Abstract
Heparan sulfate (HS) proteoglycans (PGs) interact with a number of extracellular signaling proteins thereby playing an essential role in the regulation of many physiological processes. One major function of HS is to interact with fibroblast growth factors (FGFs) and their receptors (FGFRs) and form FGF•HS•FGFR signaling complexes. Past studies primarily examined the selectivity of HS for FGF or FGFR. In present report, we used a new strategy to study the structural specificity of HS binding to 10 different FGF•FGFR complexes. Oligosaccharide libraries prepared from heparin, 6-desufated heparin and HS were used for the interaction studies by solution competition surface plasmon resonance (SPR) and using filter trapping assays. Specific oligosaccharides binding to FGF•FGFR complexes were subjected to polyacrylamide gel electrophoresis (PAGE) analysis and disaccharide compositional analysis using liquid chromatography-mass spectrometry. The competition SPR studies using sized oligosaccharide mixtures showed that binding of each of the tested FGFs or FGF•FGFR complexes to heparin immobilized to an SPR chip were size dependent. The 6-desulfated heparin oligosaccharides showed reduced inhibition of FGF and FGF•FGFR binding to heparin in the competition experiments. Heparin and the 6-desulfated heparin showed higher inhibition to FGF•FGFR complex binding to heparin than to FGF binding to heparin. In the filter trapping experiments, PAGE analysis showed different affinities between the FGF•FGFR complexes and oligosaccharides. Disaccharide analysis showed HS disaccharides degree of polymerization (dp) 10 had high binding selectively, while heparin dp10 and 6-desulfated heparin dp10 showed reduced or no selectivity to the different FGF•FGFR complexes tested.
Heparan sulfate proteoglycans (HSPGs) are essential components of both the extracellular matrix (ECM) as well as the cell surface membrane. Heparan sulfate (HS) is a linear sulfated glycosaminoglycan (GAG), consisting predominantly of repeating disaccharide motif comprised of β-D-glucuronic acid and N-acetyl-α-D-glucosamine residues connected through 1→4 glycosidic linkages. Each disaccharide unit can be differentially substituted with 2-O-sulfo groups in the uronic acid residue and 6-O-, 3-O- and N-sulfo groups in the glucosamine residue (1, 2). Each biosynthetic modification is incomplete, thus, resulting in sequence heterogeneity thought to serve as an important mechanism in the regulation of HS interaction specificity with cellular proteins including various growth and differentiation factors and morphogens, extracellular matrix components, protease inhibitors, protease, lipoprotein lipase, and various pathogens (2, 3). These interactions have been shown to play a pivotal role in various patho-physiological phenomena as well as in tissue morphogenesis. For example, genetic studies in flies and more recently in mice demonstrate that HS are indispensable for proper development (4–6).
The fibroblast growth factor (FGF) family consists of 22 structurally related proteins with a core region of homology of 100–120 residues known as a β–trefoil core, in addition to variable N- and C-terminal regions (7, 8). FGF signaling plays ubiquitous roles throughout the human life cycle (7). In the embryo, FGFs are master regulators of mesenchymal-epithelial communication and thus are required for organogenesis and pattern formation (8). In the adult, FGFs continue to regulate tissue homeostasis but also play important roles in wound healing, tissue repair, cholesterol metabolism and serum phosphate regulation (7). FGFs execute their diverse functions by binding and activating cell surface FGF receptors (FGFRs) that form a subfamily within the receptor tyrosine kinase (RTK) superfamily (9). FGF•FGFR binding specificity/promiscuity is essential for the regulation of FGF signaling and is determined by primary sequence differences among the 18 FGFs (10) and 7 FGFRs (11, 12). Receptor dimerization is a mandatory event in FGF signaling and, in addition to the FGF ligand, requires the presence of the highly sulfated heparin/HS polysaccharide chains of HSPGs. Aberrant FGF signaling is responsible for a wide spectrum of human pathological conditions including skeletal syndromes, olfactory syndromes, phosphate-wasting disorders, reproductive disorders, and cancer (13).
FGF signaling begins with the formation of a ternary complex of FGF, FGFR, and heparan sulfate. Early models suggested that heparin/HS serves primarily as a template for FGF dimerization with two molecules of FGF bound to the heparin helix in either a cis or trans orientation (14). Heparin/HS bind tightly to FGFs having dissociation constants ranging from 100 nM to 10 μM (15). Cellular studies with selectively desulfated heparins show that different types of sulfo groups can be required for promotion of FGF signaling (16–19). FGF1 and FGF2, the most studied members of the family, bind to specific sulfo groups in heparin oligosaccharides (15, 20–21). FGF2 recognizes a heparin/HS pentasaccharide containing an iduronic acid (IdoA) 2-O-sulfo residue (22) with no requirement for 6-O-sulfo groups in its glucosamine (GlcN) residue (20, 23) but requiring larger 6-O-sulfo group containing sequences for signaling (24, 25). FGF1 recognizes a specific octasaccharide (26) containing an internal IdoA2SGlcNS6SIdoA2S (where S is sulfo) trisaccharide motif (22) and also requires 6-O-sulfo groups for signaling (22, 25, 27). Early studies were focused exclusively on the interaction of FGF with heparin/HS. However, biological data clearly established heparin/HS interacts with both growth factor and receptor, thus requiring the study heparin/HS binding to the FGF•FGFR complex, the subject of the current study. In present report, an oligosaccharide library prepared from heparin, 6-desufated heparin and HS were used to analyze heparin/heparan sulfate sequences that interacted with FGF•FGFR complexes by solution competition using surface plasmon resonance (SPR) and filter trapping. Specific oligosaccharides binding to FGF•FGFR complexes were subjected to polyacrylamide gel electrophoresis (PAGE) analysis and disaccharide analysis.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
All FGFRs were refolded and purified from inclusion bodies as previously described (28). The purification procedure for FGF1 (29), FGF8 and FGF17 (30), FGF9 (31), and FGF10 (12) have all been published previously. Full length FGF3 was expressed in pET30a, refolded, and purified by heparin affinity, nickel affinity, and size exclusion chromatography. Full length FGF4 was expressed in pET28a, and the ligand was obtained from inclusion bodies via salt extraction with 2M NaCl, 25mM Hepes pH7.5–10% glycerol. FGF4 was then purified by heparin affinity and size exclusion chromatography. Full length FGF5 and FGF6 were both expressed in pET28a, refolded, and then purified by heparin affinity and size exclusion chromatography. All proteins are of human origin except FGF3, which is from mouse, all proteins are expressed in BL21 DE3 cells, and refolding protocols for all ligands follow that previously described (28).The FGFRs and some of the FGFs were refolded using slow dialysis as follows: Bacterial cells transformed with expression vectors for the D2-D3 fragments of FGFR1c, FGFR2c, and FGFR2b were induced with isopropyl-β-D-thiogalactoside (IPTG) for 5 h, centrifuged, and the bacterial pellet was lysed in 25 mM Hepes buffer (pH 7.5) containing 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), and 10% glycerol using a French press. Following centrifugation, the pellets containing ectodomains were dissolved in 6 M guanidinium hydrochloride and 10 mM dithiothreitol (DTT) in 100 mM tris(hydroxymethyl)aminomethane (Tris)-HCl buffer (pH 8.0). The solubilized ectodomains were refolded by dialysis against 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) or Tris buffer (pH 7.5) containing 150 mM NaCl, 10% glycerol, and 1 mM L-cysteine. The refolded FGFR1 and FGFR2 proteins were purified by heparin Sepharose affinity chromatography followed by size exclusion chromatography on a Superdex 200 (Pharmacia) column equilibrated with 25 mM Tris-HCl buffer (pH 7.5) containing 1.0 M NaCl. To generate the desired complexes purified ectodomains were mixed with different FGFs in 1:1 ratio, and concentrated using Centricon 30 (Amicon). Purified FGFR and purified FGF proteins were mixed in 1:1 molar ratios and then run over a Sephadex 200 size exclusion column in 1M NaCl 25 mM Hepes pH 7.5 to prepare the FGF-FGFR complexes.
