Summary
The TOR kinase, which is present in the functionally distinct complexes TORC1 and TORC2, is essential for growth but associated with disease and aging. Elucidation of how TOR modulates lifespan will identify mechanisms of fundamental importance in aging, and TOR functions. Here we show that when TORC1 is inhibited genetically in C. elegans, SKN-1/Nrf and DAF-16/FoxO activate protective genes, and increase stress resistance and longevity. SKN-1 also upregulates TORC1 pathway gene expression in a feedback loop. Rapamycin triggers a similar protective response in C. elegans and mice but increases worm lifespan dependent upon SKN-1 and not DAF-16, apparently by interfering with TORC2 along with TORC1. TORC1, TORC2, and insulin/IGF-1-like signaling regulate SKN-1 activity through different mechanisms. We conclude that modulation of SKN-1/Nrf and DAF-16/FoxO may be generally important in the effects of TOR signaling in vivo, and that these transcription factors mediate an opposing relationship between growth signals and longevity.
Introduction
The TOR (target of rapamycin) kinase integrates nutrient and anabolic signals to promote growth (Ma and Blenis, 2009; Wullschleger et al., 2006; Zoncu et al., 2011). TOR is found in two distinct complexes, TORC1 and TORC2 (Figure S1). Amino acid, oxygen, energy, and growth signals activate TORC1, which phosphorylates a set of well-characterized substrates to increase mRNA translation and inhibit autophagy. The amino acid signal is transduced by heterodimeric Rag GTPases that recruit TORC1 to lysosomal membranes, where it is activated by Rheb. Growth signals and interaction with the ribosome activate TORC2, which in turn activates AKT, SGK, and related kinases (Oh et al., 2010; Zinzalla et al., 2011; Zoncu et al., 2011).
TOR signaling is essential for growth and development, but has also been implicated in diabetes, cardiac hypertrophy, malignancies, neurodegenerative syndromes, other diseases, and aging (Stanfel et al., 2009; Wullschleger et al., 2006; Zoncu et al., 2011). TOR inhibitors have been approved or are under investigation for treatment of several conditions, including various cancers. These inhibitors include the immunosuppressant rapamycin, which is widely used to combat kidney rejection after transplantation. In model organisms ranging from yeast to mice, inhibition of TOR signaling increases lifespan (Bjedov et al., 2010; Harrison et al., 2009; Kapahi et al., 2010; Kenyon, 2010; Selman et al., 2009; Stanfel et al., 2009). TOR has also been implicated in dietary restriction (DR), an intervention that extends lifespan and protects against chronic disease. Rapamycin increases mouse lifespan even when administered late in life (Harrison et al., 2009), suggesting that pharmacological targeting of TOR signaling might be a promising anti-aging strategy. However, rapamycin is associated with insulin resistance as well as immunosuppression (Zoncu, et al., 2011; Lamming, et al., 2012), making it important to identify specific mechanisms downstream of rapamycin that affect aging.
TOR inhibition, rapamycin, and DR seem to promote longevity at least in part by reducing mRNA translation (Bjedov et al., 2010; Kapahi et al., 2010; Kenyon, 2010; Stanfel et al., 2009; Zid et al., 2009). A lower level of translation might be beneficial simply because the burden of protein synthesis is decreased, but recent evidence indicates that when translation is reduced protective mechanisms are mobilized, through translation of particular genes being preserved or even increased (Kapahi et al., 2010; Rogers et al., 2011; Stanfel et al., 2009; Zid et al., 2009). Genetic interference with translation initiation increases lifespan in C. elegans, and some studies indicate that this effect requires the conserved transcription factors DAF-16/FoxO (Hansen et al., 2007; Henderson et al., 2006; Rogers et al., 2011; Tohyama et al., 2008; Wang et al., 2010) and SKN-1/Nrf (Wang et al., 2010). This suggests that the benefits of reduced protein synthesis might also involve modulation of transcription. DAF-16 and SKN-1 regulate genes that protect against environmental, metabolic, and proteotoxic stress, and promote longevity in various species (Kenyon, 2010; Kwon et al., 2010; Li et al., 2011; Oliveira et al., 2009; Sykiotis and Bohmann, 2010; Tullet et al., 2008). In C. elegans, they are each inhibited by insulin/IGF-1-like signaling (IIS), another growth-related pathway that influences aging across metazoa (Kenyon, 2010; Tullet et al., 2008).
The genetic evidence that DAF-16 and SKN-1 are important for the benefits of reducing translation raises the question of whether TOR signaling might actually regulate these transcription factors. It is unknown whether TOR affects SKN-1/Nrf activity, but it is generally accepted that TOR influences aging by acting independently from DAF-16/FoxO and IIS, in large part because daf-16 is not needed for lifespan to be extended by reduced TOR kinase (LET-363) activity or most DR regimens (Bishop and Guarente, 2007; Hansen et al., 2007; Honjoh et al., 2009; Kapahi et al., 2010; Kenyon, 2010; Panowski et al., 2007; Sheaffer et al., 2008; Vellai et al., 2003). Lack of the TOR kinase would eliminate both TORC1 and TORC2, however, making it critical to establish how TORC1 and TORC2 affect longevity independently of each other.
Here we have investigated how longevity is affected by genetic TORC1 or TORC2 inhibition and, for the first time in C. elegans, rapamycin. We find that lifespan is increased in a SKN-1-dependent manner when either TOR pathway is inhibited. DAF-16 is also required for lifespan to be extended by genetic inhibition of TORC1, but not TORC2. Genetic interference with TORC1 results in a SKN-1- and DAF-16-mediated transcriptional response that may be triggered by lower levels of translation. Rapamycin induces a similar response but extends lifespan independently of daf-16, apparently by inhibiting both TORC1 and TORC2. We conclude that both SKN-1/Nrf and DAF-16/FoxO are opposed by TOR signaling, and have a central role in its influence on aging.
Results
SKN-1/Nrf and DAF-16/FoxO are required for stress resistance and longevity from reduced TORC1 activity
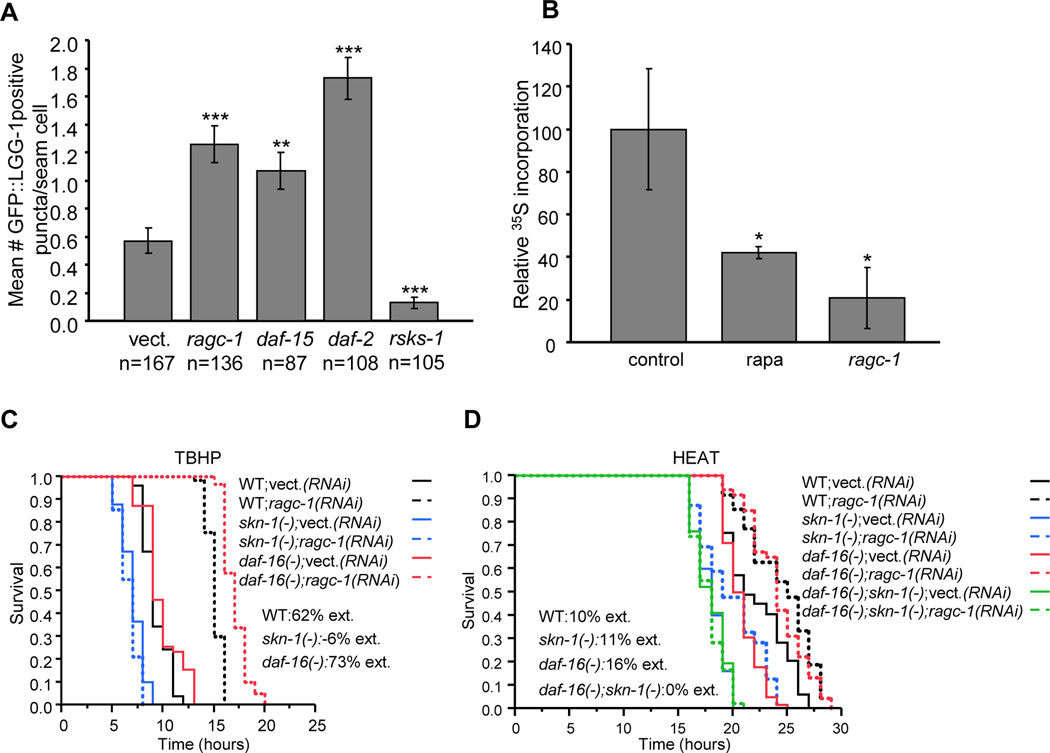
To inhibit TORC1 but not TORC2, and to obtain results that were not confounded by its developmental functions (Jia et al., 2004; Schreiber et al., 2010), we used RNA interference (RNAi) to knock down the TORC1-specific genes daf-15 (Raptor) and rheb-1 (Rheb), and the conserved Rag GTPases raga-1 and ragc-1 (Figure S1) only during adulthood. We monitored TORC1 activity by examining autophagy and translation. The GFP-fused vacuolar protein LGG-1 marks autophagic vesicles. LGG-1 puncta were increased by knockdown of the insulin receptor DAF-2, as described (Melendez et al., 2003), and by ragc-1 or daf-15 RNAi, as predicted (Figure 1A and Figure S1). In contrast, autophagy was reduced by knockdown of the S6 kinase RSKS-1, which increases translation downstream of TORC1 but also promotes autophagy (Scott et al., 2004). As expected, ragc-1 RNAi decreased overall mRNA translation, as measured by 35S methionine incorporation (Figure 1B).
