Abstract
DJ-1 is a conserved, disease-associated protein that protects against oxidative stress and mitochondrial damage in multiple organisms. Human DJ-1 contains a functionally essential cysteine residue (Cys106) whose oxidation is important for regulating protein function by an unknown mechanism. This residue is well conserved in other DJ-1 homologues, including two (DJ-1α and DJ-1β) in Drosophila melanogaster. Because D. melanogaster is a powerful model system for studying DJ-1 function, we have determined the crystal structure and impact of cysteine oxidation on Drosophila DJ-1β. The structure of D. melanogaster DJ-1β is similar to that of human DJ-1, although two important residues in the human protein, Met26 and His126, are not conserved in DJ-1β. His126 in human DJ-1 is substituted with a tyrosine in DJ-1β, and this residue is not able to compose a putative catalytic dyad with Cys106 that was proposed to be important in the human protein. The reactive cysteine in DJ-1 is oxidized readily to the cysteine-sulfinic acid in both flies and humans and this may regulate the cytoprotective function of the protein. We show that the oxidation of this conserved cysteine residue to its sulfinate form (Cys-SO2−) results in considerable thermal stabilization of both Drosophila DJ-1β and human DJ-1. Therefore, protein stabilization is one potential mechanism by which cysteine oxidation may regulate DJ-1 function in vivo. More generally, most close DJ-1 homologues are likely stabilized by cysteine-sulfinic acid formation but destabilized by further oxidation, suggesting that they are biphasically regulated by oxidative modification.
Keywords: DJ-1, cysteine oxidation, X-ray crystallography, Parkinson’s disease, protein stability
The DJ-1 superfamily is a large and functionally varied group of proteins with homologues in many organisms. Human DJ-1 has been independently shown to be an oncogene with elevated expression in many types of cancer (1) and a cytoprotective protein whose deficiency is a rare cause of heritable parkinsonism (2). DJ-1 enhances eukaryotic cell survival in response to various stressors, particularly oxidative stress and mitochondrial toxins (3–6). Several activities have been demonstrated for DJ-1, including participation in multiple signaling pathways involving mitochondrial uncoupling proteins (7), ASK1 (8, 9), AKT/PI3K/PTEN (10, 11), JNK (12, 13), ERK (14, 15), p53 (16–19), NF-κB (20), and Nrf2 (21). DJ-1 has also been proposed to act as a redox-sensitive chaperone that inhibits α-synuclein aggregation (22, 23), an RNA binding protein (24), a regulator of glutathione synthesis (25, 26), and a transcriptional regulator (27). Despite strong evidence for a role in stress response in eukaryotes and an abundance of proposed activities, the mechanistic details of DJ-1’s function remain poorly understood.
Drosophila melanogaster provides a versatile animal model system for the study of neurodegenerative diseases, including parkinsonism (10, 28, 29). D. melanogaster has two orthologs of human DJ-1, DJ-1α and DJ-1β, both of which share ~50% sequence identity with the human protein. These orthologs differ in tissue distribution, with DJ-1α being primarily expressed in the testes of male flies while DJ-1β is expressed in most tissues (5, 29, 30). Although both DJ-1 orthologs appear to play a significant role in Drosophila biology, DJ-1β has the larger role in organismal defense against oxidative stress (5, 30). Flies lacking both DJ-1 homologues have reduced survival when exposed to compounds that induce oxidative stress and display motor deficits without frank dopaminergic neurogeneration (5, 29–31). Consistent with results of several mammalian DJ-1 studies, the Drosophila DJ-1 homologues are important for the maintenance of proper mitochondrial function, which may be a key site of DJ-1 action (30, 32). Both proteins contain a highly conserved, oxidation-prone cysteine residue (Cys104 in DJ-1β; Cys106 in human DJ-1) that is critical for DJ-1’s protective function against oxidative stress in Drosophila and several other model systems (3, 9, 33). Furthermore, human DJ-1 can complement Drosophila DJ-1β knockout phenotypes, underscoring the similarity between the human and fly proteins (33). Therefore, D. melanogaster provides a powerful and established animal model system for studying DJ-1 function.
Despite intensive study, a number of questions remain about the role of the conserved Cys106 residue (human residue numbering) in DJ-1 function. Multiple studies indicate that the oxidation of this residue to the sulfinate (Cys106-SO2−) is important for DJ-1 function in eukaryotes (3, 34–36), although the detailed mechanism by which oxidation regulates DJ-1 function is not fully understood. One model holds that modification of Cys106 allows DJ-1 to function as a redox sensor, implying that both the reduced and oxidized forms of the residue may have functional significance and perhaps perform distinct cellular functions (3, 37). However, an unusual plant homologue of DJ-1 that lacks this cysteine residue is required for chloroplast development and thus viability, suggesting that some aspects of DJ-1 function may be redox-insensitive, at least in Arabidopsis thaliana (38). Oxidation of human DJ-1 has been shown to destabilize the protein and is correlated with disease (39, 40), although it is unclear if this destabilization is due to the formation of the mildly oxidized Cys106-SO2− isoform, the more extensively oxidized Cys106-SO3− (sulfonate) modification, or other oxidative modifications in DJ-1.
In addition to its susceptibility to oxidative modification, reduced Cys106 itself might play a role in DJ-1 function. In human DJ-1, Cys106 has a low pKa value of 5.4 and thus could serve as a catalytic nucleophile, although no definitive enzymatic activity for human DJ-1 has been demonstrated to date (41). One proposed enzymatic activity for DJ-1 has been as a cysteine protease that uses a Cys106-His126 dyad in the active site (42). However, His126 in human DJ-1 is not well-conserved in other homologues, including D. melanogaster DJ-1β (where it is a tyrosine), raising questions about the relevance of this proposed dyad for DJ-1 function. As human DJ-1 and Drosophila DJ-1β appear to have similar protective functions, it is unlikely that there is a large divergence in the molecular activity responsible for this function. Assessing the likelihood of a unconventional Cys104-Tyr124 catalytic dyad in DJ-1β has been hampered by the absence of an available structure for the Drosophila homologue.
We report the 2.0 Å resolution crystal structure of D. melanogaster DJ-1β. As expected, DJ-1β is structurally similar to human DJ-1, and most differences are confined to regions on the periphery of the protein. Tyr124 (equivalent to His126 in human DJ-1) is in a conformation that prohibits hydrogen bonding with Cys104 and thus is unlikely to compose an active site dyad that has been proposed to be important in the human protein. Therefore, if DJ-1 possesses an enzymatic activity, it is not likely to involve a dyad between the reactive cysteine and the histidine/tyrosine residue. The observed oxidation of Cys104 in DJ-1β to the cysteine-sulfinate thermally stabilizes the protein by 11.5°C, similar to human DJ-1. This substantial oxidative stabilization upon cysteine-sulfinic acid formation may contribute to enhancement of DJ-1 function under oxidative stress conditions and is likely common to other DJ-1 homologues.
Experimental Procedures
Protein Expression and Purification
D. melanogaster DJ-1β contains an N-terminal extension that is not found in many homologues of the protein and is a possible targeting sequence. Consequently, the mature coding sequence corresponding to the region of the protein that aligns well with other DJ-1 homologues (residues 19-205) was cloned between the NdeI and XhoI sites of the bacterial expression vector pET15b. BL21(DE3) E. coli were grown in Luria-Bertani (LB) medium supplemented with 100 μg/mL ampicillin at 37 °C with shaking until OD600 reached 0.5–0.7. The temperature of the culture was reduced to 20°C and equilibrated for 2 h with shaking. Protein expression was induced by the addition isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM to the culture, which was incubated at 20 °C with shaking overnight and harvested by centrifugation the next day. For some preparations, a same-day induction protocol was followed where protein expression was induced with 0.5 mM IPTG for 3–4 hours at 37 °C. Cell pellets were frozen and stored at −80 °C.
