SUMMARY
Human pluripotent stem cells (hPSCs) are potential sources of cells for modeling disease and development, drug discovery, and regenerative medicine. However, it is important to identify factors that may impact the utility of hPSCs for these applications. In an unbiased analysis of 205 hPSC and 130 somatic samples, we identified hPSC-specific epigenetic and transcriptional aberrations in genes subject to X chromosome inactivation (XCI) and genomic imprinting, which were not corrected during directed differentiation. We also found that specific tissue types were distinguished by unique patterns of DNA hypomethylation, which were recapitulated by DNA demethylation during in vitro directed differentiation. Our results suggest that verification of baseline epigenetic status is critical for hPSC-based disease models in which the observed phenotype depends on proper XCI or imprinting, and that tissue-specific DNA methylation patterns can be accurately modeled during directed differentiation of hPSCs, even in the presence of variations in XCI or imprinting.
INTRODUCTION
hPSCs maintain the ability to self-renew indefinitely and can be differentiated into a wide range of cell types, making them an excellent source of differentiated cells for preclinical and clinical applications. However, several studies have reported genetic, epigenetic and transcriptional variation among hPSC cultures (Bock et al., 2011; Chin et al., 2009; Feng et al., 2010; Gore et al., 2011; Hough et al., 2009; Hussein et al., 2011; Kim et al., 2007; Laurent et al., 2011; Lister et al., 2011; Marchetto et al., 2009; Ohi et al., 2011), which may affect their differentiation propensities and utility for disease modeling, cell therapy, and drug development (Bock et al., 2011; Pomp et al., 2011; Tchieu et al., 2010; Urbach et al., 2010).
Epigenetic processes, including DNA methylation, histone modifications, and non-coding RNA expression, act coordinately to regulate cellular differentiation and homeostasis. During development, different cell types acquire distinct DNA methylation profiles that reflect their developmental stage and functional identity. For most genes, the pattern of DNA methylation is identical on both alleles; at more evolutionarily complex loci, including imprinted and X chromosome genes, however, only a single allele is normally methylated.
Genomic imprinting is the mechanism by which monoallelic expression is achieved in a parent-of-origin-specific fashion. At least 60 human genes are known to be imprinted (geneimprint.org) and can be further classified as “gametic” when the imprints are established in the germline or “somatic” when they arise during early embryonic development as a result of spreading of gametic imprints (reviewed in (John and Lefebvre, 2011)). Genomic imprints are particularly susceptible to environmental factors (Dolinoy et al., 2007; Odom and Segars, 2010) and imprinting defects are associated with developmental disorders, including Silver-Russell, Beckwith-Wiedemann, and Prader-Willi syndromes, as well as several human cancers (Bhusari et al., 2011; Uribe-Lewis et al., 2011). Variability in imprinting status has been reported for hPSCs (Adewumi et al., 2007; Frost et al., 2011; Kim et al., 2007; Rugg-Gunn et al., 2007), but the extent of this variation is unclear due to the limited number of imprinted genes, cell lines and cell types assayed in those studies.
X chromosome inactivation (XCI) refers to the transcriptional repression of one of the two X chromosomes in female cells, and mediates dosage compensation between XY males and XX females (reviewed in (Kim et al., 2011)). Transcription of a long non-coding RNA, XIST (X-inactive specific transcript), has a role in initiating and maintaining XCI. In mice, female PSCs do not express Xist and have two active X chromosomes (XaXa); upon differentiation, Xist transcription is de-repressed on a single X chromosome, resulting in inactivation of that chromosome (XaXi). The process of XCI in humans also involves XIST, but the mechanisms controlling its expression are fundamentally different than those regulating Xist in mice (Migeon et al., 2002). While the “normal” state of XCI in hPSCs remains controversial, almost all reported female hPSC lines display some degree of XCI (Dvash et al., 2010; Hall et al., 2008; Hoffman et al., 2005; Pomp et al., 2011; Shen et al., 2008; Tchieu et al., 2010) with few exceptions (Lengner et al., 2010; Marchetto et al., 2010) (Hanna et al., 2010).
Previous studies of epigenetic stability and variation in hPSCs have been limited in scope and resolution. Most have used allele-specific expression of selected imprinted genes (Adewumi et al., 2007; Frost et al., 2011; Kim et al., 2007; Rugg-Gunn et al., 2007), restriction landmark genome scanning of a small portion of the genome (Allegrucci et al., 2007), or XIST expression to infer the overall epigenetic status of a small number of hESC samples (Hall et al., 2008; Shen et al., 2008; Silva et al., 2008). To obtain a comprehensive view of hPSC-specific epigenomic patterns, we collected 136 hESC and 69 hiPSC samples representing more than 100 cell lines for analysis. In order to establish expected variation in human tissues, we collected 80 high-quality and well-replicated samples representing 17 distinct tissue types from multiple individuals. Finally, we selected 50 additional samples from primary cell lines of diverse origin to control for any aberrations that may arise as a general, non-hPSC-specific, consequence of in vitro manipulation. With these samples, we performed genome-wide DNA methylation and mRNA expression profiling using the Illumina Infinium 27K and 450K DNA Methylation BeadChips (27K and 450K DNA Methylation array) as well as the Illumina HT12v3 Gene Expression BeadArray. These platforms interrogate DNA methylation at 27,578 CpG sites associated with ~14,500 well-annotated genes (27K DNA Methylation array), >450,000 CpG sites associated with both coding and non-coding genes (450K DNA Methylation array) (Sandoval et al., 2011), and the expression of over 30,000 mRNA transcripts (HT12v3). A summary of samples and analyses performed are detailed in Table S1.
RESULTS
Differential global DNA methylation in pluripotent and somatic cells
Our initial goal was to analyze the data in an unbiased manner, in order to identify variations in DNA methylation that were demonstrated by the data, rather than our preconceptions about the samples and/or DNA methylation. Preliminary clustering of all samples and all probes resulted in separation of male and female samples based on the methylation state of the X chromosome, consistent with our previous findings (Bibikova et al., 2006). We therefore examined X chromosome and autosomal probes separately.
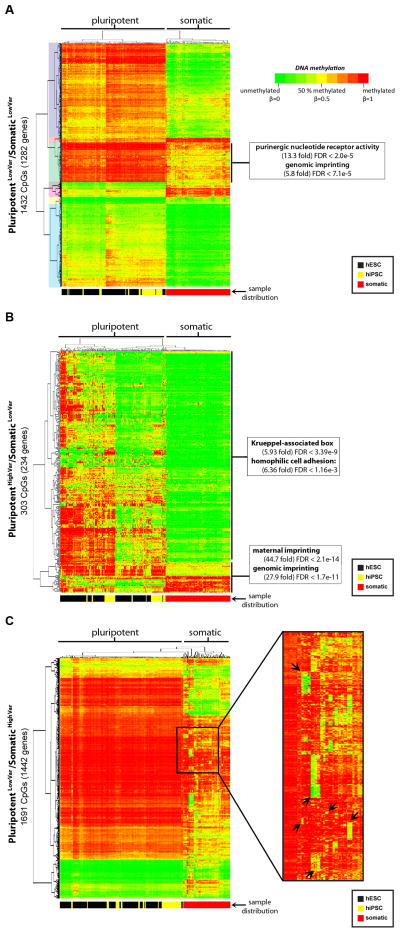
We identified 3,499 autosomal CpG sites on the 27K DNA Methylation array that were differentially methylated (Δβ > 0.2, FDR < 0.01) between pluripotent and somatic (tissue and primary) samples. In our initial analyses, we noticed that there was a large degree of variability in DNA methylation both between the pluripotent and somatic groups, and within each group. In order to dissect out these differences in detail, we divided the CpG sites into three categories, which we clustered separately: PluripotentLowVar/SomaticLowVar, where the variability was low in both the pluripotent and somatic groups (standard deviation [s.d.] <0.2, Figure 1A, Table S2A); PluripotentHighVar/SomaticLowVar, where variability was specific to hPSCs (s.d. >0.2 in hPSCs, s.d. <0.2 in somatic cells, Figure 1B, Table S2B); and PluripotentLowVar/SomaticHighVar, where variability was present in the somatic samples, but not the pluripotent samples (s.d. >0.2 in somatic cells and <0.2 in hPSCs, Figure 1C, Table S2C).
