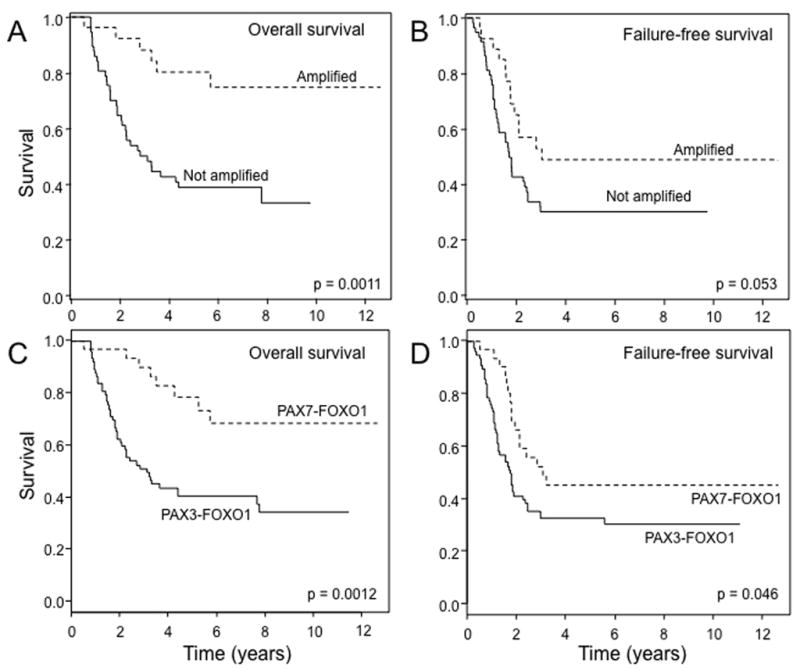
A. Comparison of overall survival between fusion-positive RMS cases with and without 13q14 amplification. Amplified, n = 28; non-amplified, n = 59. B. Comparison of failure-free survival between fusion-positive RMS cases with and without 13q14 amplification. Amplified, n = 28; non-amplified, n = 59. C. Comparison of overall survival between PAX3-FOXO1- and PAX7-FOXO1-positive RMS cases. PAX3-FOXO1, n = 76; PAX7-FOXO1, n = 30. D. Comparison of failure-free survival between PAX3-FOXO1- and PAX7-FOXO1-positive RMS cases. PAX3-FOXO1, n = 76; PAX7-FOXO1, n = 30.
In the next step, we used the Cox proportional hazards model to analyze these molecular parameters under a multivariate setting. After adjusting for age, sex, group, node status, primary site, and tumor size, 13q14 amplification status was a significant independent predictor of overall survival (p = 0.018, HR = 0.3, 95% CI: 0.11–0.81) (Table 3). A similar multivariate analysis examining fusion status instead of amplification also demonstrated an independent role for PAX7-FOXO1 fusion status in predicting overall survival (p = 0.035, HR = 0.39, 95% CI: 0.16–0.94). When both 13q14 amplification status and fusion status were included together in this multivariate analysis, the significance of both parameters decreased and neither was a significant independent predictor of overall outcome. This result was not unexpected given the high level of correlation between these two molecular parameters and the limited number of cases. In contrast to these predictive findings for overall survival, a comparable multivariate analysis indicated that neither 13q14 amplification status nor fusion status was an independent predictor of failure-free survival.
Table 3.