N-terminally hexa-histidine tagged DJ-1β was purified using metal affinity chromatography with His-Select resin (Sigma). The cell pellet was thawed and then resuspended in extraction buffer (50 mM HEPES pH=7.5, 300 mM NaCl, 10 mM imidazole, 2 mM DTT), supplemented with 1 mg/mL lysozyme and incubated on ice for 30 minutes, then sonicated to complete lysis. Centrifugally cleared lysate was mixed with His-Select resin (Sigma) in batch at 4°C and then poured into a column and washed with 20 mM imidazole-supplemented extraction buffer until no protein was detected in the flow through using Bradford reagent. The bound recombinant protein was eluted using 250 mM imidazole-supplemented extraction buffer. The N-terminal hexahistidine tag was removed by cleavage with 1 unit of thrombin per milligram of DJ-1β for 2 h at 22 °C followed by dialysis against storage buffer (25 mM HEPES pH=7.5, 100 mM KCl, 2 mM DTT) at 4 °C overnight. The protein was passed again over His-Select resin to remove any protein that retained the tag and then incubated at 4 °C with benzamidine-Sepharose resin to remove thrombin. Purified protein was concentrated using a centrifugal concentrator (Millipore) with a 10-kDa cutoff to 21 mg/mL as determined by absorbance at 280 nm using a calculated extinction coefficient at 280 nm of 7,500 M−1 cm−1. The purified protein in storage buffer was snap-frozen in 50–100 μl aliquots on liquid nitrogen and stored at −80 °C.
Crystallization and Data Collection
DJ-1β (21 mg/mL) in storage buffer was crystallized using the sitting drop vapor diffusion method with drops containing 2 μl of protein and 2 μl of reservoir solution. Rhombohedrally shaped crystals of DJ-1β formed in space group P3221 in two days at room temperature against a reservoir solution of 25% polyethylene glycol 4000, 0.2 M ammonium sulfate, 0.1 M sodium acetate pH=4.6. Crystals typically grew from dense precipitate that was carefully removed using a nylon loop during crystal harvest. The crystals were cryoprotected by serial transfer through reservoir solutions supplemented with ethylene glycol increasing in 5% increments to a final concentration of 25% v/v. Cryoprotected crystals were cooled in liquid nitrogen.
Diffraction data were collected from a single crystal at 100K at the Advanced Photon Source, GM/CA-CAT beamline 23 ID-D. The crystal was exposed to 12.66 KeV (0.98 Å) X-rays for 2 s per 1 degree of oscillation for a total of 180 degrees, and the resulting diffraction data were recorded on a MARmosaic 300 CCD detector. Data were integrated and scaled using HKL2000 (43) and final data statistics for each data set are provided in Table 1.
Table 1.
Data collection and refinement statistics
| Data collection | |
| X-ray source | APS GM/CA 23ID-D |
| X-ray wavelength (Å) | 0.98 |
| Space group | P3221 |
| Cell dimensions | |
| a=b, c (Å) | 52.52; 227.17 |
| Molecules in asymmetric unit | 2 |
| Wilson B-factor (Å2) | 26.9 |
| Resolution (Å) | 75.7–2.0 |
| Rmergea,b | 0.098 (0.587) |
| 〈I〉/〈σ(I)〉 | 21.1 (1.8) |
| Completeness (%) | 97.3 (83.9) |
| Redundancy | 8.5 (5.1) |
| Refinement | |
| PDB code | 4E08 |
| Program | Refmac5.6.0116 |
| No. unique reflections | 23814 |
| Rwork; Rfree(%)c,d | 19.3; 24.6 |
| Average B-factor (Å2) | |
| Chain A; B | 30.8; 31.4 |
| Solvent | 37.3 |
| r.m.s.d | |
| Bond length (Å) | 0.016 |
| Bond angles (deg.) | 1.503 |
| Ramachandran plot: favored; allowed; forbidden (%) | 99.2; 99.7; 0.27 |
Values in parentheses are for highest-resolution shell (2.07–2.00 Å).
, where i is the ith observation of a reflection with indices h,k,l and angle brackets indicate the average over all i observations.
, where Fchkl is the calculated structure factor amplitude with index h,k,l and Fohkl is the observed structure factor amplitude with index h,k,l.
Rfree is calculated as Rwork, where the Fohkl are taken from a test set comprising 5% of the data that were excluded from the refinement.
Structure Determination, Refinement, and Validation
Phases for DJ-1β were determined by molecular replacement using the human DJ-1 dimer (PBD code 2OR3)(41) as a search model in the CCP4 (44) implementation of PHASER (45). The initial model for DJ-1β was autobuilt using Phenix (46), inspected and manually improved in COOT (47), and subsequently refined in Refmac5 (48). Because the functional dimer of DJ-1β is present in the asymmetric unit, non-crystallographic symmetry restraints were applied to each monomer. Once maximum likelihood refinement of coordinates and isotropic displacement parameters had converged as judged by the absence of significant changes in the R factors or log likelihood gradient upon refinement, a translation-libration-screw (TLS) model with each monomer defined as a separate rigid group was refined (49). The final model was validated using MolProbity (50) and COOT (47). Final model statistics are provided in Table 1. Structural figures were made with PyMOL (Schrodinger).
Thermofluor assay of DJ-1 stability
Recombinant reduced human DJ-1 was purified and was oxidized to the cysteine-sulfinate form as previously described (34). Drosophila DJ-1β was oxidized to the cysteine-sulfinate form by desalting the purified protein into water using P6-DG resin (Bio-Rad), adding hydrogen peroxide (Fisher) to final molar ratio of 7:1 with the monomeric protein, and incubation on ice for 45 minutes. Unreacted peroxide was removed by centrifugal desalting using P6-DG resin. The oxidation states of both proteins were confirmed using electrospray mass spectrometry (see below).
The thermal stabilities of reduced and oxidized DJ-1 were measured using the thermofluor assay (51) in 25 mM HEPES pH=7.5, 100 mM KCl, 1 mM DTT. A 5000X stock of Sypro Orange in DMSO (Sigma-Aldrich) was added to protein (1–5 mg/ml) in concentrations ranging from 50–250X in optically clear PCR tube strips (Bio-Rad). The samples were heated from 20 to 95 °C at a rate of 2 °C/minute while fluorescence was excited at 490 nm and monitored at 575 nm using an iCycler iQ real-time thermal cycler (Bio-Rad). Data were plotted as the first derivative of fluorescence as a function of temperature, whose peak corresponds to the reported melting temperature (Tm). All measurements were made in triplicate at multiple protein concentrations.