Figure 1. Genetic TORC1 inhibition increases stress resistance through SKN-1 and DAF-16.
(A) Increased autophagy after TORC1-pathway gene knockdown. LGG- 1::GFP puncta were counted in seam cells (n) in day 3 adults. ***P≤0.0001, **P<0.001, unpaired t-test. (B) Decreased protein synthesis after genetic TORC1 inhibition or rapamycin treatment. *P≤0.005, Student’s one-sided t-test. Error bars represent ± SEM (C) TORC1 inhibition by ragc-1 RNAi increased oxidative stress (TBHP) resistance dependent upon skn-1 but not daf-16. The skn-1(zu67) and daf-16(mgDf47) alleles were analyzed in all experiments unless otherwise indicated. In all survival plots, ext. refers to mean survival extension associated with the indicated intervention, and WT to the wild type. The y-axis indicates proportion surviving. (D) Increased resistance to heat (35°C) is mediated by both skn-1 and daf-16. Representative experiments are shown in (A-D). Statistics and stress resistance analyses of additional TORC1 pathway genes are provided in Tables S1 and S2.
Knockdown of each TORC1 pathway gene that we examined (raga-1, ragc-1, rheb-1, daf-15) dramatically enhanced stress tolerance. skn-1 but not daf-16 was required for increased resistance to the oxidizing agent tert-butyl hydrogen peroxide (TBHP)(Figure 1C and Table S1). This result mimicked the effect of inhibiting translation initiation (Wang et al., 2010). Interference with TORC1 by ragc-1 RNAi increased heat resistance in daf-16 and skn-1 mutants, but not a skn-1; daf-16 double mutant, indicating involvement of both daf-16 and skn-1 (Figure 1D and Table S2). The increases in stress resistance that result from genetic TORC1 inhibition are therefore mediated by both SKN-1 and DAF-16, with SKN-1 being essential under TBHP oxidative stress conditions.
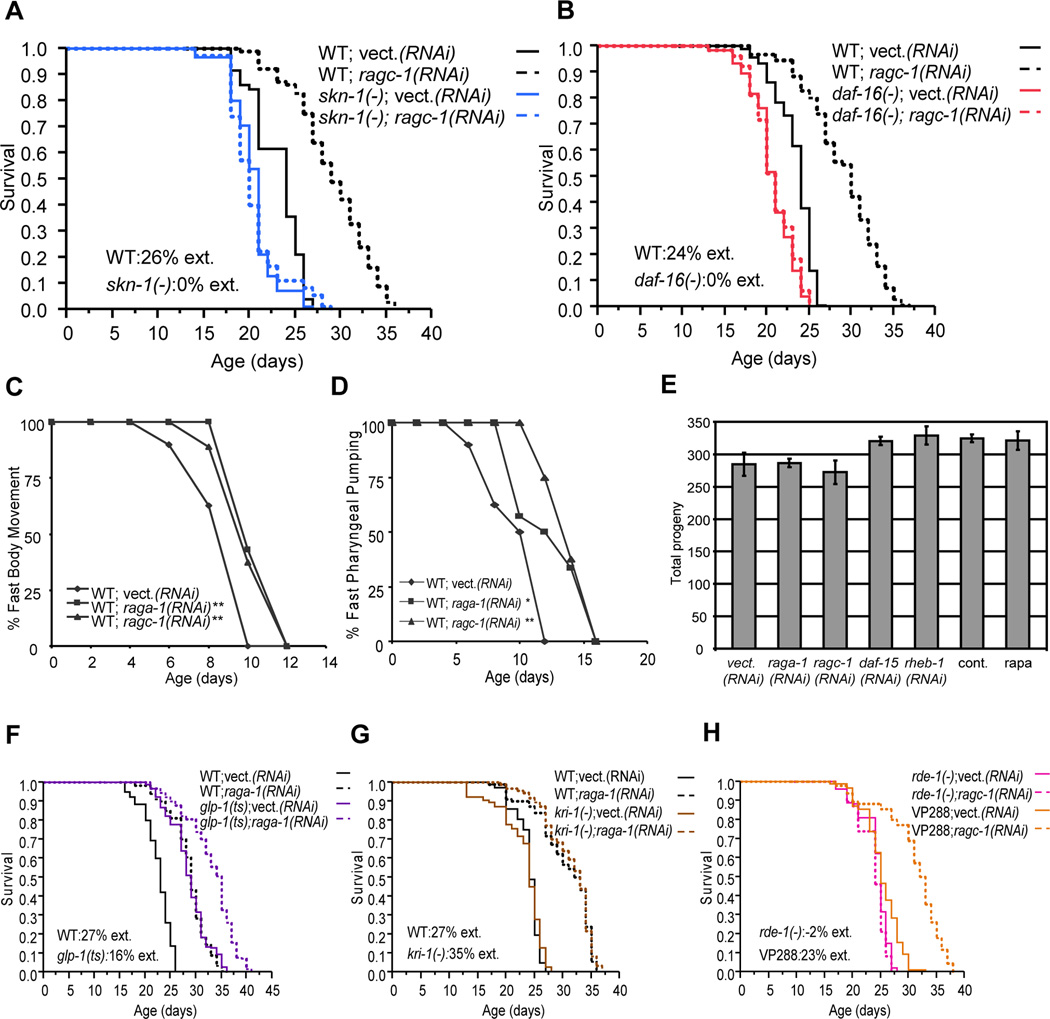
Adulthood knockdown of each TORC1 pathway gene that we tested increased lifespan (Figures 2A and 2B, Tables S3 and S4). Reduced TORC1 activity also delayed the age-associated decline in two healthspan indicators, fast movements and pharyngeal pumping, indicating that aging was slowed (Figure 2C and 2D). In most experiments, knockdown of TORC1 pathway genes failed to increase lifespan in a skn-1 mutant (Figure 2A, Tables S3 and S4). Surprisingly, RNAi against these four genes also did not increase lifespan in daf-16 mutants (Figure 2B, Tables S3 and S4), in contrast to the daf-16-independent longevity associated with TOR kinase inhibition (Hansen et al., 2007; Sheaffer et al., 2008; Vellai et al., 2003). We obtained similar findings with or without inclusion of fluorodeoxyuridine (FUdR) to prevent offspring production (Table S3 and S4). Lifespan of the skn-1; daf-16 mutant was unaffected by raga-1 RNAi but increased by inhibition of mitochondrial genes (cco-1, cyc-1; Table S3 and S4), which involves a distinct longevity pathway (Durieux et al., 2011; Kenyon, 2010). This indicates that animals that lack SKN-1 and DAF-16 are not simply refractory to lifespan extension. Both SKN-1 and DAF-16 are therefore required for the delays in aging that result from genetic TORC1 inhibition.