Figure 1. Differential DNA methylation in pluripotent and somatic cells.
Data for CpG sites differentially methylated between pluripotent and somatic cells (Δβ > 0.2) on the 27K DNA Methylation array are shown. A. PluripotentLowVar/SomaticLowVar: 1432 CpGs for 1282 genes with low variation (s.d. < 0.2) within both the pluripotent and somatic sample groups. The seven clusters of CpGs that were examined using the GREAT algorithm are shaded on the left. B. PluripotentHighVar/SomaticLowVar: 303 CpGs for 234 genes with variable methylation only in the pluripotent group (s.d. > 0.2). C. PluripotentLowVar/SomaticHighVar: 1691 CpGs for 1442 genes with variable methylation only in the somatic group. The color scale for the β values is shown. The distribution of sample types are indicated below each heatmap, with hESCs in black, hiPSCs in yellow, and somatic cells in red. See also Figure S1 and Table S2.
The CpG sites in the PluripotentLowVar/SomaticLowVar category were separated into seven clusters by hierarchical clustering (Figure 1A). Each cluster was tested for functional enrichments using the Genomic Regions Enrichment of Annotations Tool (GREAT (McLean et al., 2010)), but only one cluster showed significant enrichments. This cluster was fully methylated in pluripotent samples, partially methylated in somatic samples and was significantly enriched for genes associated with purinergic nucleotide receptor activity and genomic imprinting. The enrichment of imprinted regions in this group demonstrates that a subset of imprinted genes is consistently hypermethylated in hPSCs relative to somatic samples, suggesting a difference in the regulation of imprinted genes between hPSCs and somatic cells. DNA methylation and gene expression were anti-correlated (R < −0.50) for many of these genes (Figure S1A–B).
The CpG sites in the PluripotentHighVar/SomaticLowVar category clustered into two groups. One group was enriched for Krueppel-associated box and homophilic cell adhesion genes (Figure 1B). This enrichment (i.e. genes with KLF binding sites) is interesting because of the use of KLF4 in reprogramming (Takahashi et al., 2007) and KLF2 in converting hESCs to a mouse ESC-like phenotype (Hanna et al., 2010). The second group was also enriched for genomic imprinting, demonstrating that a second subset of imprinted genes is variable specifically in hPSCs.
The PluripotentLowVar/SomaticHighVar category contained several clusters of CpG sites that were hypermethylated in all of the hPSCs and the majority of somatic samples, but unmethylated in a small number of somatic samples containing related cell types (Figure 1C). The genes associated with each cluster of CpGs were enriched for functional categories related to the known functions of the corresponding samples (Figure S1C–G). For example, CpG sites that were uniquely hypomethylated in the blood, spleen, and lymph node samples were enriched for the immune system process, immune response, and defense response categories (Figure S1G). Since these genes were uniformly hypermethylated in the pluripotent state and in unrelated somatic cell types, it appeared that cell type-specific genes underwent selective DNA demethylation during differentiation, and led us to explore this phenomenon at higher resolution.
DNA hypomethylation distinguishes human tissues
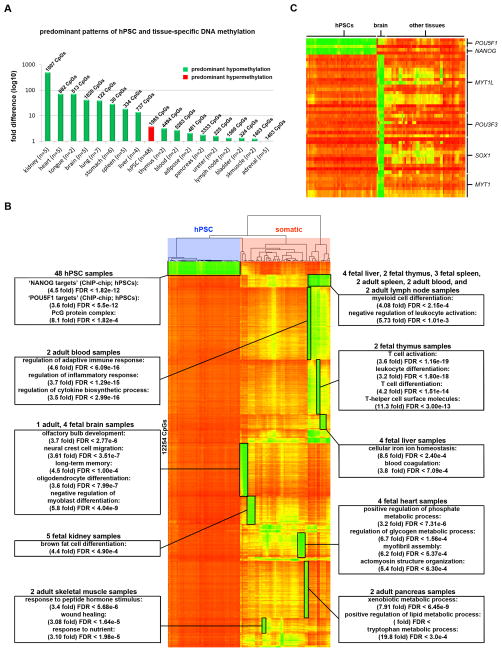
To achieve higher resolution, we analyzed a subset of 153 hPSC and tissue samples using the 450K DNA Methylation array. In order to identify unique epigenetic features in 17 distinct tissue types (e.g. brain, heart, kidney) and hPSCs, we filtered for CpGs that were differentially methylated in each tissue or cell type compared to all other samples with a Δβ > 0.5 (p < 0.05).
Consistent with our previous results, DNA hypomethylation was the most discriminate epigenetic feature of any given tissue (Figure 2A, Table S3). For a majority of these tissue-specific groups of hypomethylated genes, functional enrichments using GREAT were also consistent with the particular tissue’s function and/or cellular composition (Figure 2B). Interestingly, approximately 20% (2,554/12,254) of these hypomethylated CpGs were associated with transcription factors (according to region-gene associations in GREAT). Among these, CpGs associated with POU5F1 and NANOG, which are known master regulators of pluripotency and are among the six transcription factors commonly used in reprogramming, were hypomethylated specifically in hPSCs (Figure 2C). Additionally, CpGs associated with the neural lineage transcription factors MYT1L, POU3F3, SOX1, and MYT1 were specifically hypomethylated in brain samples. In fact, MYT1L is one of four required factors for the direct conversion of fibroblasts into neurons (Pang et al., 2011) and POU3F3 (BRN1) is a closely related functional homolog of another neuronal transdifferentiation factor, POU3F2 (BRN2) (Figure 2C).
Figure 2. Tissue-specific patterns of DNA methylation.
Data for CpG sites on the 450K DNA Methylation array that were differentially methylated between samples from a given tissue and all other samples (Δβ > 0.5) are shown. A. The histogram shows the fold difference in total number of uniquely hypomethylated and hypermethylated CpGs for a given tissue (listed in Table S3). If hypomethylated CpGs predominate, the bar is green; if hypermethylated CpGs predominate, the bar is red. The total number of unique CpGs that were differentially methylated in the given tissue type is shown above each bar, and the total number of samples per tissue type is shown on the X-axis. B. 12,254 CpGs on the 450K DNA Methylation array with uniquely hypomethylated CpGs in specific tissue types. Functional enrichments for tissue-specific hypomethylated clusters are identified with boxes. Samples are grouped according to hierarchical clustering and CpGs are rank-ordered for each tissue (see also Table S3). C. DNA methylation of pluripotency- and neural-specific transcription factor genes.