Preparation of oligosaccharide libraries
The porcine intestinal heparin, and porcine intestinal heparan sulfate were from Celsus (Celsus Laboratories, Cincinnati, OH). 6-desulfated heparin was prepared by the method with silylating reagent, N-methyl-N-(trimethylsilyl)-trifluoro acetamide (MTSTFA) (19). The oligosaccharide libraries from heparin, 6-desulfated heparin and HS were prepared using enzymatic depolymerization by using the combination heparin lyase I, II and III digestion. Undigested saccharides and enzymes were removed by ultra-filtration with a membrane molecular weight cut-off (MWCO) of 5 KD. The low molecular weight (MW) oligosaccharides (MW< 5000) obtained were fractionated on a Bio-Gel P-6 column. Individual fractions consisting of hexasaccharides, octasaccharides and decasaccharides were collected, desalted and used as oligosaccharide libraries in this study.
Preparation of heparin biochip
Albumin-heparin (Sigma) was covalently immobilized to the sensor surface (Fc2) through its primary amino groups (32). Briefly, the carboxymethyl groups on C1 Chip (GE Healthcare, Uppsala, Sweden) surface was first activated using a injection pulse 10 min (50 μl, with the flow rate 5 μl/min) of an equimolar mix of N-hydroxysuccinimide (NHS) and N-ethyl-N-(dimethyaminopropyl) carbodiimide (EDC) (final concentration is 0.05 M, mixed immediately prior to injection). A solution of albumin-heparin (200 μg/ml in sodium acetate buffer with the addition of 2 M guanidine, pH 4.0), was then applied (20 μl) by manual injection. Excess unreacted sites on the sensor surface were blocked with a 50 μl injection of 1 M ethanolamine. The successful immobilization of albumin-heparin was confirmed by the observation of a ~300 response unit (RU) increase in the sensor chip. To prepare the control flow cell (Fc1), bovine serum albumin was immobilized on the surface with a similar amine coupling procedure. After the surface was activated with NHS/EDS, 5 μl of albumin (20 μg/ml in sodium acetate buffer, pH 4.0) solution was then injected by manual injection to get ~300 RU immobilized.
Solution competition SPR study
Solution/surface competition experiments were performed by SPR (BIAcore 3000, GE Healthcare, Uppsala, Sweden) to examine the effect of saccharide chain size/structure of different heparin /HS on the heparin-FGF/FGFR interaction. Proteins (FGFs, or FGF•FGFR complex, 1000 nM) pre-mixed with certain concentration (2000 nM) of hexasaccharides (dp6), octasaccharides (dp8), decasaccharides (dp10) were injected over heparin chip at a flow rate of 30 μl/min, respectively. For each set of competition experiments on SPR, a control experiment (only protein without added oligosaccharides) was performed to make certain the surface was completely regenerated and that the results obtained between runs were comparable. The response was monitored as a function of time (sensorgram) at 25 °C.
“Fishing” for specific oligosaccharides binding to FGF•FGFR complex from oligosaccharide libraries
To characterize of heparin or HS structure required to bind FGF•FGFR complex, initially three different FGF•FGFR complexes (FGF1•FGFR1, FGF2•FGFR1, and FGF2•FGFR2) were used to bind decasaccharide libraries from heparin, 6-desulfated heparin and HS. FGF•FGFR complexes (225 μg in 75 μl buffer: 25 mM Hepes buffer, with 1M NaCl, pH 7.5) were mixed with 30 μg of each different decasaccharide library in 100 μl buffer (25 mM Hepes, with 150 mM NaCl, pH 7.5) and incubated at room temperature for one hour. The non-binding oligosaccharides were removed from the mixture using ultracentrifugation with nanosep tubes (MWCO 30 KD) and remaining complexes were washed 3-times with buffer. The ternary complexes of FGF•FGFR-oligosaccharide obtained were heated to 100°C to break the complex and then protein was removed from each sample using a centrifugal membrane filter (MWCO 10 KD). The high affinity oligosaccharides were subjected to structural analysis by gradient PAGE and disaccharide compositional analysis. Next, seven additional FGF•FGFR complexes (FGF3•FGFR1c, FGF4•FGFR2c, FGF5•FGFR1c, FGF6•FGFR2c, FGF8b•FGFR2c, FGF10•FGFR2b, and FGF17•FGFR1c) were similarly used to “fish” specific HS structures from HS dp 10 using the same approach.
Structural analysis on the specific oligosaccharides binding to FGF•FGFR complex
PAGE analysis
Polyacrylamide gel electrophoresis (PAGE) was applied to analyze the molecular weight and polydispersity of the oligosaccharides. To each lane ~5 μg oligosaccharide was subjected to electrophoresis against a standard composed of heparin oligosaccharides prepared enzymatically from bovine lung heparin, the gel was visualized with Alcian blue.