Figure 2. Lifespan extensions from genetic TORC1 inhibition require SKN-1 and DAF-16.
skn-1 (A) and daf-16 (B) are required for genetic TORC1 inhibition to increase longevity. (C, D) Rag GTPase knockdown increases healthspan. raga-1 or ragc-1 RNAi preserves fast body movements (C) and fast pharyngeal pumping (D). **P≤0.008, log rank for (C); **P< 0.007, *P< 0.08, log rank for (D). (E) Brood size is unaffected by adulthood TORC1 pathway gene RNAi or rapamycin. n=3-7 worms. Error bars represent ± SEM (F) Genetic TORC1 inhibition extends lifespan in glp-1(ts) animals independently of the GCS pathway. WT or glp-1(ts) animals were placed at the non-permissive temperature (25°C) from the L2 stage until adulthood, then maintained at 20°C, a protocol that prevents germ cell proliferation in glp-1(ts). raga-1 or control RNAi was initiated at the beginning of adulthood. (G) TORC1 inhibition by raga-1 RNAi extends lifespan in kri-1(ok1251) mutants, in which germ cell arrest fails to extend lifespan. (H) TORC1 inhibition by intestinal ragc-1 RNAi. In VP288, rde-1 is rescued using the intestine-specific promoter nhx-2 (Durieux et al., 2011; Qadota et al., 2007). Survival plots show composite or individual experiments that were performed in parallel. Corresponding data, analyses of additional TORC1 pathway genes, and statistics are shown in Table S3, and individual experiments in Table S4.
Somatic effects of TORC1 on longevity
Given that TORC1 promotes protein synthesis, and that inhibition of translation reduces fecundity (Hansen et al., 2007; Pan et al., 2007), it is possible that TORC1 inhibition might increase longevity indirectly, through effects on the germline. Reductions in germline stem cell (GSC) number delay aging, presumably to preserve the organism during adversity (Kenyon, 2010). When the number of C. elegans GSCs is reduced, DAF-16 accumulates in intestinal nuclei. The accompanying longevity extensions require DAF-16 and the ankyrin repeat protein KRI-1 (Kenyon, 2010).
Several lines of evidence indicate that genetic interference with TORC1 or translation extends lifespan independently of the GSC pathway. Fecundity and presumably GSC proliferation were not impaired by adulthood knockdown of TORC1 pathway genes (Figure 2E). Knockdown of raga-1 or the translation initiation factors ifg-1 and eif-1 increased lifespan in the temperature-sensitive (ts) germline-defective mutant glp-1(bn18), under conditions where germ cell proliferation was blocked and lifespan was extended (Figure 2F, Tables S3 and S4). Finally, knockdown of either raga-1 or eif-1 extended the lifespan of kri-1 mutants (Figure 2G, Tables S3 and S4), in which the GSC pathway is blocked (Kenyon, 2010). In C. elegans, tissue-specific RNAi can be performed in strains where the RNAi-defective mutation rde-1 is rescued in individual tissues (Qadota et al., 2007). ragc-1 RNAi increased lifespan in a strain in which RNAi is restricted to the intestine (VP288; Figure 2H, Table S3 and Table S4). This suggests that aging is delayed when amino acid signaling to TORC1 is impaired specifically in the intestine, which corresponds to the mammalian liver, gut, and adipose tissues, and plays a central role in metabolism and longevity (Kenyon, 2010).
TORC1 inhibits SKN-1/Nrf-and DAF-16/FoxO-driven transcription
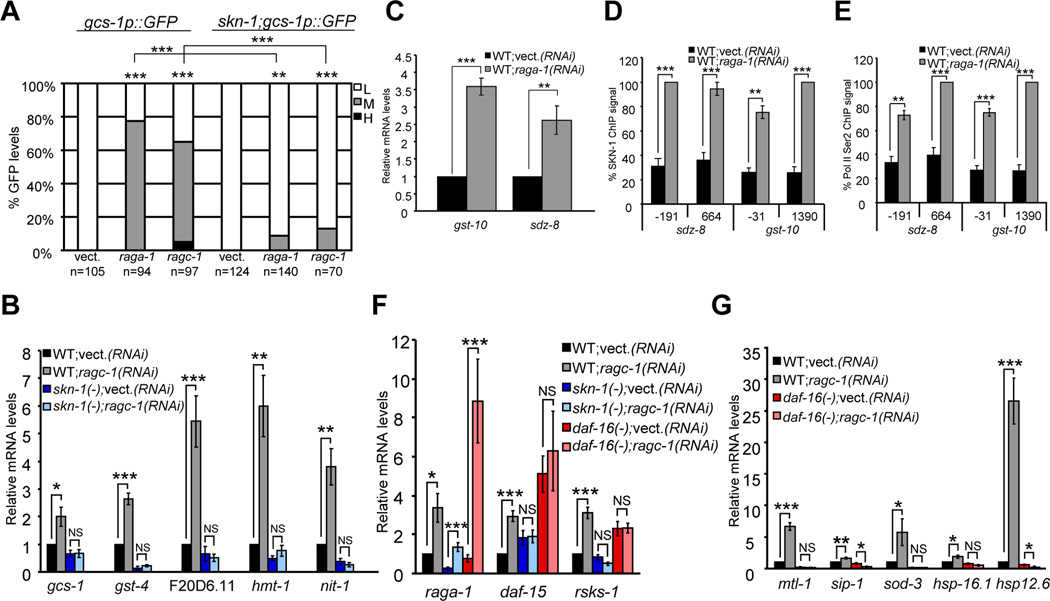
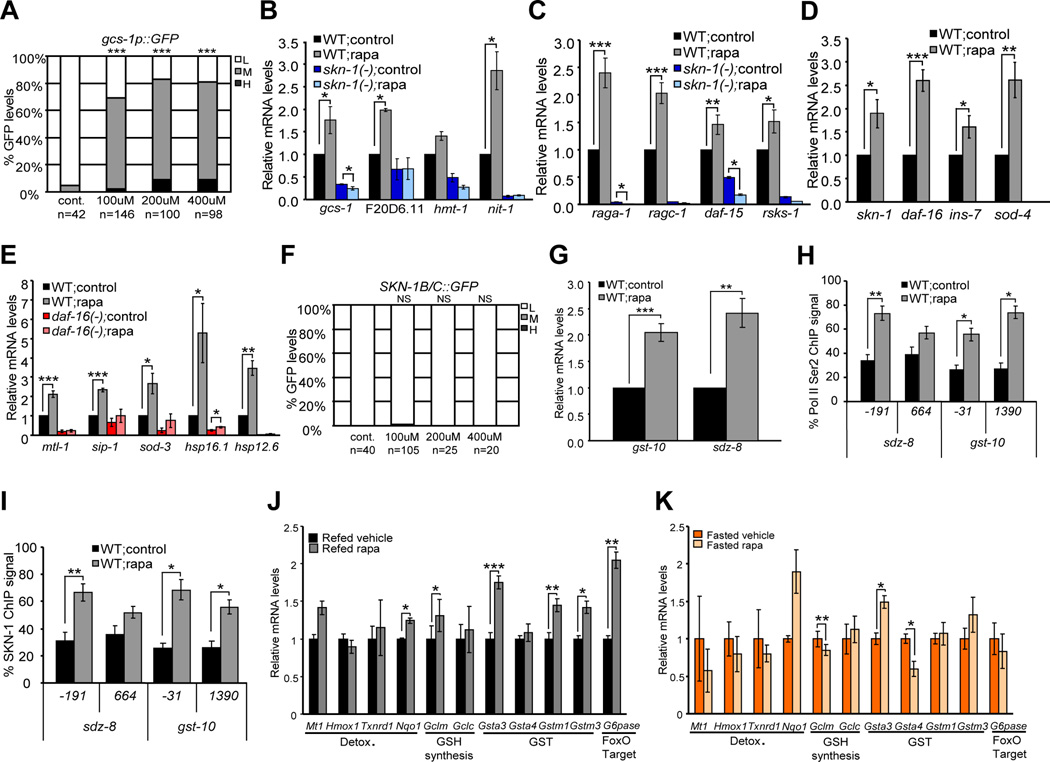
Our results raise an important question: are activities of SKN-1 and DAF-16 actually controlled by TORC1, or does longevity associated with reduced TORC1 activity simply depend upon constitutive functions of these transcription factors? The finding that TORC1 regulates SKN-1 and DAF-16 would strongly support the model that they play central roles in its effects on aging. When translation initiation factors are knocked down by RNAi, reporters in which promoters of the conserved SKN-1/Nrf targets gcs-1 and gst-4 are fused to GFP (gcs-1p::GFP and gst-4::GFP) are activated in the intestine, and several SKN-1 target genes are upregulated (Wang et al., 2010). If SKN-1 targets are activated by interference with translation, then genetic inhibition of TORC1 might induce a similar response. Accordingly, knockdown of TORC1 pathway genes increased intestinal gcs-1p::GFP and gst-4::GFP expression (Figure 3A, Figures S2A and S2B). We next analyzed how expression of endogenous SKN-1 target genes (Oliveira et al., 2009; Wang et al., 2010) was affected when TORC1 was inhibited through Rag gene RNAi (Figure 3B, Figure S2C and S2D). These and other SKN-1 targets that we analyzed in this study represent various stress-defense functions (glutathione synthesis, glutathione S-transferase, ABC transporter, oxidoreductase, lysosomal protease). Rag gene RNAi increased expression of these genes in a skn-1-dependent manner, but did not activate control genes (Figure 3B, Figure S2C and S2D). We conclude that when TORC1 is inhibited, SKN-1 induces a protective transcriptional response.