Mass spectrometry analysis of oxidative modifications in DJ-1
Protein samples were digested with endopeptidase GluC for six hours according to the manufacturer’s instructions (Promega) followed by digestion with trypsin (Roche) at 37 °C without prior protein reduction or alkylation. The tryptic peptides were dried in a SpeedVac (Savant) and then desalted and concentrated using a monolithic PepMap C18 trap column (300 μm inner diameter, 1 mm length, 5 μm particle size, 100 Å pore size) loaded at 300 nl/min flow rate. Eluted peptides were separated on a second C18 PepMap column (75 μm inner diameter, 15 cm length, 3 μm particle, 100 Å pore) with an acetonitrile/0.1% formic acid gradient. The tryptic peptides were subjected to LC-MS/MS tandem mass spectrometry analysis using an Ultimate 3000 Dionex MDLC system (Dionex Corporation) integrated with a nanospray source and LCQ Fleet Ion Trap mass spectrometer (Thermofinnigan). The LCQ Fleet mass spectrometer was operated with a 2 kV nanospray voltage, a capillary temperature of 200 °C, and a full scan m/z range of 400–2000. The mass spectrometer was operated in data-dependent mode with four MS/MS spectra collected for every full scan, five microscan averaging for full and MS/MS scans, three m/z unit isolation width, and 35% collision energy for collision induced dissociation. Dynamic exclusion was enabled with an exclusion duration of 1 minute. The MS/MS spectra were searched against sequences for human DJ-1 and Drosophila DJ-1β that had been imported into the IPI human protein database using MASCOT (Version 2.2 Matrix Science). Database search criteria were as follows: GluC/Trypsin cleavage (i.e. peptides cleaved at Lys, Arg, Asp, Glu), two missed cleavages permitted, monoisotropic masses, variable modifications that included cysteine-sulfenic acid (Cys-SOH), -sulfinic acid (Cys-SO2− ),-sulfonic acid (Cys-SO3−), and methionine sulfoxide, a peptide mass tolerance of 1.5 Da and a MS/MS fragment ion tolerance of 1 Da.
Results
Structure of Drosophila DJ-1β
The DJ-1β construct used in this work results in a 187 amino acid monomer (19 kDa) and the numbering used in this paper assigns the first amino acid in the protein to amino acid 19 in the deposited sequence (GenBank AAF57086.2, GI:28381503, see Experimental Procedures). Electron density maps calculated after density modification of molecular replacement phases in Phenix allowed for the automatic placement of all residues in DJ-1β except for two amino acids each at the N- and C-termini, which are disordered. Like other close DJ-1 homologues, DJ-1β is a dimer with extensive intersubunit interactions, resulting in 1278 Å2 of buried surface area per monomer at the dimer interface as calculated by the ePDB PISA webserver (52). This amount of buried surface area is comparable to that of human DJ-1 (PDB 2OR3; 1349 Å2), indicating that DJ-1β is also an obligate homodimer. The only Ramachandran outlier in the refined model is the conserved Cys104 residue, which is often found in marginal or unfavorable regions of Ramachandran space.
Structural comparison of DJ-1β and human DJ-1
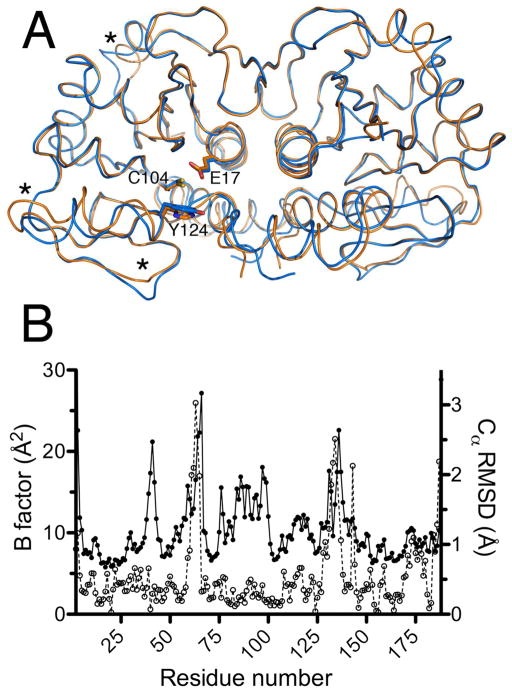
The backbone structure of Drosophila DJ-1β is highly similar to that of human DJ-1 (PDB code 2OR3) with a core Cα rmsd value for the dimers of 0.69 Å (Fig. 1A). Differences between the two structures are most pronounced at the termini and in two solvent exposed helical regions including amino acids 58-64 and 125-134 (DJ-1β numbering). These regions also have elevated atomic displacement parameters (ADPs) for Cα atoms in human DJ-1, suggesting that they are intrinsically flexible in close DJ-1 homologues (Fig. 1B). However, the converse is not true: there are regions with elevated ADPs in human DJ-1 that do not correspond to regions of high Cα RMSD for DJ-1β (Fig. 1B), likely due to crystal packing contacts. Most amino acids that have been shown to be important for human DJ-1 function or have been identified in patients with DJ-1-related parkinsonism are conserved in DJ-1β, including Leu10, Glu18, Ala104, Cys106, Asp149, Glu163, and Leu166 (human DJ-1 numbering).
Figure 1.
DJ-1β and human DJ-1 are structurally similar. Panel A: The physiologically relevant dimeric forms of human DJ-1 (orange) and Drosophila DJ-1β (blue) are superimposed, showing nearly identical backbone structures. The functionally significant residues Cys104 (Cys106), Glu17 (Glu18) and Tyr124 (His126) are shown (human DJ-1 residue numbering in parenthesis). Regions with the largest variation between the human and fly proteins are marked with asterisks. Panel B: The areas of greatest structural variation between the human and Drosophila proteins correlate with elevated atomic displacement parameter values in the human protein. Filled circles represent the average equivalent isotropic B factors for the Cα atoms of human DJ-1 (PDB 2OR3), while open circles represent the r.m.s.d. of the Cα atoms of the Drosophila and human DJ-1 crystal structures. The areas marked with asterisks in panel A correspond to the r.m.s.d. peaks from residues 60-70 and 125-150. In both plots, only data for chain A of the dimeric proteins are shown.
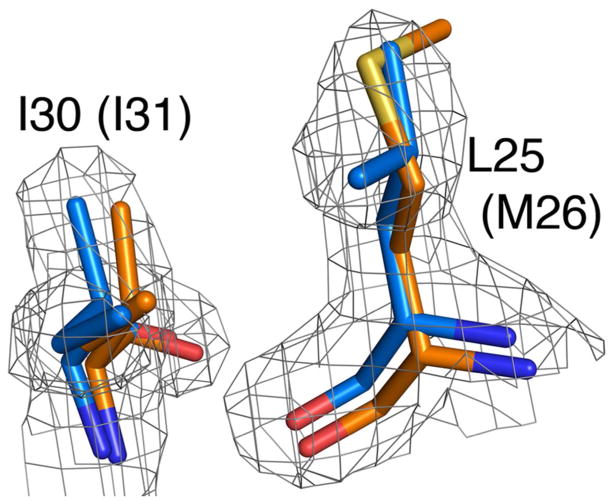
However, one position where a functionally important residue in human DJ-1 is not conserved in Drosophila DJ-1β is Met26. The M26I mutation in human DJ-1 was identified in an Ashkenazi Jewish patient with early onset parkinsonism (53) and has been shown to be detrimental to DJ-1 function in cell culture (9, 54). Therefore, this clinical variant is likely a bona fide pathological mutation and not a polymorphism. The structures of DJ-1β and human DJ-1 are highly similar in this region (Fig. 2), despite the Met to Leu substitution at position 25. Importantly, Drosophila DJ-1β does not exhibit the displaced I31 residue that is found in human M26I DJ-1 (55), indicating that the modest perturbation at this region in human M26I DJ-1 is a good candidate for the structural disruption that results in M26I DJ-1-linked pathogenesis.