Disaccharides compositional analysis using liquid chromatography-mass spectrometry (LC-MS)
A mixture of recombinant heparinase I, II, III (a generous gift from Professor Jian Liu of the University of North Carolina) was added into the ternary complex of FGF•FGFR-oligosaccharide and incubated at 37°C overnight. The products were filtered by the centrifugal filter devices (3 KD MWCO, Millipore), through which the heparin/HS disaccharides were obtained. A set of unsaturated disaccharides standards of heparin/HS (Seikagaku, Japan) including: 0S, ΔUA-GlcNAc (where ΔUA is 4-deoxy-a-L-threo-hex-4-enopyranosyluronic acid, and Ac is acetyl); NS, ΔUA-GlcNS (where S is sulfo); 6S, ΔUA-GlcNAc6S; 2S, ΔUA2S-GlcNAc; 2SNS, ΔUA2S-GlcNS; NS6S, ΔUA-GlcNS6S; 2S6S, ΔUA2S-GlcNAc6S; and triS, ΔUA2S-GlcNS6S were used in the analysis. Solution A and B for high performance liquid chromatography (HPLC) were 15% and 70% acetonitrile respectively, containing the same concentration of 37.5 mM NH4HCO3 and 11.25 mM tributylamine. The pH values of them were adjusted to 6.5 with acetic acid. The flow rate was 10 μl /min. The separation was performed on a C-18 column (Agilent) using solution A for 20 min, followed by a linear gradient from 20 to 45 min of 0% to 50% solution B. The column effluent entered the source of the electrospray ionization (ESI)-MS for continuous detection by MS (Agilent) (33).
RESULTS
Solution competition SPR study
Competitive binding studies between heparin (immobilized on the SPR chip) and soluble sized oligosaccharides, derived from heparin and 6-desulfated heparin, were performed using SPR. FGF or FGF•FGFR complexes (1 μM), with or without bound oligosaccharide, were flowed over the surface of a SPR biochip on which heparin was immobilized (Figure 1 and 2 and Table 1). Different oligosaccharides of defined length (from hexasaccharide (dp6) to decasaccharide (dp10)) were used in the competition study. The results showed that: 1) the disassociation rates of the FGF•FGFR complex injections were much slower than those observed when FGF alone was injected, based on the overall shapes of SPR sensorgrams, demonstrating that the ternary FGF•FGFR•HS complexes are considerably more stable than the binary complexes of FGF•HS; 2) heparin-derived, sized oligosaccharide mixtures inhibit the binding of FGF1, FGF2, and their complexes (FGF1•FGFR1, and FGF2•FGFR1) to immobilized heparin and the inhibition decreased with oligosaccharide size, demonstrating a chain length dependence; 3) 6-desulfated heparin oligosaccharides showed reduced inhibition in the competition experiments, demonstrating the importance of either the 6-O-sulfo groups or overall sulfation level for binding to all of the FGF1 and 2 and their FGF•FGFR complexes; 4) the 6-desulfated heparin was a better inhibitor of FGF•FGFR complex binding to heparin than to FGF binding to heparin, indicating that the 6-O-sulfo group or overall sulfation level was less critical for high affinity binding to FGF•FGFR complex than to FGF, and that the ternary FGF•FGFR•heparin complex is more stable.
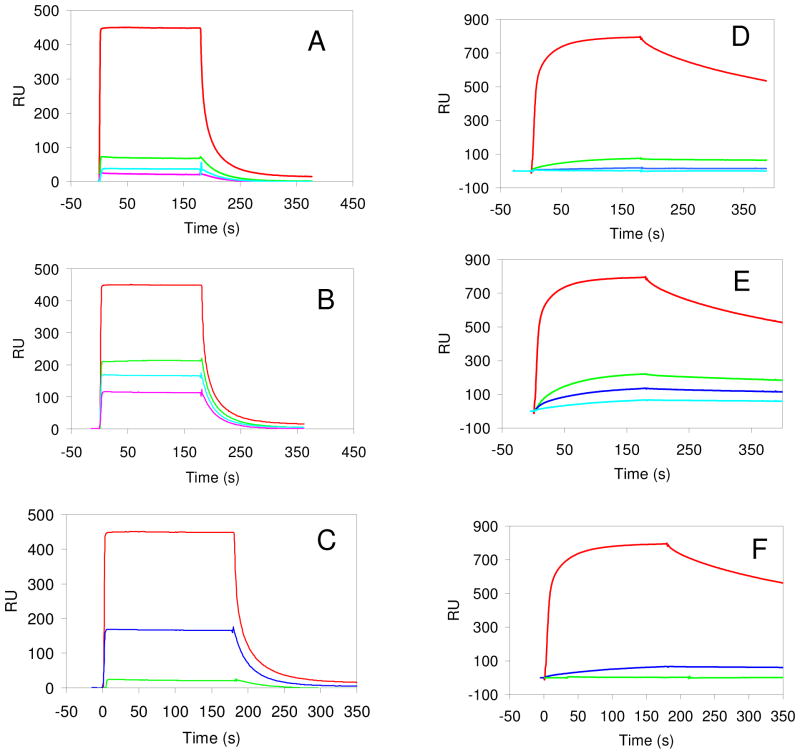
Figure 1.
A. The inhibition of FGF1 binding to immobilized heparin by sized heparin oligosaccharide mixture, control (no oligosaccharides) in red, dp6 in green, dp8 in blue, and dp10 in turquoise. B. The inhibition of FGF1 binding to immobilized heparin by sized 6-desulfated heparin oligo saccharide mixtures, control (no oligosaccharides) in red, dp6 in green, dp8 in blue, and dp10 in turquoise. C. Comparison of the inhibition of FGF1 binding to immobilized heparin by dp10 oligosaccharide mixtures: red, control; blue, heparin 6-desulfated heparin dp10; green: heparin dp10. D. The inhibition of FGF1•FGFR1 binding to immobilized heparin by sized heparin oligosaccharide mixtures, control (no oligosaccharides) in red, dp6 in green, dp8 in blue, and dp10 in turquoise. E. The inhibition of FGF1•FGFR1 binding to immobilized heparin by sized 6-desulfated heparin oligosaccharide mixtures, control (no oligosaccharides) in red, dp6 in green, dp8 in blue, and dp10 in turquoise. F. Comparison of the inhibition of FGF1•FGFR1 binding to immobilized heparin by dp10 oligosaccharide mixtures, red, control; blue, 6-desulfated heparin dp10; green: heparin dp10. The concentrations of FGF1, FGF1•FGFR1 complex, and the oligosaccharide were 1000, 500, and 2000 nM, respectively.