Figure 3. Genetic TORC1 inhibition induces SKN-1- and DAF-16-mediated transcription.
(A) skn-1-dependent induction of the SKN-1 target promoter gcs-1 by Rag GTPase RNAi. ***P< 0.0001, ** P < 0.001, chi2 method. L=Low, M=Medium, H=High. Scoring is described in Experimental Procedures. (B) Activation of endogenous SKN-1 target genes by ragc-1 RNAi, measured by quantitative (q)RT-PCR in WT animals, or skn-1 mutants. Fold induction relative to WT vector control is shown for all qRT-PCR data. (C-E) Transcriptional activation of the SKN-1 targets gst-10 and sdz-8 by raga-1 RNAi, detected by qRT-PCR (C) in lysates that were analyzed by ChIP in (D, E). The relative ChIP signal is shown for endogenous SKN-1 (D) and Serine 2-phosphorylated Pol II CTD (Ser2) (E) along each gene. Positions indicated below the graphs correspond to the middle of each qPCR amplicon relative to the predicted transcription start site. %ChIP signal is relative to input, and normalized to the highest signal in each qPCR run (Glover-Cutter et al., 2008). Analyses of intergenic regions and control genes (not shown) indicated that signals of 25% and 10% represent thresholds for specific presence of SKN-1 and P-Ser2, respectively. (F) Activation of endogenous TORC1 pathway genes by ragc-1 RNAi. (G) Induction of endogenous DAF-16 target genes in response to ragc-1 RNAi. ***P<0.001,**P<0.01, *P<0.05, NS=not significant. All qRT-PCR and ChIP p-values in this study were calculated by one- or two-sided t-test, as appropriate. Error bars represent ± SEM
To investigate further whether TORC1 regulates transcription of SKN-1 target genes, we used chromatin immunoprecipitation (ChIP) to detect endogenous SKN-1 and markers of transcription activity at the SKN-1 targets gst-10, sdz-8, and gcs-1. TORC1 inhibition (raga-1 RNAi) increased expression of these genes (Figure 3C and Figure S2E), accompanied by binding of endogenous SKN-1 to their regulatory regions (Figure 3D and Figure S2F). During transcription initiation, RNA Polymerase II (Pol II) is phosphorylated on Ser 5 of its C-terminal domain (CTD) repeat (P-Ser5) (Bentley, 2005). Phosphorylation of the CTD repeat on Ser 2 (P-Ser2) also marks transcription, particularly the elongation step. TORC1 inhibition significantly increased P-Ser5 levels 5’ of the gcs-1 coding region (Figure S2G). At gst-10 and sdz-8 we assayed P-Ser2, levels of which were similarly increased (Figure 3E). Together, the data indicate that TORC1 suppresses SKN-1-mediated transcription of protective genes.
A genome-scale ChIP analysis revealed that transgenically-expressed SKN-1 is also present at potential regulatory regions of the TORC1 pathway genes daf-15, rsks-1, raga-1, and ragc-1 (Niu et al., 2010), suggesting that SKN-1 and TORC1 might regulate their expression. Supporting this idea, ragc-1 RNAi led to an increase in TORC1 pathway gene expression that was largely skn-1-dependent (Figure 3F). A reduction in TORC1 signaling therefore directs SKN-1 not only to activate protective genes, but also to increase TORC1 pathway gene expression in a feedback loop.
We also investigated whether DAF-16 might be regulated by TORC1 and translation inhibition. Knockdown of either Rag gene led to daf-16-dependent activation of the conserved DAF-16/FoxO target sod-3 (superoxide dismutase) in the intestine, and upregulation of multiple endogenous DAF-16 target genes (Kwon et al., 2010; Murphy et al., 2003)(Figure 3G, Figure S2H and S2I). These DAF-16 targets included small heat-shock protein genes that have been implicated in longevity (sip-1, hsp-16.1, hsp-12.6). Knockdown of the translation factor ifg-1 also activated many of these endogenous DAF-16 target genes, consistent with the idea that translation suppression is involved (Figure S2J). DAF-16 reduced expression of rsks-1 and daf-15, consistent with a previous report (Jia et al., 2004), and was not required for genetic TORC1 inhibition to increase raga-1 expression (Figure 3F), indicating a distinct function from SKN-1 in TORC1 pathway autoregulation. The data indicate that both SKN-1 and DAF-16 induce protective gene expression when TORC1 activity is reduced, and therefore that TORC1 signaling opposes each of these two transcription factors.
Distinct transcriptional responses regulated by TORC1 and IIS
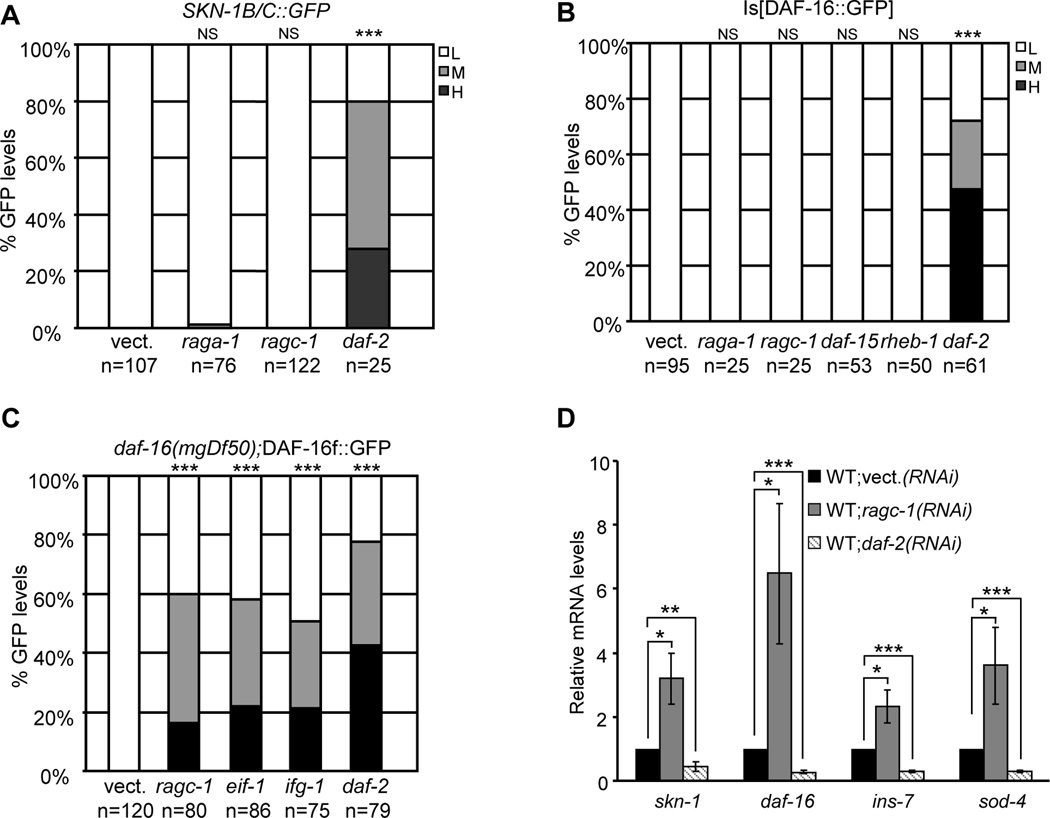
The essential role played by SKN-1 in TORC1-associated lifespan extension was surprising, because SKN-1 is not as important as DAF-16 for longevity that is associated with reduced IIS (Tullet et al., 2008). Our results could be explained if TORC1 and IIS have distinct effects on SKN-1, DAF-16, and their downstream target genes. IIS inhibits SKN-1 and DAF-16 through phosphorylation, so that they accumulate in nuclei when IIS is reduced (Kenyon, 2010; Tullet et al., 2008)(i.e. daf-2 RNAi, Figures 4A-4C). In contrast, SKN-1 generally did not accumulate in intestinal nuclei in response to genetic inhibition of either TORC1 (Figure 4A, and Figures S3A and S3B), or translation initiation (Wang et al., 2010). TORC1 might influence whether SKN-1 that is already in the nucleus is recruited to promoters, a mechanism that is feasible because SKN-1 occupies many promoters even under non-stressed conditions (Niu et al., 2010). Multiple DAF-16 isoforms accumulate in nuclei when IIS is reduced (Kenyon, 2010; Kwon et al., 2010), but only a single DAF-16 isoform (DAF-16f) localized to intestinal nuclei after genetic inhibition of TORC1 or translation initiation (Figures 4B and 4C, and Figures S3C-S3F). This DAF-16 isoform appears to be particularly important for regulating longevity (Kwon et al., 2010).