Cox proportional hazards modeling to predict overall and failure-free survival from clinical, pathologic, and molecular predictors in fusion-positive cases
| Predictor | Level | Overall Survival | Failure-free Survival | ||
|---|---|---|---|---|---|
|
| |||||
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| 13q14 amplification and clinical variables (n=83) | |||||
|
| |||||
| Age | (continuous, in year) | 1.03 (0.96, 1.11) | 0.42 | 1.01 (0.94, 1.07) | 0.84 |
|
| |||||
| Sex | Male | RL* | - | RL | - |
| Female | 0.47 (0.24, 0.92) | 0.028 | 0.45 (0.25, 0.81) | 0.0074 | |
|
| |||||
| Group | 4 | RL | - | RL | - |
| 1,2,3 | 0.42 (0.2, 0.88) | 0.022 | 0.4 (0.2, 0.79) | 0.0091 | |
|
| |||||
| Node Status | N-0 | RL | - | RL | - |
| N-1 | 1.57 (0.78, 3.17) | 0.21 | 1.04 (0.55, 1.95) | 0.9 | |
|
| |||||
| Tumor size | ≤5 cm | RL | - | RL | - |
| >5 cm | 3.33 (1.28, 8.64) | 0.013 | 2.04 (0.97, 4.31) | 0.062 | |
|
| |||||
| Extremity | No | RL | - | RL | - |
| Yes | 0.75 (0.38, 1.5) | 0.41 | 0.78( 0.43, 1.42) | 0.42 | |
|
| |||||
| 13q14 Amplification | No | RL | - | RL | - |
| Yes | 0.3 (0.11, 0.81) | 0.018 | 0.61 (0.28, 1.36) | 0.23 | |
|
| |||||
| Fusion status and clinical variables (n=102) | |||||
|
| |||||
| Age | (continuous, in year) | 1.03 (0.97, 1.1) | 0.29 | 1.01 (0.96, 1.07) | 0.64 |
|
| |||||
| Sex | Male | RL | - | RL | - |
| Female | 0.53 (0.3, 0.96) | 0.037 | 0.59 (0.35, 0.99) | 0.044 | |
|
| |||||
| Group | 4 | RL | - | RL | - |
| 1,2,3 | 0.42 (0.22, 0.8) | 0.0083 | 0.47 (0.26, 0.85) | 0.012 | |
|
| |||||
| Node Status | N-0 | RL | - | RL | - |
| N-1 | 1.54 (0.83, 2.84) | 0.17 | 1.07 (0.61, 1.86) | 0.82 | |
|
| |||||
| Tumor size | ≤5 cm | RL | - | RL | - |
| >5 cm | 3.09 (1.34, 7.14) | 0.0083 | 2.32 (1.18, 4.59) | 0.015 | |
|
| |||||
| Extremity | No | RL | - | RL | - |
| Yes | 0.76 (0.41, 1.41) | 0.38 | 0.72 (0.42, 1.25) | 0.24 | |
|
| |||||
| Fusion Status | PAX3 | RL | - | RL | - |
| PAX7 | 0.39 (0.16, 0.94) | 0.035 | 0.63 (0.32, 1.25) | 0.19 | |
|
| |||||
| 13q14 amplification + fusion status + clinical variables (n=83) | |||||
|
| |||||
| Age | (continuous, in year) | 1.03 (0.96, 1.11) | 0.4 | 1.01 (0.94, 1.07) | 0.86 |
|
| |||||
| Sex | Male | RL | - | RL | - |
| Female | 0.47 (0.24, 0.91) | 0.026 | 0.45 (0.25, 0.81) | 0.0081 | |
|
| |||||
| Group | 4 | RL | - | RL | - |
| 1,2,3 | 0.41 (0.2, 0.87) | 0.02 | 0.4 (0.2, 0.8) | 0.0096 | |
|
| |||||
| Node Status | N-0 | RL | - | RL | - |
| N-1 | 1.59 (0.79, 3.22) | 0.19 | 1.03 (0.55, 1.94) | 0.92 | |
|
| |||||
| Tumor size | <=5 cm | RL | - | RL | - |
| >5 cm | 3.35 (1.28, 8.72) | 0.013 | 2.03 (0.96, 4.29) | 0.063 | |
|
| |||||
| Extremity | No | RL | - | RL | - |
| Yes | 0.75 (0.38, 1.49) | 0.41 | 0.78 (0.43, 1.42) | 0.42 | |
|
| |||||
| Fusion Status | PAX3 | RL | - | RL | - |
| PAX7 | 1.7 (0.11, 25.72) | 0.7 | 0.72 (0.04, 12.16) | 0.82 | |
|
| |||||
| 13q14 Amplification | No | RL | - | RL | - |
| Yes | 0.19 (0.01, 2.73) | 0.22 | 0.84 (0.05, 13.69) | 0.9 | |
Reference level
Based on a previous finding that improved outcome associated with the PAX7-FOXO1 fusion was specific to metastatic cases (Sorensen et al., 2002), we studied the relationship of disease spread (locoregional or metastatic) with fusion or amplification status. In our multivariate model, there was not a statistically significant interaction of fusion (p=0.98) or amplification (p=0.095) status with disease spread. Of note, our current study shows that PAX7-FOXO1 is associated with improved overall survival for both locoregional (p = 0.032) and metastatic (p=0.0059) disease.