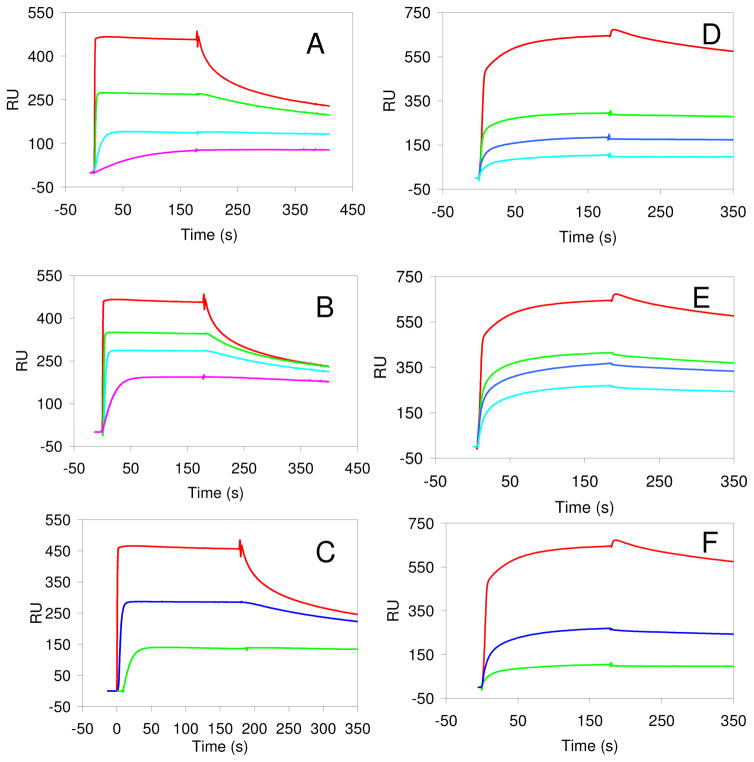
Figure 2.
A. The inhibition of FGF2 binding to immobilized heparin by sized heparin oligosaccharide mixtures, control (no oligosaccharides) in red, dp6 in green, dp8 in blue, and dp10 in turquoise. B. The inhibition of FGF2 binding to immobilized heparin chip by sized 6-desulfated heparin oligosaccharide mixtures, control (no oligosaccharides) in red, dp6 in green, dp8 in blue, and dp10 in turquoise. C. Comparison of the inhibition of FGF2 binding to immobilized heparin by dp10 oligosaccharide mixtures: red, control; blue, 6-desulfated heparin dp10; green: heparin dp10. D. The inhibition of FGF2•FGFR1 binding to immobilized heparin by sized heparin oligosaccharide mixture, control (no oligosaccharides) in red, dp6 in green, dp8 in blue, and dp10 in turquoise. E. The inhibition of FGF2•FGFR1 binding to immobilized heparin by sized 6-desulfated heparin oligosaccharide mixture, control (no oligosaccharides) in red, dp6 in green, dp8 in blue, and dp10 in turquoise. F. Comparison of the inhibition of FGF2•FGFR1 binding to immobilized heparin by dp10 oligosaccharide mixtures: red, control; blue, 6-desulfated heparin dp10; green: heparin dp10. The concentrations of FGF2, FGF2•FGFR1 complex, and the oligosaccharide were 1000, 500, and 2000 nM, respectively.
Table 1.
Summary of the inhibition percentage of dp 10 oligo on FGF or FGF•FGFR complex binding to heparin based on the solution competition SPR
| FGF1 | FGF2 | FGF1•FGFR1 Complex | FGF2•FGFR1 Complex | |
|---|---|---|---|---|
| Heparin dp10 | 95% | 70% | 99.7% | 84% |
| 6-Desulfated heparin dp10 | 63% | 38% | 92% | 58% |
Structural analysis on the specific oligosaccharides binding to FGF•FGFR complexes
In the first set of “fishing” experiments three FGF•FGFR complexes (FGF1•FGFR1, FGF2•FGFR1, FGF2•FGFR1 in 1 M NaCl) were examined for their binding to heparin or HS-derived oligosaccharide mixtures. FGF-FGFRs have poor solubility in low salt buffers and therefore they need to be stored in high (1 M) salt buffers. High salt can weaken interactions of the HS/heparin with FGF-FGFR complex. Thus, in the binding experiments, the salt was diluted when we added the oligosaccharide mixture. After mixing with the oligosaccharide (in 150 mM NaCl), the salt concentration of the FGF-FGFR complex was reduced to ~ 500 mM, which kept the complex soluble while promoting protein-oligosaccharide binding. A molar excess of each of the three sized decasaccharide mixtures (dp 10 from heparin, 6-desulfated heparin and HS) was incubated in HBS buffer with each of the three FGF•FGFR complexes, FGF1•FGFR1, FGF2•FGFR1 and FGF2•FGFR2. The complexes each had a MW ~ 45 KD, while the individual oligosaccharides had a MW < 3.3 KD (calculated for the fully sulfated heparin decasaccharide). The non-binding oligosaccharides were removed from the mixtures using ultracentrifugation (MWCO 30KD). PAGE analysis was used to examine affinity differences between the complex and decasaccharides. All the three complexes showed similar band intensities for the high affinity decasaccharides suggesting that there was either little selectivity or that PAGE was not able to detect subtle differences in oligosaccharide selectivity. The overall band intensity (total staining in each lane) of the interacting HS and 6- desulfated heparin decasaccharides showed a similar pattern indicating the order of affinity (FGF-FGFR complex to HS or 6-desulfated heparin) is FGF1•FGFR1 > FGF2•FGFR1 >FGF2•FGFR2. In contrast the overall band intensity for heparin decasaccharides was similar for all three complexes.