Figure 4. Distinct transcriptional responses to inhibition of TORC1 and IIS.
(A) Genetic TORC1 inhibition generally does not increase intestinal SKN-1 nuclear occupancy. SKN-1B/C::GFP encodes two of three SKN-1 isoforms (An and Blackwell, 2003). Scoring is described in Experimental Procedures. (B) Analysis of DAF-16 nuclear occupancy in the intestine. This transgene encodes the DAF-16a isoform (Henderson et al., 2006). (C) Accumulation of DAF-16f (Kwon et al., 2010) in intestinal nuclei after genetic inhibition of TORC1 or translation. (D) Transcriptional activation of certain genes after genetic inhibition of TORC1 but not IIS, analyzed by qRT-PCR. ***P<0.0001, NS= not significant, chi2 method L=Low, M=Medium, H=High for (A-C); ***P<0.001, **P<0.01, *P<0.05 for (D). Error bars represent ± SEM
Genetic inhibition of TORC1 or translation initiation also induced transcription of various genes that are not activated by reduced IIS. These included skn-1 and daf-16 themselves, the superoxide dismutase sod-4, which is upregulated by DR (Panowski et al., 2007), and the insulin peptide ins-7, which is inhibited by SKN-1 and DAF-16 under normal and reduced IIS conditions, respectively (Murphy et al., 2003; Oliveira et al., 2009)(Figure 4D and Figure S3G). We conclude that TORC1 and IIS act through different mechanisms to regulate SKN-1 and DAF-16, and thereby modulate transcription of overlapping but distinct sets of downstream genes.
SKN-1/Nrf- and DAF-16/FoxO-mediated transcriptional responses to rapamycin
Rapamycin has not been reported to affect C. elegans longevity or stress defense, but our findings provided a readout of TORC1 inhibition that allowed us to optimize rapamycin concentration. Exposure of adults to 100 µM rapamycin upregulated gcs-1p::GFP in a skn-1-dependent manner, and reduced translation (Figures 1B and 5A, and Figure S4A). This rapamycin concentration is higher than that used in cultured cells, but high concentrations of compounds are often required for bioavailability in C. elegans (Kokel et al., 2006). Importantly, rapamycin upregulated genes that were activated by genetic TORC1 inhibition, including skn-1, daf-16, and genes that encode TORC1 pathway components (Figures 5B-5E, Figures S4B and S4C). Rapamycin did not interfere with bacterial proliferation, indicating that these effects did not derive artifactually from food limitation (Figure S4D). As seen with genetic TORC1 inhibition, rapamycin did not increase the levels of SKN-1 in nuclei, and affected only the DAF-16f isoform (Figure 5F, Figures S4E and S4F). SKN-1 target activation was accompanied by increased presence of SKN-1 and transcription markers at these genes, however, indicating direct regulation by SKN-1 (Figures 5G-5I, Figures S4G-S4L). Rapamycin-induced activation of SKN-1 target genes was, in general, SKN- 1-dependent (Figure 5B), and DAF-16 target gene induction was largely abolished in a daf-16 mutant (Figure 5E). We conclude that rapamycin induces transcription of SKN- 1- and DAF-16-regulated protective genes.
Figure 5. Rapamycin activates SKN-1- and DAF-16-mediated transcription.
(A) Activation of gcs-1::GFP by rapamycin. 100 µM (in the agar) was the lowest concentration at which near-maximal induction occurred. ***P< 0.0001, NS= not significant, chi2 method. L=Low, M=Medium, H=High. (B-D) Rapamycin-induced activation of endogenous SKN-1 target (B), TORC1 pathway (C) other TORC1-regulated (D), and DAF-16 target (E) genes, analyzed by qRT-PCR. (F) Rapamycin does not induce nuclear accumulation of SKN-1B/C::GFP in worms grown on OP50. (G-I) Activation of SKN-1 target gene transcription by rapamycin, with accumulation of SKN-1 (H) and Serine 2-phosphorylated Pol II CTD (Ser2) (I) detected by ChIP as in Figure 3C-3E. (J, K) Rapamycin increases Nrf/SKN-1 target gene expression in mouse liver. Vehicle- or rapamycin-treated male mice were fasted (K), or fasted then re-fed (J)(n=5 for each) prior to analysis of RNA by qRT-PCR. Genes that are regulated by Nrf1 (Mt1)(Ohtsuji et al., 2008) or Nrf2 (others)(Malhotra et al., 2010; Wu et al., 2011) were assayed along with the FoxO target G6Pase (Mihaylova et al., 2011). ***P<0.001, **P<0.01, *P<0.05, NS=not significant. Error bars represent ± SEM
We investigated whether the transcriptional responses to rapamycin we have detected might be conserved in mammals. Rapamycin-treated or control mice were fasted overnight to control for feeding status and its effects on TORC1, and then were either re-fed or not. In the liver, rapamycin increased expression of the FoxO target G6Pase under re-feeding but not fasting conditions, as described (Figure 5J and 5K)(Lamming et al., 2012). Similarly, in the re-fed group rapamycin upregulated most of the ten Nrf/SKN-1 targets we tested, each of which is involved in stress defense, with five genes reaching statistical significance (Figure 5J). These effects were also largely attenuated by fasting (Figure 5K). We conclude that rapamycin treatment activates SKN-1/Nrf-regulated protective genes in vivo in mammals, as we observed in C. elegans.
SKN-1/Nrf but not DAF-16/FoxO is required for longevity from rapamycin or TORC2 inhibition
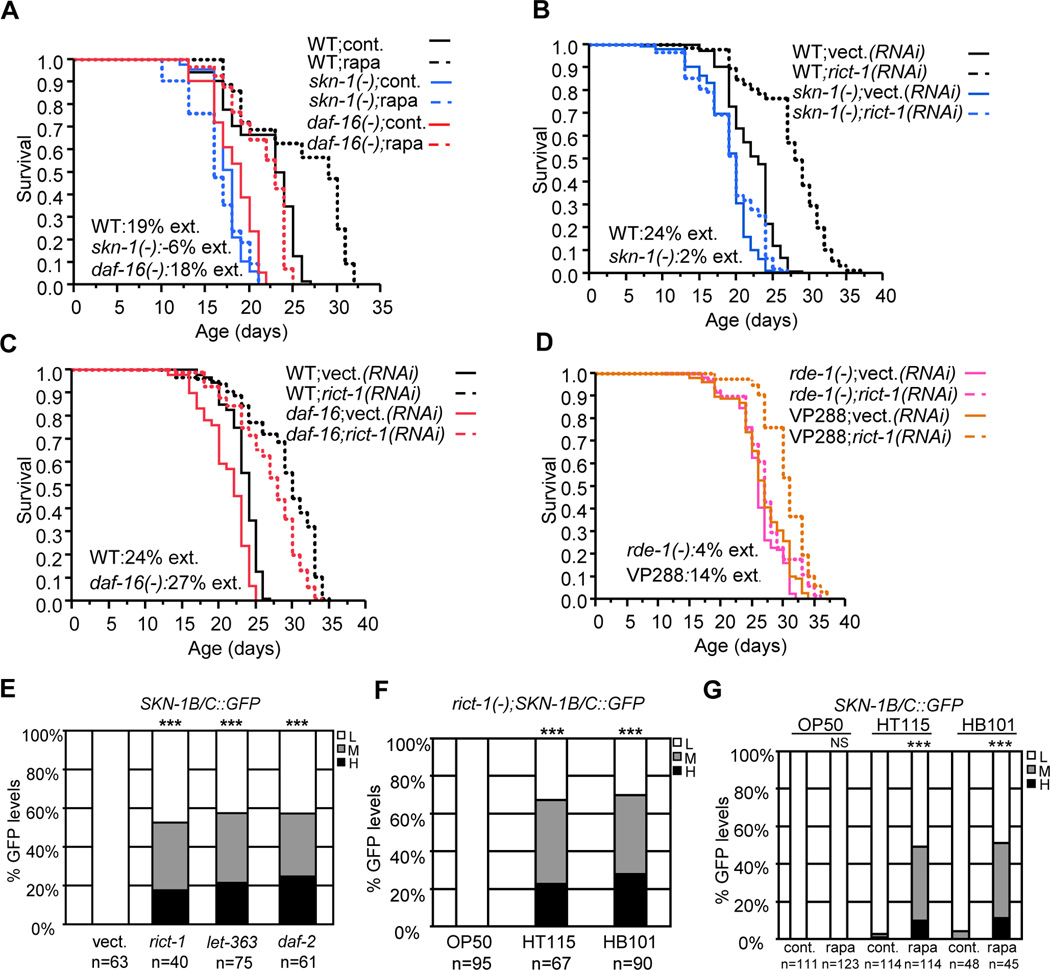
Exposure of adult C. elegans to rapamycin dramatically increased both stress resistance and lifespan. Rapamycin increased oxidative stress (TBHP) resistance dependent upon skn-1 but not daf-16, and increased lifespan in a skn-1-dependent manner, as seen with genetic TORC1 inhibition (Figure 6A, Table S1 and S5). In contrast, rapamycin robustly increased lifespan in two daf-16 mutants (mgDf47 and mu86). Similar results were obtained with or without FUdR, and with growth on either the standard strain OP50 or the feeding RNAi strain HT115 (Table S5). The daf-16-independence of longevity deriving from adulthood rapamycin treatment, together with our finding that rapamycin did not impair fecundity (Figure 2E), argued that rapamycin does not increase lifespan through the GSC pathway.