Figure 2.
The parkinsonism-associated Met26 residue is replaced with a structurally conservative leucine residue in DJ-1β. A stick representation of the superimposed structures of human DJ-1 (orange) and Drosophila DJ-1β (blue) shows that the substitution of leucine for methionine causes minimal structural disruption in that area, in contrast to packing defects created by the M26I mutation in human DJ-1. Drosophila DJ-1β numbering is shown with human residue numbering in parenthesis.
Cys104 is reactive and partially oxidized to the sulfinate in the crystal
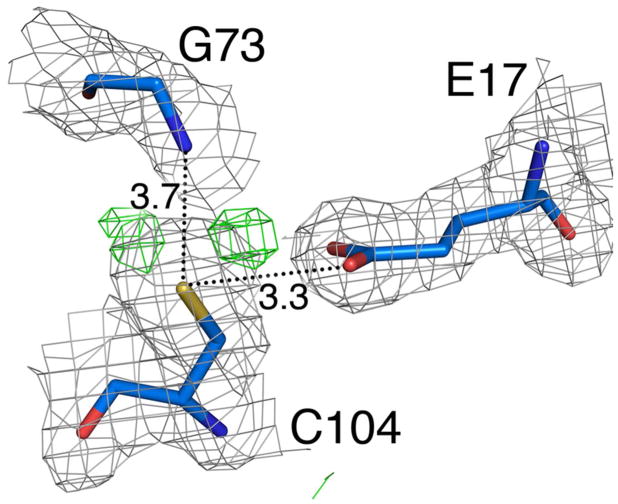
Nearly all members of the DJ-1 superfamily contain a cysteine residue at the juncture between a β-strand and an α-helix called the “nucleophile elbow” in the α/β hydrolases. This cysteine serves a catalytic role in some members of the DJ-1 superfamily, including validated peptidases, glyoxalases, and isocyanide hydratases (56–59). In the close homologues of human DJ-1, this residue is highly reactive and oxidation-prone, although no unambiguous enzymatic activity has been identified in these proteins. The environment surrounding Cys104 in DJ-1β is similar to that of human DJ-1, including the presence of a functionally important glutamic acid (Glu18 in human DJ-1, Glu17 in Drosophila DJ-1β). The 2mFo-DFc electron density for Cys104 in DJ-1β is consistent with a predominantly reduced thiol(ate), although difference electron density indicates some minor amount of modification has occurred (Fig. 3). The two peaks in the difference electron density are consistent with partial oxidation to the cysteine-sulfinate, which has been directly observed in the crystal structure of the human protein and inferred on the basis of acidic pI shifts for Drosophila DJ-1β. We note that these two difference electron density peaks could alternatively be modeled as a cysteine-sulfenic acid (Cys-SOH) sampling two alternative conformations. However, electrospray mass spectrometry of intentionally oxidized DJ-1β (see Experimental Procedures) confirms that the peptide containing Cys104 is oxidized to the cysteine-sulfinate in solution (Supporting Information Table 1).
Figure 3.
Cys104 is reactive and makes hydrogen bonds with several surrounding residues. The hydrogen bonding environment of Cys104 is shown, with 2mFo-DFc electron density (1σ) shown in grey and mFo-DFc difference electron density (3.5σ) shown in green and red. The small positive peaks in difference electron density (green) around Cys104 indicate a minor amount of oxidative modification of the residue, likely to the cysteine-sulfinate. Dotted lines indicate hydrogen bonds with distances given in Ångstroms.
A proposed catalytic dyad is absent in DJ-1β
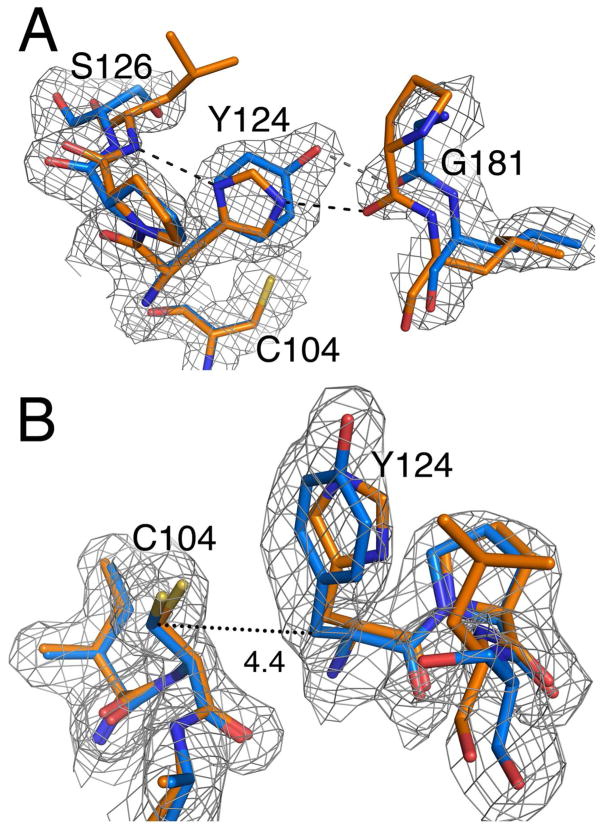
The presence of a reactive cysteine suggests the possibility that DJ-1β may be an enzyme. Members of the PfpI and Hsp31 clades of the DJ-1 superfamily have the conserved reactive cysteine in a putative catalytic triad consisting of Cys-His-Glu/Asp where the histidine immediately follows the cysteine residue (60, 61). Some of these proteins are validated proteases or peptidases (56, 62), although others appear to lack this activity (63, 64). Early speculation about a function for human DJ-1 suggested that it might also be a protease, although a catalytic triad is absent in human DJ-1 (65). A different but nearby histidine residue (His126) in human DJ-1 was proposed to compose an alternative catalytic dyad with Cys106 (42). Because the imidazole sidechain of His126 is not oriented properly to form a hydrogen bond with Cys106 in full-length DJ-1 (Fig. 4A), it has been suggested that DJ-1 may be a pro-protein that is activated by the proteolytic cleavage of the C-terminal α-helix (residues 174-189) of DJ-1. This cleavage would allow rotation of the His126 sidechain and completion of the dyad with Cys106, and truncated recombinant DJ-1 lacking this C-terminal peptide does have elevated proteolytic activity in vitro (42). Drosophila DJ-1β has a tyrosine at the equivalent position (Tyr124), which is a less compelling candidate for a second member of a catalytic dyad. The crystal structure of DJ-1β shows that the aromatic ring of Tyr124 superimposes well with the imidazole ring of His126 in human DJ-1, which positions its phenolic oxygen away from Cys104 (Fig. 4A). Because any functional dyad would require a hydrogen bond donor to interact with Cys104, this orientation of Tyr124 prohibits dyad formation with Cys104. The proposed removal of the C-terminal peptide of DJ-1β could result in rotation of Tyr124, but the size of the Tyr124 sidechain and its preferred rotameric states prohibit a hydrogen bonding interaction with Cys104, which is only 4.4 Å away from Tyr124 as measured between Cβ atoms (Fig. 4B).
Figure 4.