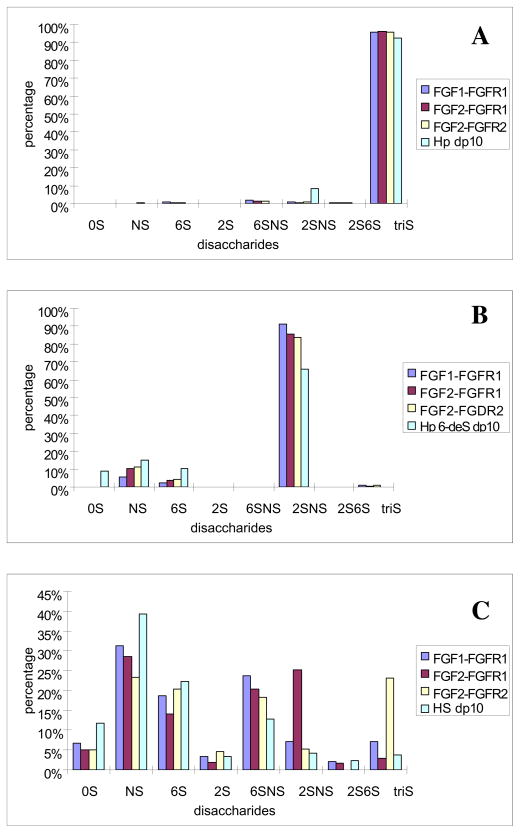
Next, the disaccharide composition of decasaccharides (dp 10) with high affinity for FGF1•FGFR1, FGF2•FGFR1 and FGF2•FGFR2 complexes were determined. The results showed little FGF•FGFR binding selectivity for heparin and 6-desulfated heparin decasaccharides (Figure 4A and 4B). This is undoubtedly due to the highly uniform repeating structures in both heparin, the tri-S disaccharide, and in the 6-desulfated heparin, the 2SNS disaccharide. Therefore, the more highly variable HS decasaccharide mixture was examined. The results (Figure 4C) showed major composition differences in the HS dp10 oligosaccharides binding to the different complexes, suggesting a very high level of selectively (diversity of the disaccharide compositional structures). The HS dp 10 that bound to FGF2•FGFR1 complex, for example, contained substantially more 2SNS disaccharide than did the HS dp 10 that bound to FGF•FGFR1 and FGF2•FGFR2. In addition, the HS dp 10 that bound to FGF2•FGFR2 complex contained substantially more tri-S disaccharide than did the HS dp 10 that bound to FGF1•FGFR1 and FGF2•FGFR1. Binding studies using the FGF1 and FGF2, in the absence of FGFR2, were next examined as a control experiment to ensure that the FGF•FGFR complexes remained intact in the oligosaccharide binding studies. The results (Table 1, Supporting Information) show clear differences between growth factor and complex binding to HS decassacharides. In particular, HS decassacharides binding FGF1 and FGF1 FGFR1 had remarkably different compositions. Moreover, the affinity of FGF1 for decasaccharides rich in TriS and FGF2 for decasaccharides rich NS2S is consistent with literature reports (20, 21, 22, 34).
Figure 4.
Disaccharide compositional determination of decasaccharides binding to FGF1•FGFR1, FGF2•FGFR1 and FGF2•FGFR2 complexes using LC-MS analysis. A: heparin dp 10 to different FGF•FGFR complex, B: heparin 6-desulfated heparin dp 10 to different FGF•FGFR complex, C: HS dp 10 to different FGF•FGFR complexes.
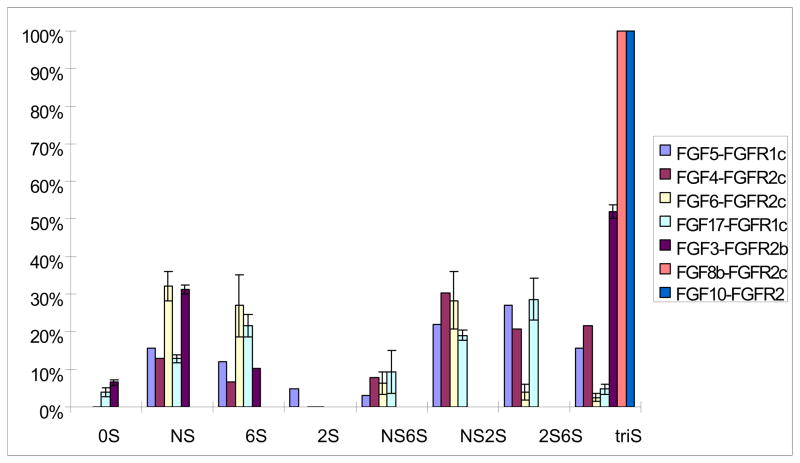
Since the HS dp10 showed highest binding selectivity to the first three FGF•FGFR complexes we examined, seven additional FGF•FGFR complexes (FGF3•FGFR1c, FGF4•FGFR2c, FGF5•FGFR1c, FGF6•FGFR2c, FGF8b•FGFR2c, FGF10•FGFR2b, and FGF17•FGFR1c) were similarly studied using the same approach. Again, the binding decasaccharides obtained were determined by disaccharide compositional analysis. The disaccharide compositional analysis (Figure 5) showed that triS disaccharide was the only interacting structure in with FGF8b•FGFR2c, and FGF10•FGFR2b complexes. This triS disaccharide was the dominant interacting structure with the FGF3•FGFR2b complex, but a small fraction of 0S, NS and 6S disaccharide were also observed. The remaining four complexes (FGF4•FGFR2c, FGF5•FGFR1c, FGF6•FGFR2c, and FGF17•FGFR1c), more diverse composition in their interacting disaccharides (Figure 5) was detected.
Figure 5.
Disaccharides compositional analysis of HS dp10 binding to seven different complexes (FGF3•FGFR1c, FGF4•FGFR2c, FGF5•FGFR1c, FGF6•FGFR2c, FGF8b•FGFR2c, FGF10•FGFR2b, and FGF17•FGFR1c).
DISCUSSION
Information regarding the structural specificity of protein-HS interactions has been afforded by technical improvement in the methods for the structural analysis of HS oligosaccharides, mutational analysis of protein HS-binding sites, molecular modeling, and, recently, crystal or NMR structures of protein-HS complexes (35). Interactions between heparin/HS and proteins have been characterized quantitatively using a number of techniques, including trapping and quantifying HS-protein complexes on surfaces, affinity co electrophoresis, optical biosensors, and isothermal titration calorimetry. The sequences in HS that interact with FGF-1 or FGF-2 have been studied by biochemical and x-ray crystallographic analysis (20, 28, 36). It was concluded from initial studies, that heparin/HS needs to interact with both FGF and FGFR for the signalling (37). In addition to the studies on FGF-1 and FGF-2, the HS sequences that mediate binding and/or activation of some HBGFs have been reported in the systems including FGF-4 (38, 39), FGF-8b (18), hepatocyte growth factor (4, 5, 19), and platelet-derived growth factor (6). These studies on the binding structures in HS appear to support the idea that each heparin-binding growth factor may specifically recognize unique structures in HS. A recent systematic study (18) using sequences modified with specific sulfotransferases show that there were at least five classes of HS octasaccharide recognition sites for FGFs: 1. requiring a 2-O-sulfo group, FGF2; 2. requiring a 6-O-sulfo group, FGF10; 3. requiring a 2-O-sulfo, with a partial requirement for a 6-O-sulfo group, FGF18; 4. requiring both 2-O-sulfo and 6-O-sulfo groups, FGF4 and FGF7; and 5. no binding to an octasaccharides sequence, FGF8. Although the importance of HS in FGF signaling has been well documented over the past decade, the heparin/HS structures involved in the interaction with most FGFs is still largely undetermined. Most important, HS binds to both FGF and FGFR to form a signal transduction complex and structural analysis of HS with binding activity to FGF•FGFR complexes has not been studied. In current study, we provide a new strategy for the study the structural specificity of HS binding to FGF•FGFR complexes rather than FGFs or FGFRs alone.