In a final analysis, we used the Cox regression to study cases with fusion gene amplification and analyze whether the fraction of amplified cells within the tumor has predictive value. From the 87 fusion-positive cases with clinical information, our FISH analysis identified 28 cases with fusion gene amplification. In a univariate analysis of these 28 cases, the fraction of amplified cells was not significant in predicting overall (p = 0.99) or failure-free survival (p = 0.65) (Supplemental Table 2). After adjusting for other clinical parameters, this fraction was significant for predicting failure free survival (p=0.029), with an increased fraction being associated with improved outcome. However, the association with better overall survival (HR=0.07) was not significant (p=0.13).
DISCUSSION
In this manuscript, we performed a comprehensive examination of the fusion gene amplification events that occur in the pediatric soft tissue cancer RMS. Our analysis of a large number of RMS cases confirmed that the PAX7-FOXO1 fusion is usually amplified while the PAX3-FOXO1 fusion is rarely amplified. The copy number array analysis indicated that PAX7 was the only consistently amplified gene in the 1p36 region and thus co-amplification of neighboring genes in the 1p36 region does not appear to explain the high frequency of PAX7-FOXO1 amplification. Though the copy number array analysis of the 13q14 amplicon did identify consistent co-amplification of FOXO1 and the neighboring LOC646982 gene, this gene could be amplified in either PAX7-FOXO1 or PAX3-FOXO1 amplicons and thus does not immediately provide an explanation for the increased frequency of PAX7-FOXO1 amplification.
Another relevant genomic feature that was assessed in our genomic data was the orientation of the three genes with respect to the chromosomes. Our earlier studies of PAX3 and FOXO1 suggested that both genes are oriented with their 5′ regions closer to the telomere and 3′ regions closer to the centromere (Barr et al., 1993; Galili et al., 1993). The current genomic maps of human chromosomes 2 and 13 along with our mapping of the 3′ FOXO1 and 5′ PAX3 amplicons confirmed these gene orientations. Similarly, the current genomic map of human chromosome 1 along with our mapping studies of the amplified 5′ PAX7 region indicates that PAX7 is also oriented with a 5′ region closer to the telomere and 3′ region closer to the centromere. Therefore, PAX7 is oriented similarly to PAX3, and both genes can conceivably form gene fusions with FOXO1 by simple breakage and fusion events. These findings refute earlier theories that the frequent PAX7-FOXO1 amplification may be due to a problem with the orientations of the PAX7 and FOXO1 genes on their respective chromosomes (Barr et al., 1996).
Based on our above findings, we suggest that a major difference between PAX3-FOXO1 and PAX7-FOXO1 is the intrinsic expression properties of these genes. Our earlier studies showed that the fusion genes are expressed at a higher level than the corresponding wild-type genes in human ARMS tumors (Davis and Barr, 1997). More recently, we demonstrated that fusion gene overexpression is necessary for transformation and tumorigenesis in a human myoblast system (Xia et al., 2009). Therefore, a critical issue in these fusion-positive RMS tumors appears to be selection of a mechanism for high fusion gene expression to reach a critical threshold level sufficient for oncogenic activity. In PAX3-FOXO1-expressing tumors, there is a copy number independent gain of transcriptional activity in the PAX3-FOXO1 gene relative to the wild-type PAX3 gene (Davis and Barr, 1997); we hypothesize that this gain involves synergism between the 5′ PAX3 and 3′ FOXO1 regulatory elements. Based on the different developmental expression patterns of the PAX3 and PAX7 genes, it is clear that the expression elements in these two genes are different (Mansouri, 1998). A reporter transfection comparison of the proximal promoters indicated that the PAX3 promoter has greater activity than the PAX7 promoter in various cell backgrounds (Moller et al., 2007). These findings suggest that the combination of 5′ PAX7 and 3′ FOXO1 regulatory elements is not sufficient to generate high level expression, and thus a different genomic mechanism, amplification, was selected in these PAX7-FOXO1-positive tumors to achieve high level expression.