A putative cysteine-histidine dyad in human DJ-1 is absent in the active site of DJ-1β. Panel A shows the superimposed models for human DJ-1 (orange) and DJ-1β (blue) around His126 in human DJ-1. This residue was proposed to compose a dyad with Cys106, although it is substituted with a tyrosine in DJ-1β. The 2mFo-DFc electron density map calculated for DJ-1β at 2.0 Å resolution is contoured at 1σ and shown in grey. In Panel B, the putative active site region around the reactive Cys104 in DJ-1β is shown. Although the histidine sidechain in the human protein could reorient by ~180° to form a dyad with Cys106, the limited space (4.4 Å) between Cys104 and Tyr124 precludes rotational reorientation to create a Cys-Tyr dyad required for the protease model for DJ-1 function.
Oxidation of Cys106 to the cysteine-sulfinate stabilizes DJ-1
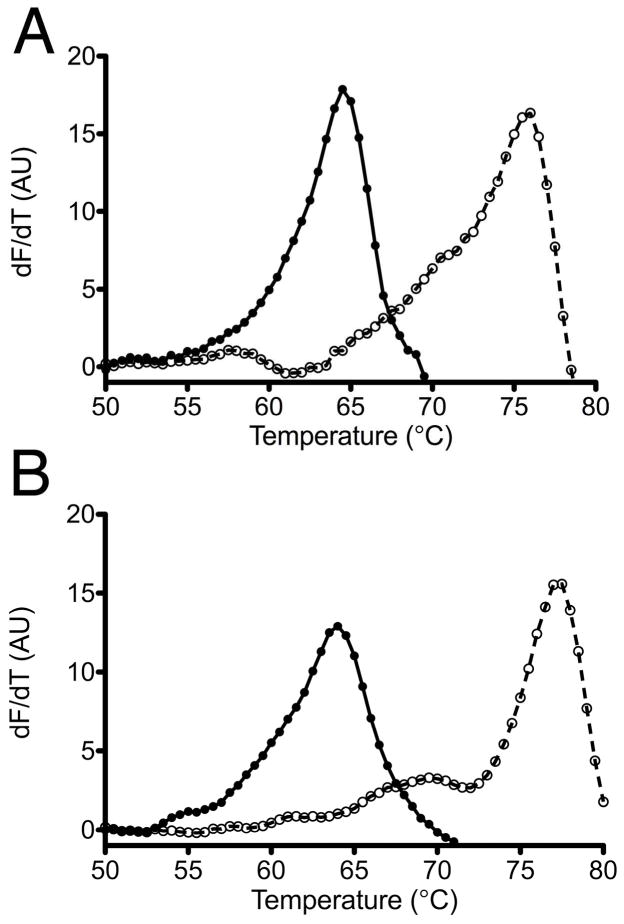
The conserved reactive cysteine residue in DJ-1 is easily oxidized to cysteine-sulfinate in vitro (3) and in vivo (36, 66). Oxidative post-translational modifications can alter protein stability, which may have functional significance in this system. The in vitro stabilities of DJ-1β and human DJ-1 in their reduced and Cys-SO2− forms were determined using the thermofluor stability assay. Proteins were oxidized using hydrogen peroxide in vitro (see Experimental Procedures) and the oxidation state of the modified cysteine was confirmed using electrospray mass spectrometry (Supporting Information Table 1). In both homologues, oxidation of the reactive cysteine to the sulfinate stabilized the proteins by ~12°C, from 64.5 to 76.0 °C for Drosophila DJ-1β (Fig. 5A) and from 64.0 to 77.3°C for human DJ-1 (Fig. 5B). We note that the melting temperature (Tm) for reduced human DJ-1 determined here (64.0°C) agrees well with that determined for the same protein by differential scanning calorimetry (66.2°C) (55), confirming that the increase in quantum yield for Sypro Orange being measured in this assay is in fact due to binding of the fluorphore in unfolded protein. The formation of three additional hydrogen bonds between Cys106-SO2− and surrounding residues is likely responsible for this stabilization, particularly the short, strong 2.5 Å hydrogen bond made between Cys106-SO2− and the protonated sidechain of Glu18 (Glu17 in DJ-1β) (Fig. 3) (41).
Figure 5.
Cysteine-sulfinate formation stabilizes DJ-1. The thermofluor assay was used to monitor the thermal denaturation of reduced DJ-1 (filled circles) or protein that was oxidized at the conserved cysteine residue to the cysteine-sulfinate (open circles). Drosophila DJ-1β is shown in panel A and human DJ-1 is in panel B. In both cases, oxidation increases the thermal stabilities of the proteins by ~12°C.
Discussion
The predicted coding sequence for D. melanogaster DJ-1β deposited with the NCBI (GenBank AAF57086.2 GI:28381503) is 205 amino acids long, including an 18 amino acid N-terminal extension that is absent in many other DJ-1 proteins from other organisms (5). This region corresponds either to an incorrectly predicted (or alternative) translational start site or a targeting sequence. The TargetP 1.1 webserver (67) predicts that this extension is a possible mitochondrial localization sequence, but the confidence of this prediction is low (reliability class 4) and thus its significance is uncertain. There is evidence that DJ-1β has a role in mitochondrial function (32) and DJ-1β partially localizes to the mitochondria of Drosophila cells (30). However, because this N-terminal region is not well conserved and native DJ-1β migrates in SDS-PAGE with a molecular mass consistent with the absence of this peptide (5), we have used a version of the protein starting from amino acid 19. However, it is possible that this N-terminal sequence has biological significance and may warrant further investigation.
In human DJ-1, Met26 is a critically important residue whose mutation to isoleucine has been convincingly associated with loss of DJ-1-mediated cytoprotection and consequent parkinsonism (9, 53). Despite the importance of Met26 for human DJ-1 stability and function, it is poorly conserved in insect and bacterial DJ-1 homologues, where is it often substituted with a leucine, as in D. melanogaster DJ-1β. The crystal structure of Drosophila DJ-1β shows that the substitution of leucine for methionine at position 26 has little structural impact on the protein and is well-tolerated by surrounding residues. In contrast, the crystal structure of human M26I DJ-1 showed that placing a β-branched isoleucine sidechain in this environment creates a steric conflict with Leu31 that can be relieved by an artificial M26L mutation (55). This packing defect, although minor, was speculated to contribute to the observed loss of stability and function of M26I DJ-1 (55). The fact that the previously designed M26L mutation in human DJ-1 relieves this steric conflict and corresponds closely with the native structure of functional Drosphila DJ-1β provides strong circumstantial evidence that the Ile26-Leu31 conflict in human M26I DJ-1 is responsible for loss of protein stability and, ultimately, pathogenesis. This structural effect may be indirectly responsible for the recently observed deficit in Cys106 oxidation in M26I DJ-1, which may compromise the protective function of this mutant protein (35).
Drosophila DJ-1β has a reactive cysteine residue (Cys104) that has been shown to be critical for its function (33). This residue is partially oxidized to the cysteine-sulfinate (Cys-SO2−) in the crystal and this modification is stabilized by hydrogen bonding with surrounding residues, consistent with previous observations for the human protein. In particular, a previously identified hydrogen bonded interaction between Glu18 and Cys106 is also seen in DJ-1β (Cys104 and Glu17; Fig. 3A). In human DJ-1, the Glu18 sidechain is protonated and donates a hydrogen bond to Cys106, depressing its pKa and stabilizing the oxidized Cys106-SO2− (41). The very similar orientation of these residues in DJ-1β suggests that Glu17 in DJ-1β is also likely a protonated carboxylic acid at physiological pH.