Figure 6. Rapamycin-induced lifespan extension may involve TORC1 and TORC2.
(A) Longevity from rapamycin requires skn-1 but not daf-16. (B, C) TORC2 inhibition (rict-1 RNAi) increases lifespan dependent upon skn-1 (B) but not daf-16 (C). (D) rict-1 RNAi extends lifespan in VP288, in which RNAi is active specifically in the intestine (see text). Representative (rapamycin) or composite (rict-1) experiments are shown, with statistics and additional analyses presented in Tables S6 and S7. (E) TORC2 or TOR kinase (let-363) inhibition by feeding RNAi increases SKN-1 nuclear occupancy. (F, G) SKN-1 nuclear accumulation in rict-1 mutants (F) and after rapamycin treatment (G) depends upon the food source. ***P< 0.0001, NS= not significant, chi2 method. L=Low, M=Medium, H=High. Error bars represent ± SEM
Our finding that rapamycin increased lifespan independently of daf-16 indicates that rapamycin influences an additional longevity pathway besides TORC1, and parallels previous analyses of the TOR kinase, in which daf-16 was not required (Hansen et al., 2007; Sheaffer et al., 2008; Vellai et al., 2003). Although rapamycin is generally considered to be a TORC1 inhibitor, in some mammalian cell lines prolonged rapamycin treatment also interferes with TORC2, apparently by physically disrupting the TORC2 complex (Zoncu et al., 2011). Recently rapamycin has also been observed to disrupt TORC2 in vivo in multiple tissues in the mouse (Lamming, et al., 2012). Our results could be reconciled with the C. elegans TOR kinase literature if: 1) rapamycin interferes with both TORC1 and TORC2 in C. elegans, as would be the case for TOR loss, 2) TORC2 inhibition increases lifespan, and 3) the lifespan extension associated with TORC2 inhibition requires skn-1 but not daf-16.
C. elegans with mutations in the TORC2 complex gene rict-1 (Rictor, Figure S1) grow slowly and have a small body size, and live slightly longer than WT when maintained on “rich” food such as the RNAi feeding strain HT115 and at elevated temperature (25°C)(Soukas et al., 2009). We investigated whether TORC2 inhibition might increase longevity at a lower temperature (20°C), and when TORC2 activity was reduced by adulthood rict-1 RNAi, a strategy that would bypass developmental functions of TORC2. Under these conditions, rict-1 RNAi increased lifespan substantially, dependent upon skn-1 (Figure 6B, Tables S6 and S7). Importantly, daf-16 was not required for lifespan to be increased by rict-1 RNAi, or when we blocked both TORC1 and TORC2 by ragc-1; rict-1 RNAi (Figure 6C, Tables S6 and S7). Our results suggest that the daf-16-independent longevity associated with TOR kinase (let-363) loss (Hansen et al., 2007; Sheaffer et al., 2008; Vellai et al., 2003) is mediated by lack of TORC2, not TORC1, and that the effects of rapamycin we observed may also involve inhibition of both TORC1 and TORC2. rict-1 RNAi extended lifespan in the intestine- specific RNAi strain VP288 (Figure 6D, Tables S6 and S7), indicating that TORC2, like TORC1, modulates lifespan at least in part by acting in the intestine.
Given the importance of SKN-1 for the lifespan increases associated with TORC2 inhibition, we investigated whether TORC2 influences SKN-1 nuclear accumulation. Genetic evidence suggests that in C. elegans, as in mammals, TORC2 increases activity of the AKT and SGK-1 kinases, which phosphorylate SKN-1 and inhibit its accumulation in intestinal nuclei (Figure S1)(Soukas et al., 2009; Tullet et al., 2008). Accordingly, SKN-1 accumulated in intestinal nuclei after rict-1 RNAi (Figure 6E). This result contrasted sharply with the failure of rapamycin to increase SKN-1 nuclear accumulation (Figure 5F), but in those earlier rapamycin experiments worms had been grown on OP50, on which the lifespan of rict-1 mutants is decreased (Soukas et al., 2009). Strikingly, either rict-1 mutation or rapamycin dramatically increased SKN-1 nuclear accumulation when worms were grown on rich food (HT115 or HB101), but not OP50 (Figure 6F and 6G). This differs from the effect of daf-2 mutation (reduced IIS), which increases SKN-1 nuclear accumulation in worms maintained on OP50 (Tullet et al., 2008). In addition, the longevity and lifespan extensions associated with rapamycin seemed to be more robust on HT115 than OP50 (Table S5). Finally, RNAi knockdown of the TOR kinase let-363 also increased SKN-1 nuclear accumulation, as would be predicted for an effect on TORC2 (Figure 6E). Taken together, our experiments suggest that rapamycin promotes longevity by interfering with both TORC1 and TORC2, with the resulting effects on SKN-1/Nrf being essential.
Discussion
Our C. elegans experiments have uncovered an important mechanism of TORC1 action: TORC1 opposes both SKN-1/Nrf and DAF-16/FoxO, so that when TORC1 is inhibited these factors increase transcription of protective genes. Previous work had identified effects of TORC1 inhibition that may enhance longevity, including decreased protein synthesis, preservation of protective gene translation, and increased autophagy (Bjedov et al., 2010; Kapahi et al., 2010; Kenyon, 2010; Stanfel et al., 2009; Zid et al., 2009). Here we have shown that these TORC1-associated mechanisms fail to increase C. elegans lifespan in the absence of either SKN-1 or DAF-16, and therefore that the SKN-1- and DAF-16-dependent transcription responses we detected are essential effectors of the longevity that results from genetic TORC1 inhibition.
We were surprised to find that DAF-16 is required for genetic TORC1 inhibition to extend lifespan (Figure 2B, Tables S3 and S4), given that lack of the TOR kinase extends lifespan independently of DAF-16 (see Introduction). It has been reported that daf-16 is needed for lifespan extensions associated with daf-15 heterozygosity or raga-1 mutation (Jia et al., 2004; Schreiber et al., 2010), but several considerations made it unclear how to interpret those findings: these mutations impair larval development, DAF-16 inhibits daf-15 expression, and longevity from TOR kinase loss does not require daf-16. Our observation that TORC2 inhibition increases lifespan independently of daf-16 (Figure 6C, Tables S6 and S7) reconciles these seemingly conflicting studies with each other and our extensive data on TORC1. It also raises the intriguing possibility that the daf-16-independent longevity associated with many C. elegans DR regimens (see Introduction) might involve TORC2, especially considering that many rict-1 phenotypes are influenced by the food source (Figure 6F)(Soukas, et al., 2009). C. elegans DR has been observed to require neuronal SKN-1 expression and the FoxA transcription factor PHA-4, which is also needed for lifespan to be extended by TOR kinase inhibition (Bishop and Guarente, 2007; Panowski et al., 2007; Sheaffer et al., 2008). Perhaps some differences among DR protocols could be explained if SKN-1, DAF-16, and possibly PHA-4 have some overlapping functions, and are affected differently by these DR regimens.