Demethylation of cell type-specific genes occurs during directed differentiation
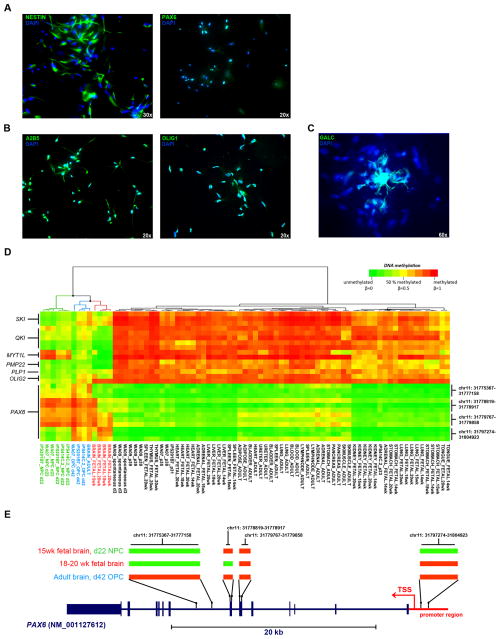
Based on the observation of tissue-specific patterns of DNA hypomethylation and the assumption that epigenetic patterns in hPSCs represent those of the early human embryo, we reasoned that DNA demethylation was a normal component of cellular differentiation. To test this hypothesis, we profiled three hPSC lines before and after in vitro directed differentiation into NESTIN/PAX6+ neural progenitor cells (NPCs; Figure 3A) and mixed populations of A2B5/OLIG1+ oligodendroctye precursor cells (OPCs; Figure 3B) and GALC+ oligodendrocytes (Figure 3C) using established methods (Harness et al., 2011; Nistor et al., 2005). Using 1,303 CpGs that were differentially methylated in OPCs or NPCs compared to hPSCs and to all non-brain tissues, hierarchical clustering of these differentiated samples, hPSCs, and tissues clearly distinguished NPCs, OPCs and brain samples from hPSCs and all other tissues (Table S4, Figure S2A). Demethylation of several genes known to regulate oligodendrocyte differentiation including SKI, QKI, and OLIG2 (Aberg et al., 2006; Atanasoski et al., 2004; Zhou and Anderson, 2002) and the myelin proteins PLP1 and PMP22 (Figure 3D) was observed and reflected in the GREAT enrichments for myelination and regulation of action potentials in neurons (Figure S2A). Methylation of MYT1L was maintained during differentiation from hPSCs to NPCs, but was subsequently lost in the more mature OPCs. In contrast to the NPCs and 15 week fetal brain, DNA methylation of the PAX6 promoter region was evident in OPCs, 18–20 week fetal brain, and adult brain (Figure 3D–E). This successive gain of methylation at the PAX6 locus was consistent with oligodendroglial commitment in OPCs and the restricted neurogenic capacity of the adult brain and led to the functional enrichments for neuron fate commitment and motor neuron cell fate specification in the GREAT analysis (Figure S2B).
Figure 3. Directed differentiation of hPSCs recapitulates epigenetic hallmarks of human tissues.
A. Immunocytochemistry showing NESTIN and PAX6 staining in WA07-derived neural progenitor cells (NPCs) on day 22 of NPC differentiation. B. Immunostaining of A2B5 and OLIG1 in WA07-derived oligodendroctye precursor cells (OPCs) on day 42 of OPC differentiation. C. Immunostaining of GALC in WA07-derived oligodendrocytes on day 42 of OPC differentiation. Magnifications are indicated. D. DNA methylation (using the 450K DNA Methylation array) of select oligodendrocyte and neuronal genes in NPCs, OPCs, hPSCs and tissues. E. Diagram of DNA methylation patterns of PAX6 in NPCs, OPCs and brain samples corresponding to the chromosomal regions listed to the right of the heatmap in Figure 3D. Segments that are green are unmethylated and those that are red are methylated in the samples listed on the left. See also Figure S2 and Table S4.
Aberrant DNA methylation at imprinted loci is prevalent in hPSCs
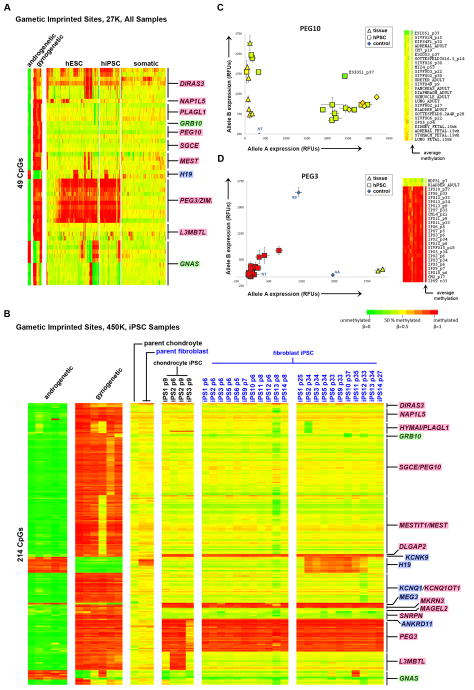
Since our unbiased analyses showed frequent differences in DNA methylation in regions of genomic imprinting between hPSCs and somatic samples, as well as variability in these regions among hPSC samples, we examined imprinted loci separately. We identified 49 CpGs from the 27K DNA Methylation array that were assigned to known imprinted genes (geneimprint.org), and also displayed methylation patterns consistent with gametic imprints (Figure 4A). These loci were partially methylated in tissue samples, and were reciprocally methylated in gynogenetic samples (our parthenogenetic hESCs and previously published data from an ovarian teratoma (Choufani et al., 2011)) and androgenetic samples (previously published data from hydatidiform moles (Choufani et al., 2011)) (Table S5A, Figure S3A–D, Experimental Procedures). Analysis of the DNA methylation status of these imprinted CpGs in pluripotent cells compared to somatic cells showed recurrent hypermethylation of CpGs associated with the genes DIRAS3, NAP1L5, MEST, H19, and ZIM2/PEG3. In a small number of hPSC samples, hypomethylation occurred in PLAGL1 and GRB10. For GNAS, some hPSCs showed a gynogenetic pattern, while other hPSCs showed an androgenetic pattern.
Figure 4. DNA methylation of imprinted genes.
Gene names highlighted in blue are paternally imprinted, pink are maternally imprinted and green are isoform dependent. Hierarachical clustering was performed for each group of samples (gynogenetic/androgenetic, hESCs, hiPSCs, somatic) independently. For each gene, CpG probes are ordered according to chromosomal position. A. DNA methylation (27K DNA Methylation array) of 49 imprinted CpG sites showing a gametic imprint pattern in parthenogenetic, androgenetic and tissue samples. B. DNA methylation (450K DNA Methylation array) of 214 gametic imprinted CpG sites in source fibroblasts and chondrocytes, early passage hiPSCs, and late passage hiPSCs. C–D. Allele-specific expression of PEG10 and PEG3 in hPSC and somatic samples. hPSC samples are represented as squares, somatic samples as triangles, and each data point is colored according to the average beta value for that gene shown in the heatmap to the right. Genomic DNA and no template (NT) controls are plotted as blue diamonds. Error bars indicate the standard error. See also Figure S3.
Hypermethylation and hypomethylation at imprinted loci in hPSCs correlate with loss of allele-specific gene expression
In order to study the effects of reprogramming and time in culture on epigenetic stability, we generated 11 hiPSC clones from fibroblasts and 4 hiPSC clones from chondrocytes. We collected samples for analysis from the parental fibroblast and chondrocyte populations, early passage samples from both chondrocyte and fibroblast-derived hiPSC clones and late passage samples from the fibroblast derived hiPSC clones. All clones were shown to be pluripotent as demonstrated by immunocytochemistry for pluripotency markers, in vitro differentiation, teratoma formation, silencing of reprogramming factors and PluriTest (Muller et al., 2011) (Figure S4, Table S1). For these analyses, we identified 214 CpGs on the 450K DNA Methylation array that had DNA methylation patterns consistent with gametic imprinting according to patterns observed in hydatidiform mole, parthenogenetic hESC and tissue samples (Experimental Procedures, Table S5B). DLGAP2, KCNK9, MEG3, MKRN3, ANKRD11 and PEG3/ZIM2, were hypermethylated in all hiPSC clones relative to the parental samples, suggesting that these aberrations occurred during reprogramming (Figure 4B).