Based initially on structural similarity to the archaeal protease PfpI (60), human DJ-1 has been considered a possible protease, although various studies have reported conflicting results about its in vitro proteolyitc activity (22, 42, 68). DJ-1 lacks a canonical catalytic dyad or triad, however His126 has been proposed to function as the second member of a catalytic dyad involving Cys106 that forms when the C-terminal 15 residues are cleaved from DJ-1 (42). The structure of Drosophila DJ-1β suggests that this mechanism is unlikely to be valid for homologues that contain a tyrosine at this postion, as the environment of this residue prohibits hydrogen bonding that is necessary to complete the dyad. Moreover, the removal of the C-terminal portion of DJ-1β is not expected to alter this, as the bulky Tyr124 residue cannot sample a conformation that would permit dyad formation with Cys104 without major structural rearrangements in the protein. Consequently, a model requiring DJ-1 to be a zymogenic protease using a Cys-His/Tyr dyad (42) is inconsistent with the detailed structural features of DJ-1β. As the human and insect proteins appear to have similar cellular functions, it seems unlikely that human DJ-1 is a physiologically relevant protease. Importantly, this does not exclude other potential enzymatic activities for DJ-1.
Cys106 is subject to modification during oxidative stress, and its oxidation has been proposed to be an important post-translational modification that may allow DJ-1 to act as a cellular redox sensor (34) or to regulate interactions with its binding partners (20, 69). However, the detailed mechanism by which cysteine oxidation alters DJ-1 function is unclear, particularly since the crystal structures of the reduced and Cys106-SO2− proteins are essentially identical (3, 70). Our results show that both the human and insect proteins are substantially stabilized by oxidation of the reactive cysteine to the cysteine-sulfinate. The stabilization contrasts with many other proteins where cysteine oxidation to the -sulfinate or -sulfonate is associated with destabilization (71, 72). When considered in the context of previous evidence that oxidation of Cys106 in human DJ-1 leads to protein destabilization (39), our observations strongly suggest that it is the formation of the more highly oxidized cysteine-sulfonate (Cys106-SO3−) that disrupts DJ-1 stability. This is consistent with some previously proposed models (37, 39) and indicates that DJ-1 stability is biphasically regulated by oxidative cysteine modification. Importantly, it is possible that Cys106-SO3− DJ-1 has a functional role, which has not been thoroughly investigated.
The melting temperatures of both reduced and oxidized DJ-1 from both species are well above physiological temperature. However, changes in thermal stability above the physiological range often correlate with changes in protein stability or half-life at 37°C (73). For example, the parkinsonian M26I mutation in DJ-1 has a modestly reduced melting temperature of ~58°C (39, 55, 74, 75) in vitro but is unstable and poorly functional in cell culture at 37°C (9, 54). Furthermore, the engineered introduction of disulfide bonds in human DJ-1 both increases its melting temperature and enhances its chaperone activity against α-synuclein in vitro at 37°C (74). Although there are likely to be several mechanisms by which cysteine-sulfinic acid formation modulates DJ-1 activity, the stabilization of the protein by this oxidative modification may contribute to DJ-1’s cytoprotective activity during oxidative stress. Moreover, because the residues that make hydrogen bonds to the cysteine-sulfinate in DJ-1 are well-conserved in close homologues, it is likely that the oxidative stabilization via cysteine-sulfinate formation is shared by many members of the DJ-1 family.
Supplementary Material
Acknowledgments
We thank Professor Nancy Bonini (University of Pennsylvania) for the DJ-1β clone and helpful discussions, Professor Joseph Barycki and Anastassia Harris (University of Nebraska-Lincoln) for help with the thermofluor assay, and Professor Melanie Simpson (University of Nebraska-Lincoln) for the use of the real-time thermal cycler. We also thank Drs. Nandakumar Madayiputhiya and Renu Nandakumar for protein modification analysis by mass spectrometry at the University of Nebraska-Lincoln Redox Biology Center Proteomic and Metabolomic Core Facility.
Funding satement: GM/CA-CAT has been funded in whole or in part with federal funds from the National Institutes of Health (NCI, Y1-CO-1020; NIGMS, Y1-GM-1104). Use of the Advanced Photon Source was supported by the United States Department of Energy, Basic Energy Sciences, Office of Science, under Contract DE-AC02–06CH11357. This work was supported, in whole or in part, by National Institutes of Health grant R01 GM092999 to MAW.
Abbreviations
- ADP
atomic displacement parameters
- CCD
charge coupled device
- Da
Dalton
- DMSO
Dimethyl sulfoxide
- DTT
dithiothreitol
- HEPES
4-(2-hydroxyethyl)-1-piperazine ethane sulfonic acid
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- KeV
kiloelectronvolt
- kV
kilovolt
- LC-MS/MS
liquid chromotagraphy-mass spectrometry/mass spectrometry
- PDB
Protein Data Bank
- r.m.s.d
root mean square deviation
- TLS
translation-libration-screw
- UV
ultraviolet
Footnotes
PDB Deposition: Refined model coordinates and experimental structure factors for Drosophila melanogaster DJ-1β have been deposited with the RCSB Protein Data Bank with accession code 4E08.
Suppporting Information Available
Supplemental Table 1 shows a mass spectrometry analysis of oxidative modifications in proteolyzed reduced and hydrogen peroxide-oxidized human DJ-1 and Drosophila DJ-1β. This material is available free of charge via the Internet at http://www.pubs.acs.org.
