Abstract
Here we show that a recent computationally designed zinc-mediated protein interface is serendipitously capable of catalyzing carboxyester and phosphoester hydrolysis. Although the original motivation was to design a de novo zinc-mediated protein-protein interaction (called MID1-zinc), we observed in the homodimer crystal structure a small cleft and open zinc coordination site. We investigated if the cleft and zinc site at the designed interface were sufficient to form a primitive active site that can perform hydrolysis. MID1-zinc hydrolyzes 4-nitrophenyl acetate (4NPA) with a rate acceleration of 105 and a kcat/KM of 630 M−1s−1, and 4-nitrophenyl phosphate (4NPP) with a rate acceleration of 104 and a kcat/KM of 14 M−1s−1. These rate accelerations by an unoptimized active site highlight the catalytic power of zinc and suggests that the clefts formed by protein-protein interactions are well-suited for creating enzyme active sites. This discovery has implications for protein evolution and engineering: from an evolutionary perspective, 3-coordinated zinc at a homodimer interface cleft represents a simple evolutionary path to nascent enzymatic activity; from a protein engineering perspective, future efforts in de novo design of enzyme active sites may benefit from exploring clefts at protein interfaces for active site placement.
Keywords: de novo, enzyme design, enzyme evolution, artificial enzyme, metalloenzyme, esterase, hydrolysis, primitive active site, computational protein interface design, protein-protein interaction, cleft
De novo protein design provides a rigorous approach for discovering the minimal determinants of protein structure and function, and helps to shape our understanding of protein evolution. Design studies can be particularly informative when the designed proteins exhibit behavior that was not explicitly encoded during the design process. For instance, most protein design algorithms do not consider folding kinetics when designing sequences, and yet many designed proteins fold with rates comparable to naturally occurring proteins (1). These results suggest that stabilizing the native state is sufficient for achieving fast folding rates, and that folding kinetics of small proteins are not a strong constraint on the sequence space sampled by evolution. In this paper, we report catalytic activity by a de novo designed zinc-mediated homodimer, even though we did not explicitly design the protein to be a catalyst. This serendipitous result highlights the intrinsic catalytic power of zinc and provides further evidence that protein-protein interfaces are fertile ground for placing active sites.
Enzyme active sites commonly occur at protein-protein interfaces: triosephosphate isomerase, thymidylate synthase, and tyrosine aminotransferase are a few examples (2). Approximately one-sixth of oligomeric enzymes are estimated to have active sites at the subunit interface (3). Generic protein-protein interactions feature clefts at the interface periphery (Figure 1A), and cavities on average are twice as common and twice as large at a protein interface compared to a monomer surface (4). Thus, protein dimerization is a high-probability evolutionary route to a cleft for a new active site (5). Despite the frequent occurrence of interface active sites in nature, most efforts in rational enzyme design have focused on placing the catalytic and substrate-binding sidechains within an existing monomeric protein scaffold (6–11). A small set of protein folds have been used for most design studies – the TIM barrel, periplasmic binding protein, lipocalin, jelly roll, and beta propeller (7) – but with the goal of using protein design to catalyze a large breadth of reactions, the diversity of scaffolds for active site design should be expanded. Native and de novo protein interfaces may provide one route for expanding the set of scaffolds for creating new active sites.
Figure 1.
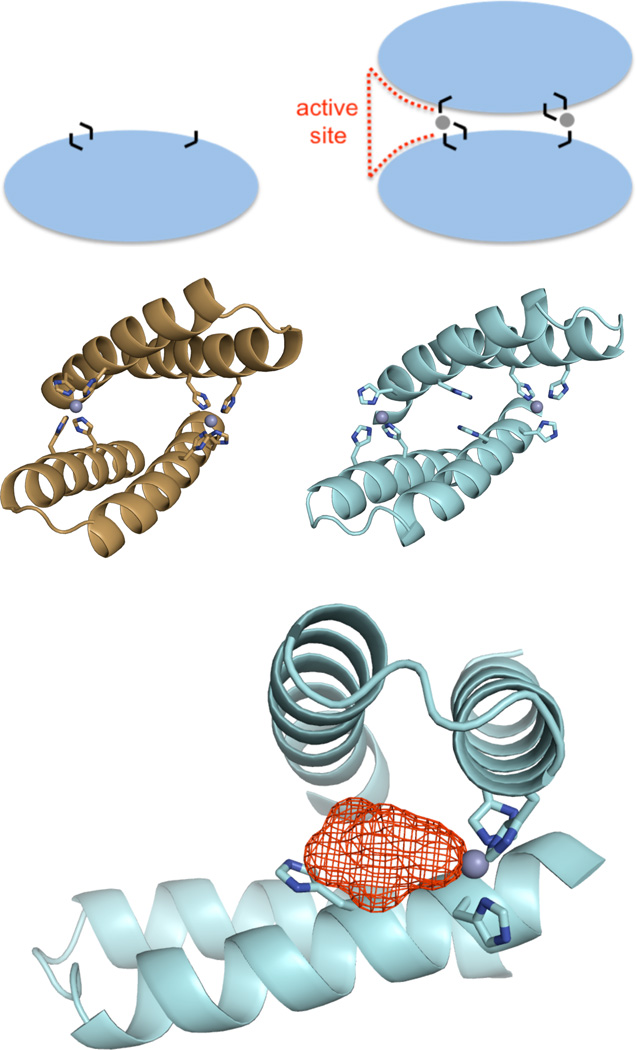
A metal-mediated protein interface as a minimalist route to a new active site. A) A zinc-mediated protein homodimer is a simple recipe for an active site: dimer interfaces naturally contain peripheral clefts (red dashes), metal binding can promote protein interactions, and metal is an effective catalytic motif. Black lines represent histidines, and gray spheres represent zinc ions. B) The computational predictive model of MID1-zinc. Design of MID1-zinc was structurally motivated, the goal was to engineer a de novo zinc-mediated protein-protein interaction. C) Crystallographyically, we observed only three of the four histidines actually participated in zinc coordination. D) Because we observed a cleft (red mesh) and an open zinc coordination site at the MID1-zinc interface, we investigated MID1-zinc as a primitive enzyme.