The competition SPR studies between heparin and sized oligosaccharides (derived from heparin and 6-desulfated heparin) using FGF or FGF•FGFR complex showed that all binding events were size dependent (Figures 1 and 2) consistent with previous reports (14). It is generally held that binding to FGFs required oligosaccharides of tetrasaccharide to hexasaccharide in length, whereas activation required larger oligosaccharide, octasaccharide to decasaccharide in length. In comparison to heparin oligosaccharide, the 6-desulfated heparin oligosaccharide showed lower inhibition in the competition, suggesting 6-desulfated heparin has lower affinity to FGF1 and FGF2 and to FGF1•FGFR1 and FGF2•FGFR1 complexes. In comparison to FGF alone (Table 1), heparin decasaccharide and the 6-desulfated heparin decasaccharide more greatly inhibited FGF•FGFR complex binding to heparin, demonstrating that the affinity to the complex is higher than to the single FGF and the ternary FGF•FGFR-heparin complex is more stable. These studies also clearly show that a 6-O-sulfo group was more important in the interaction of either oligosaccharide or GAG with FGF1 than with FGF2 consistent with literature reports (20, 22, 23, 34); and similarly, a 6-O-sulfo group was more important in the interaction of either oligosaccharide or GAG with FGF1•FGFR1 than with the FGF2•FGFR1 complex. These results also demonstrate that SPR can be utilized in competitive binding studies of FGF•FGFR complexes between GAG and GAG oligosaccharides.
In the “fishing” for specific oligosaccharides, ten FGF•FGFR complexes (FGF1•FGFR1, FGF2•FGFR1, FGF2•FGFR1, FGF3•FGFR1c, FGF4•FGFR2c, FGF5•FGFR1c, FGF6•FGFR2c, FGF8b•FGFR2c, FGF10•FGFR2b, and FGF17•FGFR1c) were used with a filter trapping method. Three sized oligosaccharides (dp 10 from heparin, 6-desulfated heparin and HS) were bound to each of three FGF•FGFR complexes in the first set of filter trapping experiment. PAGE analysis showed affinity differences between the complexes, suggesting the presence of unique high affinity decasaccharides that could be used for the sequencing studies. All the three complexes showed similar overall banding intensities for the tightly interacting heparin decasaccharides, suggesting that they have comparable high affinity to heparin. The overall banding intensity of interacting HS and 6-desulfated heparin decasaccharides suggest HS and 6-desulfated heparin show relative affinity of: FGF1•FGFR1 > FGF2•FGFR1 >FGF2•FGFR2. These studies demonstrate that FGF•FGFR complexes can be used for affinity capture of specific oligosaccharides but suggest that it is necessary to increase the structural diversity of the oligosaccharide mixture: mixture being examined in order to optimize the molar ratio of diverse components, to identify structure for high affinity binding. Since the binding of HS dp10 to different complexes displays very high selectively (diversity of the disaccharide compositional structures) by disaccharide compositional analysis (Figure 4C), the remaining filter trapping experiments examined HS dp10 binding to seven additional FGF•FGFR complexes (FGF3•FGFR1c, FGF4•FGFR2c, FGF5•FGFR1c, FGF6•FGFR2c, FGF8b•FGFR2c, FGF10•FGFR2b, and FGF17•FGFR1c). Disaccharide compositional analysis of interacting HS decasaccharide showed some of complexes (FGF3•FGFR1c, FGF8b•FGFR2c, and FGF10•FGFR2b) binding a similar disaccharide compositional pattern with dominant tri-S structure and the rest of the tested complexes (FGF1•FGFR1, FGF2•FGFR1, FGF2•FGFR2, FGF4•FGFR2c, FGF5•FGFR1c, FGF6•FGFR2c, and FGF17•FGFR1c) binding the decasaccharide having diverse composition of HS disaccharides (Figures 4 and 5). These results suggest that FGF-FGFR complex binds with diverse structures of HS, which depends on abundance of different HS available at the cell surface. These data are in agreement with a recent report (37) demonstrating the ability of HS chains to promote ternary complex formation between FGF and their receptors. These receptors may depend primarily on the abundance, length, and overall sulfation domains and possibly to a lesser degree on the selective saccharide sequence/precise location of sulfo groups. Moreover, FGF•FGFR complexes often select different HS decasaccharide binding partners than the FGF component alone (Table 1, Supporting Information).
In conclusion, SPR and filter trapping techniques were used to investigate FGF•FGFR-heparin/HS interactions and provide important structural information particular from HS decasaccharide libraries. The use of such libraries should facilitate the identification of critical structural features required for a particular interactions and can greatly simplify qualitative and quantitative analysis. The methodology described may be useful in the discovery of novel glycotherapeutics that target disease related protein-HS interactions.
Supplementary Material
Figure 3.
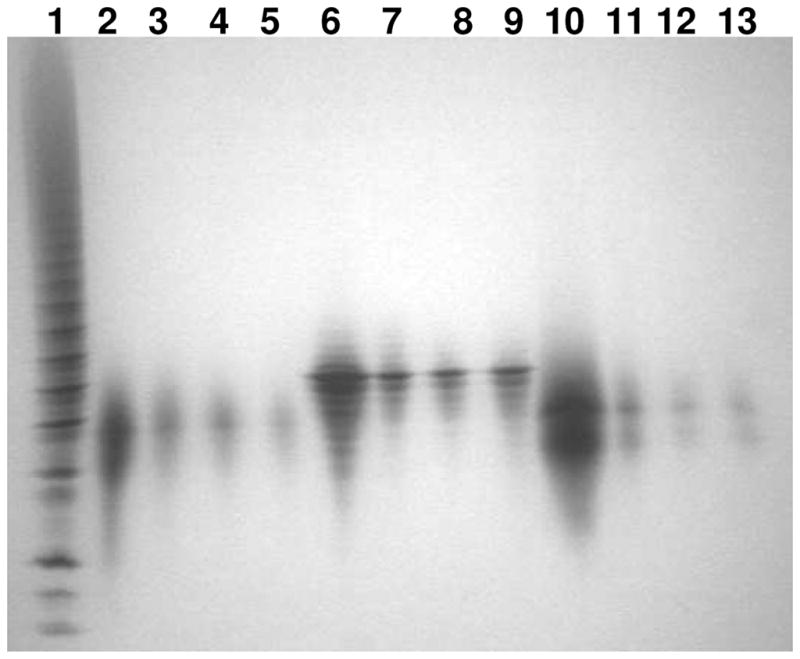
PAGE analysis on oligosaccharide released from FGF•FGFR specific binding: Lane 1, bovine lung heparin oligosaccharide standards; Lane 2: HS dp10; Lane 3 to 5, HS dp10 binding to FGF1•FGFR1, FGF2•FGFR1, and FGF2•FGFR2, respectively; Lane 6, heparin dp10; Lane 7 to 9, heparin dp10 binding to FGF1•FGFR1, FGF2•FGFR1, and FGF2•FGFR2, respectively; Lane 10, 6-desulfated heparin dp10; Lane 11 to 13, 6-desulfated heparin dp10 binding to FGF1•FGFR1, FGF2•FGFR1, and FGF2•FGFR2, respectively.