References
- 1.Nagakubo D, Taira T, Kitaura H, Ikeda M, Tamai K, IguchiAriga SMM, Ariga H. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 2.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MCJ, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 3.Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi-Niki K, Niki T, Taira T, Iguchi-Ariga SMM, Ariga H. Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson’s disease patients. Biochem Biophys Res Commun. 2004;320:389–397. doi: 10.1016/j.bbrc.2004.05.187. [DOI] [PubMed] [Google Scholar]
- 5.Meulener M, Whitworth AJ, Armstrong-Gold CE, Rizzu P, Heutink P, Wes PD, Pallanck LJ, Bonini NM. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson’s disease. Curr Biol. 2005;15:1572–1577. doi: 10.1016/j.cub.2005.07.064. [DOI] [PubMed] [Google Scholar]
- 6.Larsen NJ, Ambrosi G, Mullett SJ, Berman SB, Hinkle DA. DJ-1 knockdown impairs astrocyte mitochondrial function. Neuroscience. 2011;196:251–264. doi: 10.1016/j.neuroscience.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT, Surmeier DJ. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Im JY, Lee KW, Junn E, Mouradian MM. DJ-1 protects against oxidative damage by regulating the thioredoxin/ASK1 complex. Neurosci Res. 2010;67:203–208. doi: 10.1016/j.neures.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waak J, Weber SS, Gorner K, Schall C, Ichijo H, Stehle T, Kahle PJ. Oxidizable residues mediating protein stability and cytoprotective interaction of DJ-1 with apoptosis signal-regulating kinase 1. J Biol Chem. 2009;284:14245–14257. doi: 10.1074/jbc.M806902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim RH, Peters M, Jang Y, Shi W, Pintilie M, Fletcher GC, DeLuca C, Liepa J, Zhou L, Snow B, Binari RC, Manoukian AS, Bray MR, Liu FF, Tsao MS, Mak TW. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7:263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 11.Das F, Dey N, Venkatesan B, Kasinath BS, Ghosh-Choudhury N, Choudhury GG. High glucose upregulation of early-onset Parkinson’s disease protein DJ-1 integrates the PRAS40/TORC1 axis to mesangial cell hypertrophy. Cell Signal. 2011;23:1311–1319. doi: 10.1016/j.cellsig.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karunakaran S, Diwakar L, Saeed U, Agarwal V, Ramakrishnan S, Iyengar S, Ravindranath V. Activation of apoptosis signal regulating kinase 1 (ASK1) and translocation of death-associated protein, Daxx, in substantia nigra pars compacta in a mouse model of Parkinson’s disease: protection by alpha-lipoic acid. FASEB J. 2007;21:2226–2236. doi: 10.1096/fj.06-7580com. [DOI] [PubMed] [Google Scholar]
- 13.Ren H, Fu K, Mu C, Li B, Wang D, Wang G. DJ-1, a cancer and Parkinson’s disease associated protein, regulates autophagy through JNK pathway in cancer cells. Cancer Lett. 2010;297:101–108. doi: 10.1016/j.canlet.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Aron L, Klein P, Pham TT, Kramer ER, Wurst W, Klein R. Pro-survival role for Parkinson’s associated gene DJ-1 revealed in trophically impaired dopaminergic neurons. PLoS Biol. 2010;8:e1000349. doi: 10.1371/journal.pbio.1000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Liu J, Chen S, Wang Y, Cao L, Zhang Y, Kang W, Li H, Gui Y, Ding J. DJ-1 modulates the expression of Cu/Zn-superoxide dismutase-1 through the Erk1/2-Elk1 pathway in neuroprotection. Ann Neurol. 2011;70:591–599. doi: 10.1002/ana.22514. [DOI] [PubMed] [Google Scholar]
- 16.Fan J, Ren H, Jia N, Fei E, Zhou T, Jiang P, Wu M, Wang G. DJ-1 decreases Bax expression through repressing p53 transcriptional activity. J Biol Chem. 2008;283:4022–4030. doi: 10.1074/jbc.M707176200. [DOI] [PubMed] [Google Scholar]
- 17.Vasseur S, Afzal S, Tomasini R, Guillaumond F, Tardivel-Lacombe J, Mak TW, Iovanna JL. Consequences of DJ-1 upregulation following p53 loss and cell transformation. Oncogene. 2012;31:664–670. doi: 10.1038/onc.2011.268. [DOI] [PubMed] [Google Scholar]
- 18.Bretaud S, Allen C, Ingham PW, Bandmann O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson’s disease. J Neurochem. 2007;100:1626–1635. doi: 10.1111/j.1471-4159.2006.04291.x. [DOI] [PubMed] [Google Scholar]
- 19.Shinbo Y, Taira T, Niki T, Iguchi-Ariga SM, Ariga H. DJ-1 restores p53 transcription activity inhibited by Topors/p53BP3. Int J Oncol. 2005;26:641–648. [PubMed] [Google Scholar]
- 20.McNally RS, Davis BK, Clements CM, Accavitti-Loper MA, Mak TW, Ting JP. DJ-1 enhances cell survival through the binding of Cezanne, a negative regulator of NF-kappaB. J Biol Chem. 2011;286:4098–4106. doi: 10.1074/jbc.M110.147371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci USA. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A. DJ-1 is a redox- dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004;2:e362. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W, Zhu M, Wilson MA, Petsko GA, Fink AL. The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J Mol Biol. 2006;356:1036–1048. doi: 10.1016/j.jmb.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 24.van der Brug MP, Blackinton J, Chandran J, Hao LY, Lal A, Mazan-Mamczarz K, Martindale J, Xie C, Ahmad R, Thomas KJ, Beilina A, Gibbs JR, Ding J, Myers AJ, Zhan M, Cai H, Bonini NM, Gorospe M, Cookson MR. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci USA. 2008;105:10244–10249. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu F, Nguyen JL, Hulleman JD, Li L, Rochet JC. Mechanisms of DJ-1 neuroprotection in a cellular model of Parkinson’s disease. J Neurochem. 2008;105:2435–2453. doi: 10.1111/j.1471-4159.2008.05333.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhou W, Freed CR. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J Biol Chem. 2005;280:43150–43158. doi: 10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Zhong N, Wang H, Elias JE, Kim CY, Woldman I, Pifl C, Gygi SP, Geula C, Yankner BA. The Parkinson’s disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum Mol Genet. 2005;14:1231–1241. doi: 10.1093/hmg/ddi134. [DOI] [PubMed] [Google Scholar]
- 28.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson’s disease. Science. 2002;295:865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 29.Menzies FM, Yenisetti SC, Min KT. Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr Biol. 2005;15:1578–1582. doi: 10.1016/j.cub.2005.07.036. [DOI] [PubMed] [Google Scholar]
- 30.Park J, Kim SY, Cha GH, Lee SB, Kim S, Chung J. Drosophila DJ-1 mutants show oxidative stress-sensitive locomotive dysfunction. Gene. 2005;361:133–139. doi: 10.1016/j.gene.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 31.Lavara-Culebras E, Munoz-Soriano V, Gomez-Pastor R, Matallana E, Paricio N. Effects of pharmacological agents on the lifespan phenotype of Drosophila DJ-1beta mutants. Gene. 2010;462:26–33. doi: 10.1016/j.gene.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci USA. 2010;107:9747–9752. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meulener MC, Xu KX, Thomson L, Ischiropoulos H, Bonini NM. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging (vol 103, pg 12517, 2006) Proc Natl Acad Sci USA. 2006;103:14978–14978. doi: 10.1073/pnas.0601891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackinton J, Lakshminarasimhan M, Thomas KJ, Ahmad R, Greggio E, Raza AS, Cookson MR, Wilson MA. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J Biol Chem. 2009;284:6476–6485. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madian AG, Hindupur J, Hulleman JD, Diaz-Maldonado N, Mishra VR, Guigard E, Kay CM, Rochet JC, Regnier FE. Effect of Single Amino Acid Substitution on Oxidative Modifications of the Parkinson’s Disease-Related Protein, DJ-1. Mol Cell Proteomics. 2012;11:M111 010892. doi: 10.1074/mcp.M111.010892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitsumoto A, Nakagawa Y, Takeuchi A, Okawa K, Iwamatsu A, Takanezawa Y. Oxidized forms of peroxiredoxins and DJ-1 on two-dimensional gels increased in response to sublethal levels of paraquat. Free Radic Res. 2001;35:301–310. doi: 10.1080/10715760100300831. [DOI] [PubMed] [Google Scholar]