We developed methods for measuring PAX3-FOXO1 and PAX7-FOXO1 fusion transcript expression on a common scale. In comparisons of tumor subsets, these methods showed the same overall results. In particular, PAX7-FOXO1 mRNA is expressed at higher levels than PAX3-FOXO1 mRNA in the respective tumors. Therefore, these results indicate that, though the PAX7 promoter may be “weaker” than the PAX3 promoter, the amplification mechanism more than compensates for this difference and in fact results in generally higher final PAX7-FOXO1 mRNA levels.
In the relatively uncommon PAX3-FOXO1-positive cases with fusion gene amplification, we identified several notable differences from PAX7-FOXO1-positive amplified cases. Our FISH analysis detected rearrangements in most of PAX3-FOXO1-positive cells and amplification in only a small subset of the cells. In contrast, PAX7-FOXO1-positive tumors demonstrate amplification in the majority of the cells. These findings suggest that PAX7-FOXO1 amplification occurs early in tumorigenesis while PAX3-FOXO1 amplification occurs later in tumor progression. Alternatively, PAX3-FOXO1 amplification may occur early but has no selective advantage. Expression studies of the PAX3-FOXO1-amplified tumors did not indicate increased expression relative to non-amplified PAX3-FOXO1-positive tumors and comparable to PAX7-FOXO1-positive tumors. However, this lack of expression can be readily explained by the low fraction of cells with the PAX3-FOXO1 amplification event.
Based on our published phenotypic studies of the PAX3-FOXO1 protein (Xia and Barr, 2004; Xia et al., 2007; Xia et al., 2009), we hypothesize that higher levels of the fusion product will have phenotypic consequences in RMS cells. Our cell culture studies indicate that high fusion product levels exert toxic effects such as growth suppression in many cells types, and tolerance mechanisms are needed to attenuate these toxic effects and allow oncogenic effects to be exerted. Therefore, higher PAX7-FOXO1 levels may still be fully tolerated and exert higher oncogenic effects, or may have partially exceeded tolerance mechanisms and resulted in RMS tumor cells that are less “fit” than the PAX3-FOXO1-expressing RMS tumor cells. This latter possibility is consistent with clinical data from the current study. For example, there is a significant association of amplification status and PAX7-FOXO1 fusion status with lower nodal status and lower tumor invasion status. Both of these indices are measures of tumor progression and suggest the possibility that the amplified PAX7-FOXO1-overexpressing cells are relatively less able to undergo these processes.
A major finding in this current study is the association of fusion gene amplification as well as the PAX7-FOXO1 fusion gene with a favorable overall outcome. Though most of the data points to improved overall survival, there is also some evidence of improved failure-free survival. The high correlation of fusion gene amplification and PAX7-FOXO1 positivity is expected given the finding that >90% of PAX7-FOXO1-positive cases were amplified and >80% of amplified cases were PAX7-FOXO1-positive in our dataset. From a practical standpoint, this high correlation indicates that fusion gene amplification, which is also determined on the FISH assay for FOXO1 rearrangement, can be used as a marker of good outcome without the need for determining fusion subtype. The association of fusion gene amplification and amplification of the PAX7-FOXO1 fusion gene in particular with better survival is consistent with the above-described theme of less well-adapted tumor cells with high PAX7-FOXO1 fusion expression.