Hypermethylation of H19 and GNAS was seen only in the late passage samples (in 8/11 and 1/11 fibroblast-derived clones, respectively), pointing to instability at these loci with time in culture. Losses in DNA methylation were also observed in HYMA1/PLAGL1, GRB10, KCNQ1, SNRPN and GNAS. Aberrant methylation of L3MBTL was present in 2/4 chondrocyte hiPSC clones. Analysis of an additional 22 hPSC, 60 tissue and 19 primary samples identified additional aberrations in methylation of DIRAS3, PEG10, and MEST in hPSCs, and demonstrated the relative stability of these loci in tissues and primary cell lines (Figure S3E).
In order to determine whether the hypermethylation and hypomethylation we observed at imprinted loci resulted in loss of imprinting, we examined allele-specific gene expression at a subset of imprinted loci. We first used SNP genotyping data we had previously obtained on our samples using the HumanOmni1 SNP genotyping microarray (Laurent et al., 2011) to identify which of the samples contained informative heterozygous SNPs in the PEG10 and PEG3 mRNAs. We then performed allele-specific real time polymerase chain reaction (RT-PCR) to show that loss of DNA methylation at the PEG10 locus correlated with biallelic expression in the ESI051p37 hPSC sample and that hypermethylation of PEG3 led to a total loss of gene expression in several hPSC lines in comparison to the monoallelic expression observed in parental fibroblasts and an adult bladder sample (Figure 4C–D). Using the HT12V3 mRNA expression array, we also determined that CpG methylation and mRNA expression were anti-correlated for MEG3, PEG3/ZIM2, NAP1L5, NNAT, GNAS, NDN, H19 and SNRPN (Table S5A). For many imprinted genes, our DNA methylation data show similar frequencies of either stable or aberrant CpG methylation compared to previous studies reporting on patterns of allelic expression (Table S6, (Adewumi et al., 2007; Allegrucci et al., 2007; Frost et al., 2011; Kim et al., 2007; Rugg-Gunn et al., 2007)). However, for PEG3, MEG3 and H19, we identified frequent aberrant hypermethylation with corresponding silencing of gene expression in hPSCs, which is in contrast to these previous studies, which reported stable monoallelic expression for these genes in hPSCs (Table S6, (Adewumi et al., 2007; Allegrucci et al., 2007; Frost et al., 2011; Kim et al., 2007; Rugg-Gunn et al., 2007)).
Specific culture conditions are associated with aberrant DNA methylation of certain imprinted genes
In order to determine if the observed aberrations in genomic imprints resulted from in vitro manipulations, we selected 140 hPSC samples for which we had detailed histories of culture media, passaging techniques and growth substrates (Table S7). For each imprinted gene (dependent variables), we generated two separate multiple linear regression models, which in addition to the source lab, considered each in vitro manipulation (independent variables) either in combination with other concurrent in vitro manipulations (model 1; e.g. number of manual passages in Wicell medium on MEFs) or in isolation (model 2; e.g. number of passages in Wicell medium). Analysis of these models showed apparent lab-specific effects for DIRAS3, L3MBTL and PEG3 (Bonferroni-adjusted p<0.001, Table S7A–C). We constructed correlation matrices to investigate inter-variable relationships that could potentially explain these apparent lab-specific effects (Table S7D–E). DIRAS3 aberrations were most highly correlated with the Laslett lab samples (R=0.59), which was the only lab positively correlated with use of the original medium used to maintain hESCs (“originalES”, R=0.63) (Thomson et al., 1998). This lab-specific effect can therefore be explained by the use of originalES medium, which is the highest correlating variable with DIRAS3 aberrations (R=0.82). L3MBTL aberrations were most significantly correlated with the Keirstead lab samples (R=0.75). The samples from the Keirstead lab used in this analysis consisted of 2 isogenic clones, which were both passaged in collagenase and grown in Wicell-conditioned medium on Matrigel™ (R=0.74). As theses samples were nearly perfectly correlated with these concurrent variables (R=0.99), this lab-specific effect for L3MBTL can almost entirely be explained by technique and/or cell line of origin. The association of PEG3 aberrations with the Loring lab samples (Bonferroni adjusted p<0.001, R=0.42) could not be attributed to any particular manipulation, but most likely results from an overrepresentation of HDF51 hiPSC clones that were derived from the same fibroblast culture in the same experiment and comprise 70% of all Loring lab samples in the model.
To determine if hESC derivation methods may affect imprinted gene methylation, we performed an independent analysis on 40 samples from 34 hESC lines that were derived in the same lab from embryos of different quality, at varying days post-fertilization (D.P.F), and using two different methods (bisection vs. whole embryo plating). The only correlation that we found was that aberrations in the methylation status of PEG3 were weakly associated with earlier D.P.F and whole embryo plating (Bonferroni adjusted p<0.05) (Table S7F).
XCI and XIST expression are unstable in hPSCs
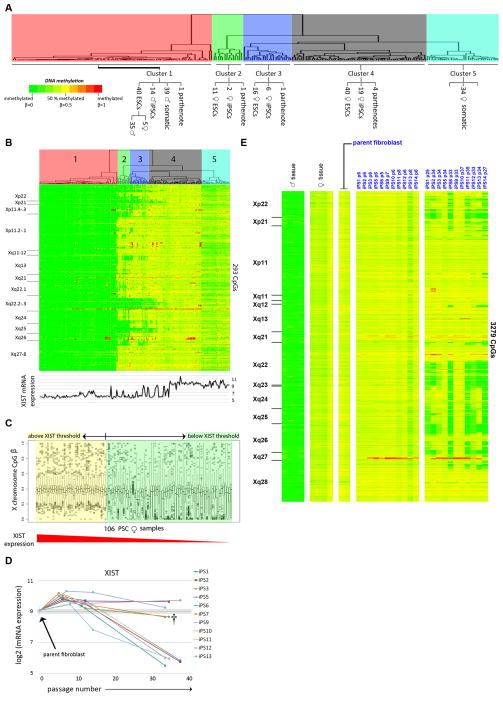
To assess the stability of XCI in our samples, we identified 293 CpGs on the X chromosome (using the 27K DNA Methylation array) that were methylated in a manner consistent with XCI in tissue samples (Experimental Procedures). Hierarchical clustering on the samples yielded five major sample clusters (X-Cluster 1 – X-Cluster 5), which are displayed with the CpGs ordered according to chromosomal location in Figure 5A–B. All of the female somatic samples were in X-Cluster 5, and had partial DNA methylation across the entire X chromosome, consistent with the expected somatic female X-inactivated (XaXi) state. X-Cluster 1 contained all of the male somatic and male hPSC samples (which were, as expected, unmethylated throughout the X chromosome), as well as one parthenogenetic hESC line (LLC15) and samples from four female hESC lines (SIVF024, SIVF028, SIVF029, and CM8) (Figure 5A–B). SIVF024 was XO by SNP genotyping as evidenced by loss-of-heterozygosity in the pseudoautosomal regions (Figure S5A), and would be expected to have a male pattern of X chromosome DNA methylation. However, SIVF028, SIVF029, and CM8 had normal heterozygous XX SNP genotypes, indicating that they contained two different X chromosomes (Figure S5A). Therefore, the lack of DNA methylation on the X chromosome seen in these samples was due to absence of XCI, rather than deletion or uniparental disomy of the X chromosome. The remaining X-Clusters 4, 3 and 2 contained female hPSC samples, with those in X-Cluster 4 showing a uniform partially methylated pattern, and possessing a slightly higher level of methylation than the female somatic samples (X-Cluster 5). The X-Clusters 2 and 3 samples lacked DNA methylation in several non-contiguous regions of the X chromosome (Figure 5B); this was specific for hPSCs, and was not seen in tissues or primary cell cultures (Figure S5C).