- 37.Wilson MA. The role of cysteine oxidation in DJ-1 function and dysfunction. Antioxid Redox Signal. 2011;15:111–122. doi: 10.1089/ars.2010.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J, Nazarenus TJ, Frey JL, Liang X, Wilson MA, Stone JM. A plant DJ-1 homolog is essential for Arabidopsis thaliana chloroplast development. PLoS One. 2011;6:e23731. doi: 10.1371/journal.pone.0023731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hulleman JD, Mirzaei H, Guigard E, Taylor KL, Ray SS, Kay CM, Regnier FE, Rochet JC. Destabilization of DJ-1 by familial substitution and oxidative modifications: implications for Parkinson’s disease. Biochemistry. 2007;46:5776–5789. doi: 10.1021/bi7001778. [DOI] [PubMed] [Google Scholar]
- 40.Choi J, Sullards MC, Olzmann JA, Rees HD, Weintraub ST, Bostwick DE, Gearing M, Levey AI, Chin LS, Li L. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J Biol Chem. 2006;281:10816–10824. doi: 10.1074/jbc.M509079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witt AC, Lakshminarasimhan M, Remington BC, Hasim S, Pozharski E, Wilson MA. Cysteine pKa depression by a protonated glutamic acid in human DJ-1. Biochemistry. 2008;47:7430–7440. doi: 10.1021/bi800282d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Li L, Chin LS. Parkinson disease protein DJ-1 converts from a zymogen to a protease by carboxyl-terminal cleavage. Hum Mol Genet. 2010;19:2395–2408. doi: 10.1093/hmg/ddq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 44.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 45.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 50.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 52.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 53.Abou-Sleiman PM, Healy DG, Quinn N, Lees AJ, Wood NW. The role of pathogenic DJ-1 mutations in Parkinson’s disease. Ann Neurol. 2003;54:283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 54.Blackinton J, Ahmad R, Miller DW, van der Brug MP, Canet-Aviles RM, Hague SM, Kaleem M, Cookson MR. Effects of DJ-1 mutations and polymorphisms on protein stability and subcellular localization. Brain Res Mol Brain Res. 2005;134:76–83. doi: 10.1016/j.molbrainres.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 55.Lakshminarasimhan M, Maldonado MT, Zhou W, Fink AL, Wilson MA. Structural impact of three Parkinsonism-associated missense mutations on human DJ-1. Biochemistry. 2008;47:1381–1392. doi: 10.1021/bi701189c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Malki A, Caldas T, Abdallah J, Kern R, Eckey V, Kim SJ, Cha SS, Mori H, Richarme G. Peptidase activity of the Escherichia coli Hsp31 chaperone. J Biol Chem. 2005;280:14420–14426. doi: 10.1074/jbc.M408296200. [DOI] [PubMed] [Google Scholar]
- 57.Subedi KP, Choi D, Kim I, Min B, Park C. Hsp31 of Escherichia coli K-12 is glyoxalase III. Mol Microbiol. 2011;81:926–936. doi: 10.1111/j.1365-2958.2011.07736.x. [DOI] [PubMed] [Google Scholar]
- 58.Goda M, Hashimoto Y, Takase M, Herai S, Iwahara Y, Higashibata H, Kobayashi M. Isonitrile hydratase from Pseudomonas putida N19-2. Cloning, sequencing, gene expression, and identification of its active acid residue. J Biol Chem. 2002;277:45860–45865. doi: 10.1074/jbc.M208571200. [DOI] [PubMed] [Google Scholar]
- 59.Halio SB, Blumentals II, Short SA, Merrill BM, Kelly RM. Sequence, expression in Escherichia coli, and analysis of the gene encoding a novel intracellular protease (PfpI) from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1996;178:2605–2612. doi: 10.1128/jb.178.9.2605-2612.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du XL, Choi IG, Kim R, Wang WR, Jancarik J, Yokota H, Kim SH. Crystal structure of an intracellular protease from Pyrococcus horikoshii at 2-angstrom resolution. Proc Natl Acad Sci USA. 2000;97:14079–14084. doi: 10.1073/pnas.260503597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quigley PM, Korotkov K, Baneyx F, Hol WG. The 1.6-A crystal structure of the class of chaperones represented by Escherichia coli Hsp31 reveals a putative catalytic triad. Proc Natl Acad Sci U S A. 2003;100:3137–3142. doi: 10.1073/pnas.0530312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee SJ, Kim SJ, Kim IK, Ko J, Jeong CS, Kim GH, Park C, Kang SO, Suh PG, Lee HS, Cha SS. Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J Biol Chem. 2003;278:44552–44559. doi: 10.1074/jbc.M304517200. [DOI] [PubMed] [Google Scholar]
- 63.Fioravanti E, Dura MA, Lascoux D, Micossi E, Franzetti B, McSweeney S. Structure of the Stress Response Protein DR1199 from Deinococcus radiodurans: A Member of the DJ-1 Superfamily. Biochemistry. 2008;47:11581–11589. doi: 10.1021/bi800882v. [DOI] [PubMed] [Google Scholar]
- 64.Abdallah J, Kern R, Malki A, Eckey V, Richarme G. Cloning, expression, and purification of the general stress protein YhbO from Escherichia coli. Protein Expr Purif. 2006;47:455–460. doi: 10.1016/j.pep.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 65.Huai Q, Sun Y, Wang H, Chin LS, Li L, Robinson H, Ke H. Crystal structure of DJ-1/RS and implication on familial Parkinson’s disease. FEBS Lett. 2003;549:171–175. doi: 10.1016/s0014-5793(03)00764-6. [DOI] [PubMed] [Google Scholar]
- 66.Knobbe CB, Revett TJ, Bai Y, Chow V, Jeon AH, Bohm C, Ehsani S, Kislinger T, Mount HT, Mak TW, St George-Hyslop P, Schmitt-Ulms G. Choice of biological source material supersedes oxidative stress in its influence on DJ-1 in vivo interactions with Hsp90. J Proteome Res. 2011;10:4388–4404. doi: 10.1021/pr200225c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 68.Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson’s disease. Proc Natl Acad Sci U S A. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ren H, Fu K, Wang D, Mu C, Wang G. Oxidized DJ-1 interacts with the mitochondrial protein BCL-XL. J Biol Chem. 2011;286:35308–35317. doi: 10.1074/jbc.M110.207134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Premkumar L, Dobaczewska MK, Riedl SJ. Identification of an artificial peptide motif that binds and stabilizes reduced human DJ-1. J Struct Biol. 2011;176:414–418. doi: 10.1016/j.jsb.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mueller M, Kratzer R, Schiller M, Slavica A, Rechberger G, Kollroser M, Nidetzky B. The role of Cys108 in Trigonopsis variabilis d-amino acid oxidase examined through chemical oxidation studies and point mutations C108S and C108D. Biochim Biophys Acta. 2010;1804:1483–1491. doi: 10.1016/j.bbapap.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 72.Fox JH, Connor T, Stiles M, Kama J, Lu Z, Dorsey K, Lieberman G, Sapp E, Cherny RA, Banks M, Volitakis I, DiFiglia M, Berezovska O, Bush AI, Hersch SM. Cysteine oxidation within N-terminal mutant huntingtin promotes oligomerization and delays clearance of soluble protein. J Biol Chem. 2011;286:18320–18330. doi: 10.1074/jbc.M110.199448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Willis MN, Liu Y, Biterova EI, Simpson MA, Kim H, Lee J, Barycki JJ. Enzymatic defects underlying hereditary glutamate cysteine ligase deficiency are mitigated by association of the catalytic and regulatory subunits. Biochemistry. 2011;50:6508–6517. doi: 10.1021/bi200708w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Logan T, Clark L, Ray SS. Engineered disulfide bonds restore chaperone-like function of DJ-1 mutants linked to familial Parkinson’s disease. Biochemistry. 2010;49:5624–5633. doi: 10.1021/bi902164h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Malgieri G, Eliezer D. Structural effects of Parkinson’s disease linked DJ-1 mutations. Protein Sci. 2008;17:855–868. doi: 10.1110/ps.073411608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.