In interpreting the clinical findings in this paper, we acknowledge that this panel of tumors constitutes a convenience sample in which there may be unknown selection biases. To optimize future investigation of these issues, we are planning to analyze these parameters in RMS cases from a single large clinical trial (D9803) of intermediate risk RMS. We also acknowledge the need to continue to accumulate fusion-amplified cases (particular PAX3-FOXO1-positive) to generate a larger sample size to verify and extend our current findings. Therefore, the present study provides the impetus for important future studies by firmly establishing the basic molecular findings related to fusion gene amplification in RMS and clearly providing strong evidence of a novel clinical marker of good outcome in RMS.
Supplementary Material
Acknowledgments
Supported by: NIH grants R01 CA104896, U10 CA098543, U10 CA098413 and U24 CA114766; The Hartwell Foundation; Alex’s Lemonade Stand Foundation; and the Alveolar Rhabdomyosarcoma Research Fund. Tissue samples were provided by the Pediatric Division of the Cooperative Human Tissue Network, which is funded by NIH grant U01 CA054021.
The authors would like to thank Lucas Kocia for his technical assistance.
References
- Barr FG, Duan F, Smith LM, Gustafson D, Pitts M, Hammond S, Gastier-Foster JM. Genomic and clinical analyses of 2p24 and 12q13-q14 amplification in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. Genes Chromosomes Cancer. 2009;48:661–672. doi: 10.1002/gcc.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FG, Galili N, Holick J, Biegel JA, Rovera G, Emanuel BS. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;3:113–117. doi: 10.1038/ng0293-113. [DOI] [PubMed] [Google Scholar]
- Barr FG, Nauta LE, Davis RJ, Schafer BW, Nycum LM, Biegel JA. In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum Mol Genet. 1996;5:15–21. doi: 10.1093/hmg/5.1.15. [DOI] [PubMed] [Google Scholar]
- Barr FG, Smith LM, Lynch JC, Strzelecki D, Parham DM, Qualman SJ, Breitfeld PP. Examination of gene fusion status in archival samples of alveolar rhabdomyosarcoma entered on the Intergroup Rhabdomyosarcoma Study-III trial: a report from the Children’s Oncology Group. J Mol Diagn. 2006;8:202–208. doi: 10.2353/jmoldx.2006.050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FG, Womer R. Rhabdomyosarcoma. In: Orkin SH, Fisher DE, Look AT, Lux SE, Ginsburg D, Nathan DG, editors. Oncology of Infancy and Childhood. Philadelphia: Saunders; 2009. pp. 743–828. [Google Scholar]
- Bridge JA, Liu J, Qualman SJ, Suijkerbuijk R, Wenger G, Zhang J, Wan X, Baker KS, Sorensen P, Barr FG. Genomic gains and losses are similar in genetic and histologic subsets of rhabdomyosarcoma, whereas amplification predominates in embryonal with anaplasia and alveolar subtypes. Genes Chromosomes Cancer. 2002;33:310–321. doi: 10.1002/gcc.10026. [DOI] [PubMed] [Google Scholar]
- Davis RJ, Barr FG. Fusion genes resulting from alternative chromosomal translocations are overexpressed by gene-specific mechanisms in alveolar rhabdomyosarcoma. Proc Natl Acad Sci USA. 1997;94:8047–8051. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJd, Emanuel BS, Rovera G, Barr FG. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- Gordon AT, Brinkschmidt C, Anderson J, Coleman N, Dockhorn-Dworniczak B, Pritchard-Jones K, Shipley J. A novel and consistent amplicon at 13q31 associated with alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 2000;28:220–226. [PubMed] [Google Scholar]
- Mansouri A. The role of Pax3 and Pax7 in development and cancer. Crit Rev Oncog. 1998;9:141–149. doi: 10.1615/critrevoncog.v9.i2.40. [DOI] [PubMed] [Google Scholar]