Figure 5. DNA methylation on the X chromosome.
CpGs are ordered by chromosomal location, with the cytobands indicated to the left of the heatmaps. A–B. Hierarchical clustering of all samples according to 27K DNA Methylation array data. Cluster assignments are shown on the enlarged dendrogram above the heatmaps. XIST expression is shown below the heatmap. C. Box and whisker plot of chrX β values in 106 female hPSC samples ordered according to decreasing XIST expression. D. XIST expression in parental fibroblasts and 11 hiPSC clones at early, intermediate, and late passages shows an increase in XIST expression following reprogramming and a subsequent tendency for loss of XIST expression over time in culture. E. DNA methylation (450K DNA Methylation array) for fibroblast, early passage hiPSC, and late passage hiPSC samples. See also Figure S4–5.
Examining the relationship between XIST expression and X chromosome DNA methylation, we noted that there was a relative threshold of XIST expression, above which we saw uniform partial DNA methylation, and below which we observed decreased methylation in at least a subset of CpG sites (Figure 5C). This result was consistent with DNA methylation on the chromosomal level (Figure 5B), where it was apparent that the absence of DNA methylation on the X chromosome occurred in a patchy fashion.
XCI is maintained during reprogramming and is subsequently lost with time in culture
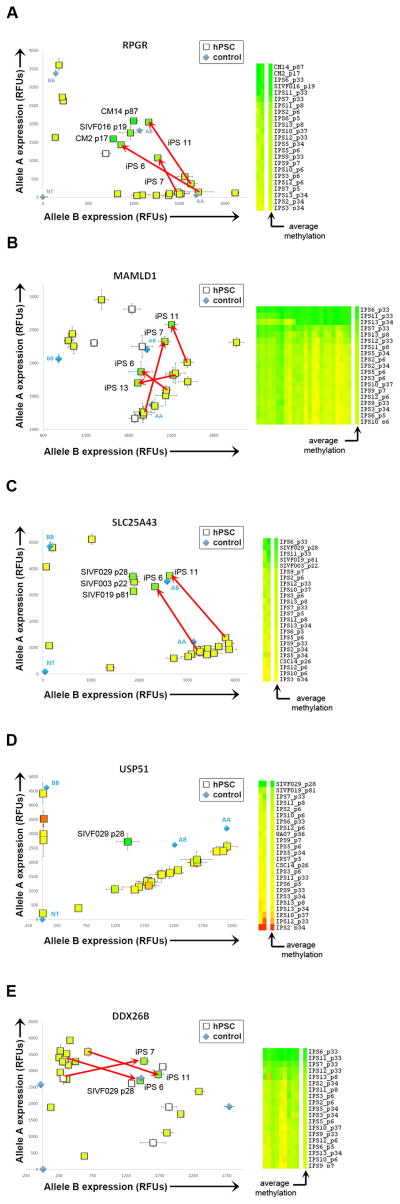
We compared matched fibroblasts and 11 hiPSC clones analyzed at early, intermediate and late passages (Figure 5D–E, Figure S5B) using both the 27K and 450K DNA Methylation arrays (Table S5C–D). Shortly after reprogramming, there was an increase in XIST expression and in overall X chromosome DNA methylation. This was consistent with the higher level of X chromosome DNA methylation seen in a subset of female hPSC samples compared to the female somatic samples (X-Cluster 4 and X-Cluster 5; Figure 5B). At later passages, 8/11 hiPSC clones showed focal loss of XCI, indicated by loss of DNA methylation and increased mRNA expression, in the same regions observed in the hPSC collection as a whole (Figure S5B). The 3/11 hiPSC clones that retained full XCI at late passage also retained high levels of XIST expression (Figure S5B). There were two hiPSC clones (iPS3 and iPS7) that had intermediate levels of XIST expression at late passage (†, Figure S5B), but showed focal loss of XCI, consistent with the XIST threshold effect suggested above (Figure 5C–D). Using allele-specific RT-PCR, we confirmed that loss of DNA methylation was associated with biallelic expression of genes located in regions of XCI (Figure 6A–E). We observed loss of XIST expression (and loss of DNA methylation on the X chromosome) in most of the hiPSC clones, and retention of XIST expression (with preservation of X chromosome DNA methylation) in a minority of the clones, even though all the clones were generated and passaged in the same manner and at the same time. In contrast to the examined imprinted genes, no significant associations for the loss of XCI with specific cell culture or derivation conditions were detected in the multiple linear regression models.
Figure 6. Allele-specific expression of genes subject to XCI.
Allele-specific expression of RPGR, MAMLD1, SLC25A43, USP51 and DDX26B in hPSC samples. hPSC samples are represented as squares and each data point is colored according to the average beta value for that gene shown in the heatmap to the right; data points without corresponding DNA methylation data are white. Red arrows identify HDF51 iPSC lines that switched from monoallelic expression at early passages to biallelic expression at late passages. Genomic DNA and no template (NT) controls are plotted as blue triangles. Error bars indicate the standard error.
Aberrations in genomic imprints and XCI genes are maintained through differentiation
If hPSC-specific aberrations in genomic imprints and XCI persist through differentiation, they may impact the utility of hPSC-derived cells for cellular transplantation and disease modeling. Therefore, we assessed the status of such aberrations in undifferentiated and differentiated hPSCs. WA09 hESCs were studied before and after a 3 day spontaneous differentiation, while WA07, iPS201B7 and iPS414C.2 were studied before and after more extensive NPC and OPC differentiations. In every case, the aberrations in imprinting and XCI that were present in the starting undifferentiated hPSC populations were maintained, and no new aberrations arose during the course of differentiation (Figure 7A–B). In our collection of eleven fibroblast-derived hiPSC clones, loss of XCI was observed in 49 X-linked disease genes by late passage (OMIM; Figure 7C). These results merit caution in the use of female hiPSCs for studies of X-linked disease modeling.
Figure 7. Implications of aberrations in XCI and genomic imprints on disease modeling.
A. DNA methylation (450K DNA Methylation array) of 214 gametic imprinted CpG sites at imprinted loci for control androgenetic and gynogenetic samples, undifferentiated (labeled with green text) and differentiated (labeled with red text) hPSC samples. Arrows indicate direction of differentiation. B. The heatmap shows DNA methylation (450K DNA Methylation array) on the X chromosome for control male tissue samples, control female tissue samples, undifferentiated samples (labeled with green text) and differentiated samples (labeled with red text) hPSC samples. Arrows indicate direction of differentiation. CpGs are ordered by chromosomal location, with the cytobands indicated to the left of the heatmap. C. Diagram indicating the frequency of loss of XCI at X-linked disease genes among 11 hiPSC clones reprogrammed from the same fibroblast culture. The number of clones showing loss of XCI at each locus is listed to the right of the gene name. Genes with loss of XCI in 1–3 clones are shown in black, in 4–5 clones in orange, and in 6–8 clones in red.