Acknowledgments
This work is supported by the National Institutes of Health Grants: GM 38060 and HL 62244 (to R.J.L), DE13686 to M.M.
Abbreviations
- HS
heparan sulfate
- PGs
proteoglycans
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptors
- SPR
surface plasmon resonance
- PAGE
polyacrylamide gel electrophoresis
- dp
degree of polymerization
- PG
proteoglycan
- ECM
extracellular matrix
- GAG
glycosaminoglycan
- RTK
receptor tyrosine kinase
- IdoA
iduronic acid
- S
sulfo
- GlcN
glucosamine
- IPTG
isopropyl-β-D-thiogalactoside
- EDTA
ethylenediaminetetraacetic acid
- DTT
dithiothreitol
- Tris
tris(hydroxymethyl)aminomethane
- Hepes
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- MTSTFA
N-methyl-N-(trimethylsilyl)-trifluoro acetamide
- MW
molecular weight
- CO
cut-off
- NHS
N-hydroxysuccinimide
- EDC
N-ethyl-N-(dimethyaminopropyl) carbodiimide
- RU
response unit
- Fc
flow cell
- LC
liquid chromatography
- MS
mass spectrometry
- ΔUA
4-deoxy-α-L-threo-hex-4-enopyranosyluronic acid
- Ac
acetyl
- 0S
ΔUA-GlcNAc
- NS
ΔUA-GlcNS
- 6S
ΔUA-GlcNAc6S
- 2S
ΔUA2S-GlcNAc
- 2SNS
ΔUA2S-GlcNS
- NS6S
ΔUA-GlcNS6S
- 2S6S
ΔUA2S-GlcNAc6S
- triS
ΔUA2S-GlcNS6S
- HPLC
high performance liquid chromatography
- ESI
electrospray ionization
Footnotes
A Table comparing the disaccharide composition of HS dp10 binding to FGF and HS dp10 binding to the related FGF-FGFR complex is provided. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Linhardt RJ, Loganathan D. Heparin, heparinoids and heparin oligosaccharides: structure and biological activities. In: Gebelein G, editor. Biomimetic Polymers. Plenum Press; New York: 1990. pp. 135–173. [Google Scholar]
- 2.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 3.Iozzo RV. Matrix proteoglycans: from molecular design to cellular function. Annu Rev of Biochem. 1998;67:609–652. doi: 10.1146/annurev.biochem.67.1.609. [DOI] [PubMed] [Google Scholar]
- 4.Bullock SL, Fletcher JM, Beddington RS, Wilson VA. Renal agenesis in mice homozygous for a gene trap mutation in the gene encoding heparan sulfate 2-sulfotransferase. Genes Dev. 12:1894–1906. doi: 10.1101/gad.12.12.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merry CL, Bullock SL, Swan DC, Backen AC, Lyon M, Beddington RS, Wilson VA, Gallagher JT. The molecular phenotype of heparan sulfate in the Hs2st−/− mutant mouse. J Biol Chem. 2001;276:35429–35434. doi: 10.1074/jbc.M100379200. [DOI] [PubMed] [Google Scholar]
- 6.Kamimura K, Fujise M, Villa F, Izumi S, Habuchi H, Kimata K, Nakato H. Drosophila heparan sulfate 6-O-sulfotransferase (dHS6ST) gene. Structure, expression, and function in the formation of the tracheal system. J Biol Chem. 2001;276:17014–21. doi: 10.1074/jbc.M011354200. [DOI] [PubMed] [Google Scholar]
- 7.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2:1–12. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ornitz DM. FGFs, heparan sulfate and FGFRs: complex interactions essential for development. BioEssays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 9.Bennasroune A, Gardin A, Aunis D, Cremel G, Hubert P. Tyrosine kinase receptors as attractive targets of cancer therapy. Crit Rev Oncol Hematol. 2004;50:23–38. doi: 10.1016/j.critrevonc.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Olsen SK, Garbi M, Zampieri N, Eliseenkova AV, Ornitz DM, Goldfarb M, Mohammadi M. Fibroblast growth factor (FGF) homologous factors share structural but not functional homology with FGFs. J Biol Chem. 2003;278:34226–34236. doi: 10.1074/jbc.M303183200. [DOI] [PubMed] [Google Scholar]
- 11.Jaye M, Schlessinger J, Dionne CA. Fibroblast growth factor receptor tyrosine kinases: molecular analysis and signal transductiom. Biochim Biophys Acta. 1992;1135:185–199. doi: 10.1016/0167-4889(92)90136-y. [DOI] [PubMed] [Google Scholar]
- 12.Yeh BK, Igarashi M, Eliseenkova AV, Plotnikov AN, Sher I, Ron D, Aaronson SA, Mohammadi M. Structural basis by which alternative splicing confers specificity in fibroblast growth factor receptors. Proc Natl Acad Sci USA. 2003;100:2266–2271. doi: 10.1073/pnas.0436500100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Wu ZL, Zhang L, Yabe T, Kuberan B, Beeler DL, Love A, Rosenberg RD. The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. J Biol Chem. 2003;278:17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- 15.Tumova S, Woods A, Couchman JR. Heparan sulfate proteoglycans on the cell surface: versatile coordinators of cellular functions. Int J Biochem Cell Biol. 2000;32:269–288. doi: 10.1016/s1357-2725(99)00116-8. [DOI] [PubMed] [Google Scholar]
- 16.Ostrovsky O, Berman B, Gallagher J, Mulloy B, Fernig DG, Delehedde M, Ron D. Differential effects of heparin saccharides on the formation of specific fibroblast growth factor (FGF) and FGF receptor complexes. J Biol Chem. 2002;277:2444–2453. doi: 10.1074/jbc.M108540200. [DOI] [PubMed] [Google Scholar]
- 17.Lundin L, Larsson H, Kreuger J, Kanda S, Lindahl U, Salmivirta M, Claesson-Welsh L. Selectively desulfated heparin inhibits fibroblast growth factor-induced mitogenicity and angiogenesis. J Biol Chem. 2000;275:24653–24660. doi: 10.1074/jbc.M908930199. [DOI] [PubMed] [Google Scholar]