- Matsumura T, Yamaguchi T, Seki K, Shimoda T, Wada T, Yamashita T, Hasegawa T. Advantage of FISH analysis using FKHR probes for an adjunct to diagnosis of rhabdomyosarcomas. Virchows Arch. 2008;452:251–258. doi: 10.1007/s00428-007-0554-9. [DOI] [PubMed] [Google Scholar]
- Moller E, Isaksson M, Mandahl N, Mertens F, Panagopoulos I. Comparison of the proximal promoter regions of the PAX3 and PAX7 genes. Cancer Genet Cytogenet. 2007;178:114–119. doi: 10.1016/j.cancergencyto.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Newton WA, Jr, Gehan EA, Webber BL, Marsden HB, van Unnik AJ, Hamoudi AB, Tsokos MG, Shimada H, Harms D, Schmidt D, Ninfo V, Cavazzana AO, Gonzalez-Crussi F, Parham DM, Reiman HM, Asmar L, Beltangady MS, Sachs NE, Triche RJ, Maurer HM. Classification of rhabdomyosarcomas and related sarcomas. Pathologic aspects and proposal for a new classification--an Intergroup Rhabdomyosarcoma Study. Cancer. 1995;76:1073–1085. doi: 10.1002/1097-0142(19950915)76:6<1073::aid-cncr2820760624>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Pandita A, Zielenska M, Thorner P, Bayani J, Godbout R, Greenberg M, Squire JA. Application of comparative genomic hybridization, spectral karyotyping, and microarray analysis in the identification of subtype-specific patterns of genomic changes in rhabdomyosarcoma. Neoplasia. 1999;1:262–275. doi: 10.1038/sj.neo.7900036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham DM, Ellison DA. Rhabdomyosarcomas in adults and children: an update. Arch Pathol Lab Med. 2006;130:1454–1465. doi: 10.5858/2006-130-1454-RIAACA. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Ogawa S, Hangaishi A, Yuji K, Izutsu K, Kunisato A, Imai Y, Wang L, Hosoya N, Nannya Y, Sato Y, Maki K, Mitani K, Hirai H. Identification of a novel fusion gene, TTL, fused to ETV6 in acute lymphoblastic leukemia with t(12;13)(p13;q14), and its implication in leukemogenesis. Leukemia. 2003;17:1112–1120. doi: 10.1038/sj.leu.2402919. [DOI] [PubMed] [Google Scholar]
- Reichek JL, Duan F, Smith LM, Gustafson DM, O’Connor RS, Zhang C, Pitts MJ, Gastier-Foster JM, Barr FG. Genomic and clinical analysis of amplification of the 13q31 chromosomal region in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. Clin Cancer Res. 2011;17:1463–1473. doi: 10.1158/1078-0432.CCR-10-0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, Bridge JA, Crist WM, Triche TJ, Barr FG. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. J Clin Oncol. 2002;20:2672–2679. doi: 10.1200/JCO.2002.03.137. [DOI] [PubMed] [Google Scholar]
- Weber-Hall S, McManus A, Anderson J, Nojima T, Abe S, Pritchard-Jones K, Shipley J. Novel formation and amplification of the PAX7-FKHR fusion gene in a case of alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 1996;17:7–13. doi: 10.1002/(SICI)1098-2264(199609)17:1<7::AID-GCC2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Xia SJ, Barr FG. Analysis of the transforming and growth suppressive activities of the PAX3-FKHR oncoprotein. Oncogene. 2004;23:6864–6871. doi: 10.1038/sj.onc.1207850. [DOI] [PubMed] [Google Scholar]
- Xia SJ, Holder DD, Pawel BR, Zhang C, Barr FG. High expression of the PAX3-FKHR oncoprotein is required to promote tumorigenesis of human myoblasts. Am J Pathol. 2009;175:2600–2608. doi: 10.2353/ajpath.2009.090192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia SJ, Rajput P, Strzelecki DM, Barr FG. Analysis of genetic events that modulate the oncogenic and growth suppressive activities of the PAX3-FKHR fusion oncoprotein. Lab Invest. 2007;87:318–325. doi: 10.1038/labinvest.3700521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