Enzyme active sites commonly contain metal ions (estimated 40%), covering all six classes of enzymes: oxidoreductases, transferases, hydrolases, lyases, isomerases, ligases (12). Most artificial metalloenzymes are oxidoreductases (13–19), but hydrolases have been recently engineered (10, 20). In addition to promoting catalysis, protein-protein interactions can also be strengthened by metal ions. For example, the Tezcan group showed that a monomer could be converted to a low-affinity tetramer by adding histidines to coordinate zinc at points of contact in the monomer crystal lattice (21). Additionally, we recently designed a homodimeric protein interface in which zinc improves the homodimer binding affinity by >100-fold (4 µM to <30 nM) (22).
Combining these observations, metal-mediated protein interfaces should be an effective recipe for natural evolution and de novo engineering of enzyme active sites (Figure 1A). The formation of clefts at protein interfaces, the intrinsic catalytic power of metals, and the ability of metals to promote protein interactions, formulate a hypothesis that mutations favoring metal binding and/or protein dimerization have a high probability for gain of catalytic function. Thus, we hypothesize that a metal-mediated protein interface has a high probability of catalytic function.
In this work, we investigate this hypothesis by examining the catalytic properties of a protein that we previously engineered to form a zinc-mediated homodimer (named MID1-zinc, for metal interface design with zinc) (22). The starting monomeric scaffold for MID1-zinc was a 5 kDa helical hairpin, and the computational model for the zinc-mediated homodimer featured two zinc-binding sites at the protein interface, each with four tetrahedrally-arranged histidines and no second-shell interactions (Figure 1B). The crystal structure of MID1-zinc (PDB code 3V1C) shows two sites of tetrahedral zinc coordination at the dimer interface, both zinc ions ligated by three histidine residues instead of four (Figure 1C). The fourth coordination site is occupied by a tartrate molecule from the crystallization buffer (Figure S1). The co-crystallization of tartrate with the protein revealed a small molecule binding pocket and a Zn(His)3O zinc binding site, a common catalytic motif. Thus, co-crystallization of tartrate suggested to us that the de novo protein interface might display esterase activity.
We investigated the ability of MID1-zinc to catalyze the hydrolysis of 4-nitrophenyl acetate (4NPA) and 4-nitrophenyl phosphate (4NPP). Zinc enzymes often catalyze hydrolysis (12), and the small cavity in MID1-zinc (~6 Å wide and ~4 Å deep) might provide a serviceable binding pocket for these substrates. Furthermore, 4NPA has been frequently employed in catalytic studies involving artificial enzymes and therefore allows for direct rate comparisons (20, 23–30). Esterase activity was observed and characterized, and here we report the serendipitous discovery of a de novo active site at a zinc-mediated protein interface that catalyzes carboxyester and phosphoester hydrolysis. The serendipitous nature of this de novo active site is a compelling illustration of evolutionary plausibility, as evolution relies on selection of happenstance mutations. In addition to providing a plausible path of natural enzyme evolution, high probability of catalysis by a metal-mediated interface is an attractive feature for future efforts in rational enzyme engineering.
MATERIALS AND METHODS
Cloning and purification of the MID1-zinc enzyme
The computational design of MID1-zinc was derived from the Rab4-binding domain of rabenosyn (PDB code 1YZM), a 46-residue helix-turn-helix. The gene for MID1-zinc was ordered from GenScript, USA with an N-terminal BamHI restriction site, a C-terminal stop codon, a C-terminal SalI restriction site, and codon-optimized for expression in E. coli. The sequence of MID1-zinc is: GSPLAQQIKNIHSFIHQAKAAGRMDEVRTLQENLHQLMHEYFQQSD.