- 18.Ashikara-Hada S, Habuchi H, Kariya Y, Itoh N, Reddi AH, Kimata K. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharides library. J Biol Chem. 2004;279:12346–12354. doi: 10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- 19.Kariya Y, Kyogashima M, Suzuki K, Isomura T, Sakamoto T, Horie K, Ishihara M, Takano R, Kamei K, Hara S. Preparation of completely 6-O-desulfated heparin and its ability to enhance activity of basic fibroblast growth factor. J Biol Chem. 2000;275:25949–25958. doi: 10.1074/jbc.M004140200. [DOI] [PubMed] [Google Scholar]
- 20.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. Heparin structure and interactions with basic fibroblast growth factor. Science. 1996;271:1116–1120. doi: 10.1126/science.271.5252.1116. [DOI] [PubMed] [Google Scholar]
- 21.DiGabriele AD, Lax I, Chen DI, Svahn CM, Jaye M, Schlessinger J, Hendrickson WA. Structure of a heparin-linked biologically active dimer of fibroblast growth factor. Nature. 1998;393:812–817. doi: 10.1038/31741. [DOI] [PubMed] [Google Scholar]
- 22.Kreuger J, Salmivirta M, Sturiale L, Giménez-Gallego G, Lindahl U. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J Biol Chem. 2001;276:30744–30752. doi: 10.1074/jbc.M102628200. [DOI] [PubMed] [Google Scholar]
- 23.Walker A, Turnbull JE, Gallagher JT. Specific heparan sulfate saccharides mediate the activity of basic fibroblast growth factor. J Biol Chem. 1994;269:931–935. [PubMed] [Google Scholar]
- 24.Lyon M, Gallagher JT. Bio-specific sequences and domains in heparan sulphate and the regulation of cell growth and adhesion. Matrix Biol. 1998;17:485–493. doi: 10.1016/s0945-053x(98)90096-8. [DOI] [PubMed] [Google Scholar]
- 25.Guimond S, Maccarana M, Olwin BB, Lindahl U, Rapraeger AC. Activating and inhibitory heparin sequences for FGF2 (basic FGF). Distinct requirements for FGF1, FGF2, and FGF4. J Biol Chem. 1993;268:23906–23914. [PubMed] [Google Scholar]
- 26.Fromm JR, Hileman RE, Weiler JM, Linhardt RJ. Interaction of fibroblast growth factor-1 and related peptides with heparan sulfate and its oligosaccharides. Arch Biochem Biophys. 1997;346:252262. doi: 10.1006/abbi.1997.0299. [DOI] [PubMed] [Google Scholar]
- 27.Ishihara M. Structural requirements in heparin for binding and activation of FGF1 and FGF4 are different from that for FGF2. Glycobiology. 1994;4:817–824. doi: 10.1093/glycob/4.6.817. [DOI] [PubMed] [Google Scholar]
- 28.Plotnikov AN, Hubbard SR, Schlessinger J, Mohammadi M. Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell. 2000;101:413–24. doi: 10.1016/s0092-8674(00)80851-x. [DOI] [PubMed] [Google Scholar]
- 29.Olsen SK, Ibrahimi OA, Raucci A, Zhang F, Eliseenkova AV, Yayon A, Basilico C, Linhardt RJ, Schlessinger J, Mohammadi M. Insights into the molecular basis for fibroblast growth factor receptor autoinhibition and ligand-binding promiscuity. Proc Natl Acad Sci U S A. 2004;101:935–40. doi: 10.1073/pnas.0307287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, Mohammadi M. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006;20:185–98. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plotnikov AN, Eliseenkova AV, Ibrahimi OA, Shriver Z, Sasisekharan R, Lemmon MA, Mohammadi M. Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition. J Biol Chem. 2001;276:4322–4329. doi: 10.1074/jbc.M006502200. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F, Fath M, Marks R, Linhardt RJ. A highly stable covalent conjugated heparin biochip for heparin-protein interactions studies. Anal Biochem. 2004;304:271–273. doi: 10.1006/abio.2002.5617. [DOI] [PubMed] [Google Scholar]
- 33.Thanawiroon C, Rice KG, Toida T, Linhardt RJ. LC/MS sequencing of highly sulfated heparin-derived oligosaccharides. J Biol Chem. 2004;279:2608–2615. doi: 10.1074/jbc.M304772200. [DOI] [PubMed] [Google Scholar]
- 34.Guglier S, Hricovíni M, Raman R, Polito L, Torri G, Casu B, Sasisekharan R, Guerrini M. Minimum FGF2 binding structural requirements of heparin and heparan sulfate oligosaccharides as determined by NMR spectroscopy. Biochemistry. 2008;47:13862–13869. doi: 10.1021/bi801007p. [DOI] [PubMed] [Google Scholar]
- 35.Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- 36.Ibrahimi OA, Eliseenkova AV, Plotnikov AN, Yu K, Ornitz DM, Mohammadi M. Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome. Proc Natl Acad Sci U S A. 2001;98:7182–7187. doi: 10.1073/pnas.121183798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jastrebova N, Vanwildemeersch M, Rapraeger AC, Giménez-Gallego G, Lindahl U, Spillmann D. Heparan Sulfate-related Oligosaccharides in Ternary Complex Formation with Fibroblast Growth Factors 1 and 2 and Their Receptors. J Biol Chem. 2006;281:26884–26892. doi: 10.1074/jbc.M600806200. [DOI] [PubMed] [Google Scholar]
- 38.Powell AK, Fernig DG, Turnbull JE. Fibroblast growth factor receptors 1 and 2 interact differently with heparin/heparan sulfate. J Biol Chem. 2002;277:28554–28563. doi: 10.1074/jbc.M111754200. [DOI] [PubMed] [Google Scholar]
- 39.Woods A, Couchman JR. Heparan sulfate proteoglycans and signaling in cell adhesion. Adv Exp Med Biol. 1992;313:87–96. doi: 10.1007/978-1-4899-2444-5_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.