It was particularly striking that SKN-1/Nrf was required for lifespan extensions associated with genetic inhibition of either TORC1 or TORC2, or rapamycin treatment, and therefore must be central to the influence of TOR pathways on aging (Figures 2A, 6A, 6B, and 7; Tables S3-S7). In contrast, SKN-1 is only partially required for lifespan extension associated with reduced IIS, in which daf-16 is essential (Kenyon, 2010; Tullet et al., 2008). We have determined that TORC1 and IIS not only act through different mechanisms to regulate SKN-1 and DAF-16, but also regulate expression of overlapping but distinct sets of their target genes (Figures 3 and 4). Regulators like SKN-1 and DAF-16 affect longevity through control of numerous genes that function in stress defense, metabolism, and other processes (Kenyon, 2010). If these transcription factors influence overlapping biological processes, it could explain why the lifespan extensions associated with genetic TORC1 and TORC2 inhibition are not additive (Table S6), and why rapamycin increases DAF-16 activity but extends lifespan independently of DAF-16 (Figures 5, S4, and 6A). It will be an important challenge to understand why the requirements for SKN-1 and DAF-16 differ with respect to longevity associated with reductions in IIS, TORC1, or TORC2 (Figure 7). This will involve identification of critical downstream genes, and analyses of how these signaling pathways 1) act in different tissues, 2) affect the action of SKN-1, DAF-16, and possibly other transcription factors, and 3) are influenced by other signals. For example, TORC2 and IIS activate kinases that regulate SKN-1 and DAF-16 (AKT and SGK)(Kenyon, 2010; Zoncu et al., 2011), but seem to mirror each other with respect to the relative importance of these transcription factors for longevity (Figure 7).
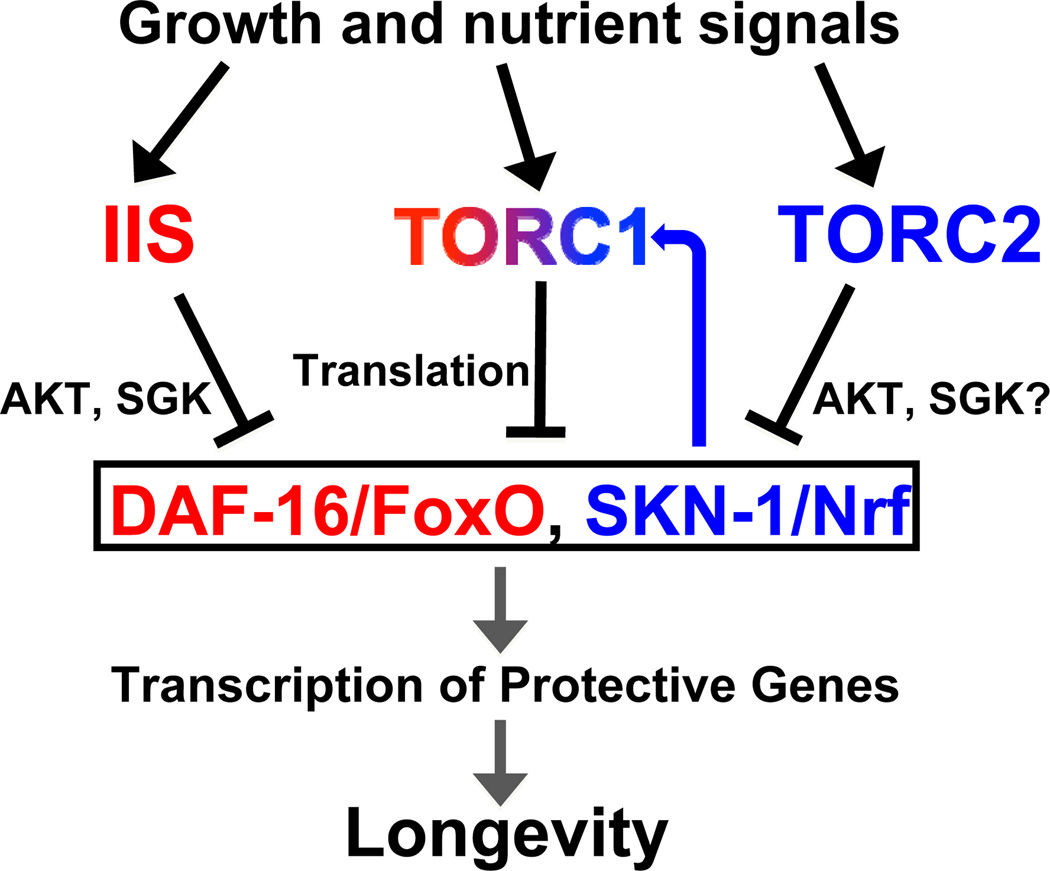
Figure 7. Regulation of SKN-1 and DAF-16 by TOR signaling.
The IIS, TORC1, and TORC2 pathways are involved in growth (see text). IIS inhibits SKN-1 and DAF-16 directly, through phosphorylation. TORC1 inhibits SKN-1 and DAF-16 expression and activity, at least in part by increasing mRNA translation. TORC2 regulates SKN-1 nuclear occupancy in a nutrient-dependent manner. DAF-16 is required for longevity that derives from inhibiting IIS or TORC1, but not TORC2. SKN-1 plays a contributory role in the effects of IIS on longevity, but is essential for TORC1 or TORC2 inhibition to extend lifespan. When TORC1 is inhibited, SKN-1 increases transcription of TORC1 pathway genes in a feedback loop.
The TORC1 and IIS pathways involve an intricate set of auto- and inter-regulatory networks. While we have shown that protective DAF-16/FoxO targets are activated when TORC1 is inhibited, in Drosophila and mammalian cells a feedback loop has been described in which high TORC1 activity leads to accumulation of reactive oxygen species (ROS), and in response FoxO increases expression of sestrins that inhibit TORC1 (Budanov et al., 2010; Hay, 2011). In mammals IIS increases TORC1 activity, but is inhibited by feedback loops emanating from TORC1 (Hsu et al., 2011; Yu et al., 2011; Zoncu et al., 2011). When mammalian TORC1 activity is low, autophagy- driven nutrient accumulation eventually drives a compensatory activation of TORC1 (Yu et al., 2010). We have described a feedback loop that may help compensate for reduced TORC1 activity: a SKN-1-driven induction of TORC1 pathway gene transcription (Figure 7). These complex inter-regulatory loops may allow for effective coordination of the TORC1 and IIS pathways with each other, and with growth factor, ROS, and nutrient levels.
Several lines of evidence suggest that TORC1 regulates SKN-1 and DAF-16 at least in part through modulating translation. Genetic inhibition of either TORC1 or translation initiation resulted in similar effects on stress resistance and lifespan, SKN-1 and DAF-16 nuclear occupancy, and regulation of downstream genes, including skn-1 and daf-16 themselves (see Results). Many genes encoding regulatory proteins are among those that are translated preferentially when translation is globally reduced, and translation of both SKN-1 and DAF-16 seems to be spared when translation initiation is suppressed (Kapahi et al., 2010; Rogers et al., 2011; Stanfel et al., 2009; Zid et al., 2009; Zoncu et al., 2011). We speculate that functional interactions among SKN-1, DAF-16, and other preferentially translated proteins may lead to the transcriptional responses that result from inhibiting either TORC1 or translation, although some of the many phosphorylation targets of TORC1 could also be involved (Hsu et al., 2011; Yu et al., 2011; Zoncu et al., 2011).