DISCUSSION
In this study, we explored epigenetic and transcriptional variation in the most comprehensive collection of hPSC and somatic samples to date. Using a combination of genome-wide DNA methylation and mRNA expression data, we identified unique epigenetic and transcriptional properties of the pluripotent state. Most distinctive among these characteristics are prevalent, but not uniform, losses of imprinting and XCI and consistent hypermethylation of somatic cell-type-specific genes in hPSCs. We observed the acquisition of the appropriate cell-type-specific DNA methylation marks during differentiation of hPSCs, despite persistence of aberrant imprinting and XCI. The scope and resolution of our study has allowed us to address many inconsistencies in the literature, which arose from the inclusion of limited numbers of cell lines and/or sparse coverage of the genome.
In order to determine which imprinted genes we could confidently analyze in our study, we identified a panel of loci that showed appropriate imprinting in normal tissue samples, as well as gynogenetic and androgenetic samples. We observed aberrations at many of the examined imprinted genes in a substantial subset of hPSCs; changes at some loci arose during reprogramming and others over time in culture. Very few studies have addressed potential causes of aberrant imprinting in hPSCS (such as culture conditions or derivation methods), although a recent study comparing the hESC line WA09 and six isogenic WA09-derived hiPSC lines reported that imprinting of NNAT (as well as XCI) was specifically lost in hiPSCs compared to hESCs (Teichroeb et al., 2011). While we identified NNAT as one of the genes with hPSC-specific variability in DNA methylation (Figure 1B and Table S2B), none of the observed variations were specific to hESCs or hiPSCs, and none of the 20 CpG sites in the promoter region of NNAT interrogated by the 27K and 450K DNA Methylation array passed our imprinted site filters. Using linear regression, we were able to correlate the imprinting status of DIRAS3, L3MBTL and PEG3 with specific in vitro manipulations, while the DNA methylation status of the other imprinted genes in our analysis were independent of the identifiable variables. The strongest association seen was between DIRAS3 hypermethylation and culture in the original hESC medium, which contained LIF and FBS, in contrast to the currently used hPSC media, which contain knockout serum replacement and purified FGF2. Given the limited numbers of samples and cell lines representing each variable in the regression models, it will be necessary in future studies to systematically test specific variables in a well replicated manner in order to identify causal relationships between specific derivation/reprogramming and culture conditions and epigenetic aberrations.
We observed a large degree of variability in X chromosome CpG methylation in female hPSCs, which appeared to be dependent on the level of XIST expression. Our results were consistent with a loss of XIST expression with time in culture, followed by erosion of DNA methylation, originating in several sub-segments of the X chromosome and spreading to involve larger regions. Our prediction that loss of XCI may affect the fidelity of hPSC-based X-linked disease models is consistent with the findings reported in an accompanying manuscript from Mekhoubad et al. In their studies, a hiPSC-based disease model of the X-linked disease, Lesch-Nyhan Syndrome, lost the ability to recapitulate hallmark biochemical characteristics of the disease with time in culture. These researchers showed that this phenomenon was due to loss of XCI and reactivation of the wild-type HPRT gene in the late passage female hiPSCs, consistent with our observations that loss of XCI at the HPRT locus was a common feature that occurred in more than half of the female hESC and hiPSC samples we analyzed.
We found that DNA hypomethylation was the most discriminate epigenetic feature of any given tissue and that tissue-specific hypomethylated genes were associated with the function of that tissue. Among these genes were transcription factors used for the transdifferentiation of fibroblasts into neurons, master regulators of oligodendrocyte differentiation, and iPSC reprogramming factors. We suggest that the identification of uniquely hypomethylated genes will permit the discovery of high-level regulators of cellular identity, and may inform the selection of factors for novel transdifferentiation protocols.
Our results suggest that an interplay between DNA methylation and demethylation regulates cellular differentiation. We observed DNA methylation changes during directed differentiation of hPSCs into NPCs and OPCs that recapitulated patterns of DNA methylation at neural and oligodendrocyte-specific genes in fetal and adult brain samples, supporting the validity of hPSCs as models of development and disease. For example, dysregulation of QKI (KH domain- containing RNA binding factor “quaking homolog”), is strongly associated with schizophrenia (Aberg et al., 2006). As QKI was uniquely hypomethylated in fetal and adult brain samples in our data, the demethylation of this gene observed during neural differentiation of hPSCs may be a necessary feature for accurate in vitro modeling of schizophrenia.
By studying genome-wide DNA methylation and gene expression profiles in a large and diverse collection of pluripotent and somatic samples, we have discovered that pluripotent cells differ from somatic cells at sites in the genome that are generally considered to be epigenetically stable: the inactivated X chromosome in female cells and imprinted loci. Among pluripotent cultures, there was a large degree of variation at these sites, and their methylation status was not changed with differentiation. These epigenetic instabilities merit a degree of caution in the interpretation of X-linked hPSC-based disease models, and indicate that hPSC derivatives destined for clinical use should be examined for aberrations in imprinting and XCI. Therefore, identification of specific culture conditions or small molecules that promote the stability of genomic imprints and XCI over long-term culture would be of great value to the stem cell community.
EXPERIMENTAL PROCEDURES
Sample collection and culture conditions
All samples were either cultured in house or obtained from collaborators (for sample details, see Table S1). Plat-A Packaging cells (Cell Biolabs, Inc.) were maintained according to the manufacturer’s instructions. Human dermal fibroblasts (HDFs, ScienCell Research Laboratories) were cultured in Dulbecco’s modified eagle medium, 2mM GlutaMax, 10% fetal bovine serum and 0.1 mM non-essential amino acids (Life Technologies, Inc.). Culture conditions for hPSCs are listed in Table S1.
Generation of iPS Cells
PLAT-A packaging cells were plated onto six well plates coated with Poly-D-Lysine at a density of 1.5×106 cells per well without antibiotics and incubated overnight. Cells were transfected with 4 μg of moloney murine leukemia-based retroviral vectors (pMXs) containing the human cDNA of POU5F1, SOX2, KLF4 or MYC (Addgene) using Lipofectamine 2000 (Life Technologies, Inc.) according to the manufacturer’s instructions. Viral supernatants were collected at 48 and 72 hours post-transfection, filtered through a 0.45 μm pore-size filter. 150,000 HDFs were seeded onto each well of a six well plate overnight. Equal volumes of fresh 48 hour and 72 hour viral polybrene-supplemented (6 μg/ml, Sigma) supernatants were added onto the cells at 24 hours and 48 hours post-seeding. On day three, the transduced cells were split onto MEFs at a density of 104 cells per well of a six well plate in hESC medium supplemented with 0.5 mM Valproic Acid (VPA, Stemgent). Cells were fed every other day with hESC medium + VPA for 14 days. iPSC colonies were picked three weeks post-transduction and transferred onto MEF plates.
In vitro Differentiation
For spontaneous embryoid body (EB) formation, hPSCs were manually passaged to low attachment plates in hESC medium without bFGF for 7 days, changing media every other day. On Day 8, EBs were transferred onto gelatin coated coverslips and cultured in the same medium for 7 more days. Directed differentiation was performed as previously described (Harness et al., 2011; Nistor et al., 2005).
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde in PBS for 15 minutes, washed 3x with PBS, and blocked in PBS with 2% BSA (Sigma); 0.1% Triton X-100 and 2% low fat milk for 30 minutes at room temperature. Primary antibodies include POU5F1, NANOG, BRACHYURY (Santa Cruz; 1:100, 1:100, 1:300); TRA1-81 (1:100, Stemgent); MAP2, AFP, NESTIN, SMA (Millipore; 1:100, 1:400, 1:2000, 1:10,000); PAX6, NESTIN, OLIG1, GALC, A2B5 (Chemicon; 1:200, 1:200, 1:200, 1:200, 1:500). Fluorescence conjugated secondary antibodies were used according to manufacturer’s protocol (Life Technologies, Molecular Probes). Images were obtained with IX51 Olympus and Nikon Eclipse Ti microscopes.
Teratoma Assay
1×106 HDF51iPS cells were harvested by Accutase treatment (Life Technologies, Inc.), re-suspended in a 1:1 mixture of DMEM/F12 and Matrigel (BD Biosciences) and injected into the right testis of a C.B-17-Prkdcscid mouse (Charles River). Six to eight weeks after injection, tumors were dissected, fixed in 4%PFA, sectioned and stained with hematoxylin and eosin.
DNA Methylation Profiling
DNA was extracted from 1×106 cells (Qiagen DNeasy Blood and Tissue Kit), quantified (Qubit dsDNA BR Assay Kits, Life Technologies, Inc.), quality controlled (DNA1000 Kit and BioAnalyzer 2100, Agilent) and bisulfite-converted (EZ DNA Methylation Kit, Zymo Research) according to each manufacturer’s protocol. Bisulfite converted DNA was hybridized to Infinium HumanMethylation27K and Infinium 450K BeadChips (Illumina, Inc.), scanned with an iScan (Illumina, Inc.) and quality controlled in GenomeStudio. For 27K data, β values for each probe were range-scaled using data collected from DNA controls that were fully methylated (SSI DNA methyltransferase treated (NEB), bisulfite converted DNA), unmethylated (untreated genomic DNA) and half-methylated (50/50 mix of methylated and unmethylated controls). 450K data was background subtracted and normalized to controls in GenomeStudio. Hierarchical clustering was performed using Cluster, using Euclidian distance and complete linkage.
Expression Profiling
Total RNA was extracted from snap-frozen sample pellets (Ambion mirVana Kit, Life Technologies, Inc.) according to the manufacturer’s protocol. RNA quantity (QubitTM RNA BR Assay Kits, Life Technologies, Inc.) and quality (RNA6000 Nano Kit and Bioanalyzer 2100, Agilent) was determined to be optimal for each sample prior to further processing. 200 ng RNA per sample was amplified using the Total PrepTM RNA Amplification Kit (Illumina, Inc.) according to manufacturer’s protocol and quantified as above. 750 ng labeled RNA/sample was hybridized to HT-12v3 Expression BeadChips (Illumina, Inc.), scanned with an iScan (Illumina, Inc.). In GenomeStudio, probes were filtered for those detected at p<0.01 in at least 1 sample and exported for normalization in R using robust spline normalization (RSN). Hierarchical clustering was performed using Cluster, using Euclidian distance and complete linkage.
Identification of DNA methylation probes for imprinted genes
CpG probes that were reciprocally methylated in gynogenetic (ovarian teratoma and parthenotes; entirely of maternal origin) and androgenetic (complete hydatidiform moles; entirely of paternal origin), partially methylated in tissue samples (stable imprinting) and associated with imprinted genes (according to geneimprint.com) were identified as gametic imprints (Figure S4). For range-scaled 27K DNA Methylation array data, we supplemented our data with published data from an ovarian teratoma and three hydatidiform mole samples46. All imprinted probes were partially methylated (0.25<β<0.75) in at least 75% of tissue samples. Maternal imprints were unmethylated (β<0.09) in at least 2 androgenetic samples and fully methylated (β>0.85) in at least 2 gynogenetic samples. Paternal imprints were unmethylated (β<0.09) in at least 2 gynogenetic samples and fully methylated (β>0.85) in at least 2 androgenetic samples (Table S5A). Due to differences in normalization, the criteria for identification of imprinted probes were slightly different for the 450K DNA Methylation array data. CpG probes were annotated to imprinted genes using the 450K DNA Methylation array manifest file, or if it fell within 5 kb upstream of the gene according to UCSC hg18. All imprinted probes were partially methylated (0.20<β<0.80) in at least 90% of tissue samples. Maternal imprints were unmethylated (β<0.20) in at least 2 androgenetic samples and fully methylated (β>0.80) in at least 2 gynogenetic samples. Paternal Imprints were unmethylated (β<0.20) in at least 2 gynogenetic samples and fully methylated (β>0.80) in at least 2 androgenetic samples (Table S5B).
Identification of DNA methylation probes for X-inactivated regions
In order to select for X-chromosome CpG sites subject to XCI on the 27K DNA Methylation array, we first removed probes for sites in the pseudoautosomal regions of the X chromosome, and then selected the X-chromosome probes that were partially methylated (β values between 0.09–0.85) in at least 75% of the female tissue samples and unmethylated (β value less than 0.09) in at least 75% of the male tissue samples. 452 X chromosome DNA methylation probes representing 289 genes passed these filters, of which 293 probes for 199 genes were anti-correlated with gene expression (Table S5C). To identify probes subject to XCI on the 450K DNA Methylation array, we filtered for probes that were partially methylated (0.2<β<0.8) in 90% of female tissues tissues and unmethylated (β<0.2) 90% male tissues (Table S5D).
Allele-Specific RT-PCR
Informative heterozygous SNPs in the mRNA region of selected X chromosome and imprinted genes were identified using microarray SNP genotyping data (Laurent et al., 2011). Total RNA was collected as above and converted to cDNA (Quantitect Reverse Transcription Kit, Qiagen) according to the manufacturer’s protocol. 40 ng of cDNA was then used as input for a Taqman qPCR SNP genotyping array selected to determine allelic expression. Quantitative expression data were acquired and analyzed using a CFX-96 Real-Time PCR Detection System (BIORAD) using the SNP genotyping taqman probes DDX26B (C_16188987_10), MAMLD1 (C_15867801_10), RPGR (C_11874860_10), SLC25A43 (C_25953804_20), USP51 (C_27476233_10), PEG10 (C_25805777_10) and PEG3 (C_25643544_10).
Functional Enrichment Analysis
Functional enrichment analysis was performed using GREAT (McLean et al., 2010). The basal+extension setting was used with all CpG probes on the 27K or 450K DNA Methylation array (except for those on X and Y) used as the background set.
Supplementary Material
Acknowledgments
We would like to gratefully acknowledge the following for contributing samples for this study: Juan-Carlos Belmonte, Eirini Papapetrou (Sadelain lab), Dongbao Chen, Jerold Chun, Martin Pera, James Shen, Scott McKercher, Timo Otonkoski, Allan Robins, Thomas Schulz, Philip Schwartz, Scott McKercher, Ralph Graichen, Jeong Beom Kim, Nils O. Schmidt, Christopher Barry, Robin Wesselschmidt, James Shen, Joel Gottesfeld, Christina Lu, the NICHD Brain and Tissue Bank for Developmental Disorders, and Planned Parenthood of the Pacific Southwest. We would also like to acknowledge Yinchun Li for generously contributing his expertise in histological evaluation of iPSC-derived teratomas. We would like to thank Trevor Leonardo, Robert Morey, Sara Abdelrahman, Inbar Friedrich Ben-Nun, Victoria Glenn, Dumitru Brinza, Dmitry Pushkarev, Tenneille Ludwig and Kristen Brennand for their valuable help in the laboratory and for useful discussions. Many thanks to Marina Bibikova and Jian-Bing Fan at Illumina for their assistance and expertise with the 450K DNA Methylation array. LCL is supported by an NIH/NICHD K12 Career Development Award and the Hartwell Foundation. GA, KLN, CL, IS, FJM, EF and JFL are supported by CIRM (CL1-00502, RT1-01108, TR1-01250, RN2-00931-1), NIH (R33MH87925), the Millipore Foundation, and the Esther O’Keefe Foundation. KLN is supported by an Autism Speaks Dennis Weatherstone fellowship. YCW is supported by the Marie Mayer Foundation. EF was supported by a Bill and Melinda Gates Grand Challenges Explorations Award and a UNCF/Merck Postdoctoral Fellowship. ALL is supported by the Australian Stem Cell Centre, Stem Cells Australia and the Victoria-California Stem Cell Alliance (CIRM grant TR1-01250). HSP and SL were supported by a grant (SC2250) from the Stem Cell Research Center of the 21st Century Frontier Research Program funded by Ministry of Educational Science and Technology. FJM is supported by an Else-Kröner Fresenius Stiftung fellowship. IS is supported by the Pew Foundation.