Our findings establish paradigms for understanding the biological effects of TORC1 signaling, and rapamycin-related TOR inhibitors. They predict that in mammals transcription programs regulated by SKN-1/Nrf and DAF-16/FOXO may be important in the effects of TORC1, including its influence on stress resistance, growth, proliferation, development, stem cell function, and TOR-associated diseases. Consistent with this idea, rapamycin treatment upregulated SKN-1/Nrf target genes in the mouse, as seen in C. elegans (Figure 5). Our C. elegans evidence indicated that rapamycin interferes with TORC2 in addition to TORC1 in vivo (Figure 6), also in agreement with recent observations in the mouse (Lamming, et al., 2012). In C. elegans, SKN-1 was required for longevity associated with genetic inhibition of TORC1 or TORC2, or rapamycin treatment, strongly predicting that Nrf proteins may mediate critical effects of rapamycin on mammalian longevity. Development of agents that promote particular SKN-1/Nrf or DAF-16/FoxO activities might therefore capture beneficial effects of TOR inhibition without interfering with critical TOR functions, or inducing unwanted effects of rapamycin on immune function or insulin sensitivity.
Together, our data show that the IIS and TOR pathways each influence aging by regulating SKN-1 and DAF-16 (Figure 7). Why would these pathways converge upon these two transcription factors? IIS, TORC1, and TORC2 are each involved in growth, and SKN-1 and DAF-16 functions include maintenance of protein homeostasis and resistance to various stresses (Kenyon, 2010; Lee et al., 2003; Li et al., 2011; McElwee et al., 2007; Murphy et al., 2003; Oliveira et al., 2009). Under anabolic conditions it might be advantageous to inhibit these defenses, which would influence regulatory processes in addition to consuming resources. For example, IIS is suppressed by redox-regulated phosphatases, and may function optimally under oxidizing conditions (Loh et al., 2009). Perhaps under conditions that do not favor growth, such as poor nutrient availability, it is important to mobilize stress defenses that are regulated by SKN-1 and DAF-16. A critical determinant of longevity seems to be the tradeoff between growth and reproduction on one hand, and mechanisms that increase stress resistance and delay aging on the other. Given that the TOR and IIS pathways influence aging by regulating SKN-1 and DAF-16, we conclude that the protective mechanisms controlled by these two transcription factors are of central importance for the balance between growth signals and longevity.
Experimental Procedures
Rapamycin treatment of C. elegans
Rapamycin (LC laboratories) was dissolved in DMSO at 50 mg/mL and added to plate agar to 100µm unless otherwise indicated. Control plates contained an appropriate DMSO concentration.
Lifespan and healthspan
Lifespans were carried out at 20°C essentially as described (Wang et al., 2010), and measured from hatching. RNAi or rapamycin treatments were continued throughout life. Mitochondrial gene RNAi was initiated at the L2 stage. Healthspan assays were performed essentially as described (Huang et al., 2004). Animals that crawled off the plate, ruptured, or died from internal hatching were excluded.
ChIP
ChIP was performed as essentially as described (Glover-Cutter et al., 2008), using a lysate protocol that was modified for C. elegans.
Transgenic reporter scoring
Expression or nuclear accumulation of transgenic GFP proteins was scored as “Low, Medium, or High” essentially as published (An and Blackwell, 2003; Tullet et al., 2008; Wang et al., 2010). Details are in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgements
We thank Javier Apfeld, Eric Greer, Marcia Haigis, and the Blackwell lab for helpful discussions, and David Bentley, Bruce Bowerman, and Heidi Tissenbaum for reagents. Supported by grants from the NIH (GM62891 to T. K. B and CA129105 to D. M. S.), the Ellison Medical Foundation (to T. K. B and to D. M. S.), the American Federation for Aging Research, Starr Foundation, and Koch Institute Frontier Research Program (to D. M. S.), and a DRC grant from the NIDDK. K. G.-C. was supported by an NIDDK T32 and an F32 NRSA, D. W. L. by an F32 NRSA and a Mentor-Based Fellowship from the ADA, and M. M. by the JSPS Excellent Young Researchers Overseas Visit Program. T. K. B. planned the project, T. K. B. and D. M. S. oversaw the work, all authors designed, carried out, and/or interpreted the experiments, and T. K. B., S. R.-S., K. G.-C., and D. W. L. wrote the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Other methods are available in the Supplemental Experimental Procedures.
No competing interests exist.
References
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov AV, Lee JH, Karin M. Stressin' Sestrins take an aging fight. EMBO Mol Med. 2010;2:388–400. doi: 10.1002/emmm.201000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N. Interplay between FOXO, TOR, Akt. Biochim Biophys Acta. 2011;1813:1965–1970. doi: 10.1016/j.bbamcr.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Bonafe M, Johnson TE. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J Gerontol A Biol Sci Med Sci. 2006;61:444–460. doi: 10.1093/gerona/61.5.444. [DOI] [PubMed] [Google Scholar]
- Honjoh S, Yamamoto T, Uno M, Nishida E. Signalling through RHEB-1 mediates intermittent fasting-induced longevity in C. elegans. Nature. 2009;457:726–730. doi: 10.1038/nature07583. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Chen D, Riddle DL. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development. 2004;131:3897–3906. doi: 10.1242/dev.01255. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Chen D, Rogers AN, Katewa SD, Li PW, Thomas EL, Kockel L. With OR less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 2010;11:453–465. doi: 10.1016/j.cmet.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kokel D, Li Y, Qin J, Xue D. The nongenotoxic carcinogens naphthalene and para-dichlorobenzene suppress apoptosis in Caenorhabditis elegans. Nat Chem Biol. 2006;2:338–345. doi: 10.1038/nchembio791. [DOI] [PubMed] [Google Scholar]
- Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA. A new DAF-16 isoform regulates longevity. Nature. 2010;466:498–502. doi: 10.1038/nature09184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Lan Y, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon A, Richardson A, Ahima RS, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 Target Genes That Control C. elegans Life-Span and Metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Li X, Matilainen O, Jin C, Glover-Cutter K, Holmberg CI, Blackwell TK. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K, Deng H, Fukushima A, Cai X, Boivin B, Galic S, Bruce C, Shields BJ, Skiba B, Ooms LM, et al. Reactive oxygen species enhance insulin sensitivity. Cell Metab. 2009;10:260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Piper MD, Thomas JH, Patel DS, Selman C, Withers DJ, Thornton JM, Partridge L, et al. Evolutionary conservation of regulated longevity assurance mechanisms. Genome Biol. 2007;8:R132. doi: 10.1186/gb-2007-8-7-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2010 doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh WJ, Wu CC, Kim SJ, Facchinetti V, Julien LA, Finlan M, Roux PP, Su B, Jacinto E. mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. Embo J. 2010;29:3939–3951. doi: 10.1038/emboj.2010.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira RP, P AJ, Dilks K, JL JA, Murphy CT, Blackwell TK. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski SH, Wolff S, Aguilaniu H, Durieux J, Dillin A. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- Qadota H, Inoue M, Hikita T, Koppen M, Hardin JD, Amano M, Moerman DG, Kaibuchi K. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 2007;400:166–173. doi: 10.1016/j.gene.2007.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson BW, Hubbard A, Melov S, Lithgow GJ, Kapahi P. Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 2011;14:55–66. doi: 10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber MA, Pierce-Shimomura JT, Chan S, Parry D, McIntire SL. Manipulation of behavioral decline in Caenorhabditis elegans with the Rag GTPase raga-1. PLoS Genet. 2010;6:e1000972. doi: 10.1371/journal.pgen.1000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009;326:140–144. doi: 10.1126/science.1177221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaffer KL, Updike DL, Mango SE. The Target of Rapamycin pathway antagonizes pha-4/FoxA to control development and aging. Curr Biol. 2008;18:1355–1364. doi: 10.1016/j.cub.2008.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfel MN, Shamieh LS, Kaeberlein M, Kennedy BK. The TOR pathway comes of age. Biochim Biophys Acta. 2009;1790:1067–1074. doi: 10.1016/j.bbagen.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohyama D, Yamaguchi A, Yamashita T. Inhibition of a eukaryotic initiation factor (eIF2Bdelta/F11A3.2) during adulthood extends lifespan in Caenorhabditis elegans. Faseb J. 2008;22:4327–4337. doi: 10.1096/fj.08-112953. [DOI] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. doi: 10.1038/426620a. [DOI] [PubMed] [Google Scholar]
- Wang J, Robida-Stubbs S, Tullet JM, Rual JF, Vidal M, Blackwell TK. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010;6:e1001048. doi: 10.1371/journal.pgen.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KC, Cui JY, Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by Association with the Ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.