Footnotes
ACCESSION NUMBERS
All DNA methylation and gene expression array data are available at the NCBI GEO database under the accession designation GSE30654. Previously published 27K DNA Methylation array data from an ovarian teratoma and three hydatidiform mole samples are available at the NCBI GEO database under the accession designation GSE22091.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci U S A. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Allegrucci C, Wu YZ, Thurston A, Denning CN, Priddle H, Mummery CL, Ward-van Oostwaard D, Andrews PW, Stojkovic M, Smith N, et al. Restriction landmark genome scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. doi: 10.1093/hmg/ddm074. [DOI] [PubMed] [Google Scholar]
- Atanasoski S, Notterpek L, Lee HY, Castagner F, Young P, Ehrengruber MU, Meijer D, Sommer L, Stavnezer E, Colmenares C, et al. The protooncogene Ski controls Schwann cell proliferation and myelination. Neuron. 2004;43:499–511. doi: 10.1016/j.neuron.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Bhusari S, Yang B, Kueck J, Huang W, Jarrard DF. Insulin-like growth factor-2 (IGF2) loss of imprinting marks a field defect within human prostates containing cancer. Prostate. 2011 doi: 10.1002/pros.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Chudin E, Wu B, Zhou L, Garcia EW, Liu Y, Shin S, Plaia TW, Auerbach JM, Arking DE, et al. Human embryonic stem cells have a unique epigenetic signature. Genome Research. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choufani S, Shapiro JS, Susiarjo M, Butcher DT, Grafodatskaya D, Lou Y, Ferreira JC, Pinto D, Scherer SW, Shaffer LG, et al. A novel approach identifies new differentially methylated regions (DMRs) associated with imprinted genes. Genome Research. 2011;21:465–476. doi: 10.1101/gr.111922.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Das R, Weidman JR, Jirtle RL. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatric Research. 2007;61:30R–37R. doi: 10.1203/pdr.0b013e31804575f7. [DOI] [PubMed] [Google Scholar]
- Dvash T, Lavon N, Fan G. Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS One. 2010;5:e11330. doi: 10.1371/journal.pone.0011330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Lu SJ, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim KS, Lanza R. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- Frost J, Monk D, Moschidou D, Guillot PV, Stanier P, Minger SL, Fisk NM, Moore HD, Moore GE. The effects of culture on genomic imprinting profiles in human embryonic and fetal mesenchymal stem cells. Epigenetics. 2011;6:52–62. doi: 10.4161/epi.6.1.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Butler J, Becker KA, Nelson A, Amit M, Itskovitz-Eldor J, Stein J, Stein G, Ware C, et al. X-inactivation reveals epigenetic anomalies in most hESC but identifies sublines that initiate as expected. Journal of Cellular Physiology. 2008;216:445–452. doi: 10.1002/jcp.21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, Cassady JP, Muffat J, Carey BW, Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harness JV, Turovets NA, Seiler MJ, Nistor G, Altun G, Agapova LS, Ferguson D, Laurent LC, Loring JF, Keirstead HS. Equivalence of conventionally-derived and parthenote-derived human embryonic stem cells. PLoS One. 2011;6:e14499. doi: 10.1371/journal.pone.0014499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LM, Hall L, Batten JL, Young H, Pardasani D, Baetge EE, Lawrence J, Carpenter MK. X-inactivation status varies in human embryonic stem cell lines. Stem Cells. 2005;23:1468–1478. doi: 10.1634/stemcells.2004-0371. [DOI] [PubMed] [Google Scholar]
- Hough SR, Laslett AL, Grimmond SB, Kolle G, Pera MF. A continuum of cell states spans pluripotency and lineage commitment in human embryonic stem cells. PLoS One. 2009;4:e7708. doi: 10.1371/journal.pone.0007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Nagy K, Nagy A. Human induced pluripotent stem cells: the past, present, and future. Clin Pharmacol Ther. 2011;89:741–745. doi: 10.1038/clpt.2011.37. [DOI] [PubMed] [Google Scholar]
- John RM, Lefebvre L. Developmental regulation of somatic imprints. Differentiation. 2011;81:270–280. doi: 10.1016/j.diff.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Kim DH, Jeon Y, Anguera MC, Lee JT. X-chromosome epigenetic reprogramming in pluripotent stem cells via noncoding genes. Semin Cell Dev Biol. 2011 doi: 10.1016/j.semcdb.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KP, Thurston A, Mummery C, Ward-van Oostwaard D, Priddle H, Allegrucci C, Denning C, Young L. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Research. 2007;17:1731–1742. doi: 10.1101/gr.6609207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic Changes in the Copy Number of Pluripotency and Cell Proliferation Genes in Human ESCs and iPSCs during Reprogramming and Time in Culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon BR, Lee CH, Chowdhury AK, Carpenter H. Species differences in TSIX/Tsix reveal the roles of these genes in X-chromosome inactivation. American Journal of Human Genetics. 2002;71:286–293. doi: 10.1086/341605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, Danner S, Goldmann JE, Herbst A, Schmidt NO, et al. A bioinformatic assay for pluripotency in human cells. Nature Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistor GI, Totoiu MO, Haque N, Carpenter MK, Keirstead HS. Human embryonic stem cells differentiate into oligodendrocytes in high purity and myelinate after spinal cord transplantation. Glia. 2005;49:385–396. doi: 10.1002/glia.20127. [DOI] [PubMed] [Google Scholar]
- Odom LN, Segars J. Imprinting disorders and assisted reproductive technology. Curr Opin Endocrinol Diabetes Obes. 2010;17:517–522. doi: 10.1097/MED.0b013e32834040a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomp O, Dreesen O, Leong DF, Meller-Pomp O, Tan TT, Zhou F, Colman A. Unexpected X Chromosome Skewing during Culture and Reprogramming of Human Somatic Cells Can Be Alleviated by Exogenous Telomerase. Cell Stem Cell. 2011;9:156–165. doi: 10.1016/j.stem.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum Mol Genet. 2007;16(Spec No 2):R243–251. doi: 10.1093/hmg/ddm245. [DOI] [PubMed] [Google Scholar]
- Sandoval J, Heyn HA, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci U S A. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, Zack JA, et al. Female human iPSCs retain an inactive X chromosome. Cell Stem Cell. 2010;7:329–342. doi: 10.1016/j.stem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichroeb JH, Betts DH, Vaziri H. Suppression of the Imprinted Gene NNAT and X-Chromosome Gene Activation in Isogenic Human iPS Cells. PLoS One. 2011;6:e23436. doi: 10.1371/journal.pone.0023436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Urbach A, Bar-Nur O, Daley GQ, Benvenisty N. Differential modeling of fragile X syndrome by human embryonic stem cells and induced pluripotent stem cells. Cell Stem Cell. 2010;6:407–411. doi: 10.1016/j.stem.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Lewis S, Woodfine K, Stojic L, Murrell A. Molecular mechanisms of genomic imprinting and clinical implications for cancer. Expert Reviews in Molecular Medicine. 2011;13:e2. doi: 10.1017/S1462399410001717. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.