The expression vector (pQE-H6MBP) was derived from the pQE-80L vector, supplemented with an N-terminal 6x-His tag and an MBP fusion with a TEV protease cleavage site. Insertion of the gene into the pQE-H6MBP vector was confirmed by DNA sequencing analysis. BL21(DE3) pLysS cells were transformed with the plasmid for gene expression. Cells were grown to OD600 = 0.6–0.8 (37 °C, LB broth, 67 mg/L ampicillin) and expression was induced with 0.3 mM IPTG and proceeded at 18 °C for 16 hours. Cells were pelleted by centrifugation at 4000 rpm for 20 minutes (Sorvall RC-3B series). Cell pellets were resuspended in lysis buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 10% glycerol, 0.5 mM DTT, 0.5 mM PMSF, and 1 mM benzamidine). Cells were lysed by sonication, and the lysate was treated with 2 units of RNase and DNase. Centrifugation at 15 000 rpm for 20 minutes (Sorvall RC-5B Plus series) cleared the lysate, which was then subjected to immobilized-metal affinity chromatography (IMAC) using a Ni-NTA HisTrap HP column (GE Healthcare). The His-column loading buffer contained 20 mM Tris, pH 8.0, 100 mM NaCl, 25 mM imidazole, and His-column elution buffer was similar except for having 500 mM imidazole. DTT and EDTA (1 mM) were added to the eluted protein, and TEV proteolysis (0.05 mg/ml TEV) occurred overnight at 4°C. The cleaved protein was concentrated for size exclusion chromatography on a Superdex-75 column (GE Healthcare, HiLoad 16/60 prep grade). Appropriate fractions were combined and concentrated (Amicon Ultra, Millipore). The column buffer contained 40 mM HEPES pH 7.5 and 50 mM NaCl, and the enzyme was stored in this buffer at 4°C. Purity near 100% was estimated using SDS-PAGE, and protein concentrations were estimated by absorbance at 280 nm using the theoretical molar extinction coefficient 2980 M−1cm−1 for the dimer. A 277 µM aqueous stock solution of MID1 was prepared containing 50 mM NaCl and 40 mM HEPES buffer at pH 7.5, and zinc sulfate was added in equimolar concentration to generate MID1-zinc for use in kinetic experiments.
Kinetics of 4NPA hydrolysis
Kinetic experiments were initiated by the addition of an aliquot of 4NPA (100 mM in CH3CN) to an aqueous solution containing 50 mM NaCl, 40 mM buffer and 2.5 µM MID1-zinc in a standard 1 cm pathlength quartz cuvette at 25°C. The buffer system for reactions run from pH 7 to 9 was HEPES, whereas potassium hydrogen carbonate was used for reactions at higher pH values. 4-nitrophenyl acetate (4NPA) hydrolysis was monitored by UV-Vis spectrophotometry at 400 nm for the production of 4-nitrophenoxide. Initial rates were determined from linear fits of the absorbance versus time data (<5% conversion) corrected for the rate of uncatalyzed hydrolysis under otherwise identical conditions. Observed rate constants were calculated from Equation 1 where the extinction coefficient of 4-nitrophenoxide was corrected for incomplete ionization under the pH conditions where appropriate. Similar methods were used to measure the kinetics of MID1-zinc catalyzed hydrolysis of 4-nitrophenyl phosphate (4NPP). pH-dependent profiles of catalytic activity for 4NPA and 4NPP hydrolysis were fit to Equation 2, giving values for a pH-independent kmax and a pKa.
| (Equation 1) |
| (Equation 2) |
RESULTS
The kinetic parameters of MID1-zinc hydrolysis of 4NPA were determined spectrophotometrically by the method of initial rates (Equations 1 and S1) following 4-nitrophenoxide production at 400 nm (Figure S2). Notably, MID1-zinc is capable of multiple turnovers as indicated by 4-nitrophenoxide production to a turnover number >50 (Figure S2). Control experiments demonstrate a lack of observable buffer effects throughout this series of experiments. Shown in Figure 2 is a plot of kobs versus [4NPA] determined at 2.5 µM MID1-zinc dimer, which is fully formed at this concentration (Figures S3 and S4). A fit of the data at pH 8.5 to the Michaelis-Menten equation yielded the kinetic parameters kcat = 0.22 s−1 and KM = 0.47 mM.
Figure 2.
Michaelis-Menten kinetics of MID1-zinc hydrolysis of 4NPA. A fit of kobs versus [4NPA] to a standard binding equation indicates a kcat of 0.22 ± 0.01 s−1 and a KM of 0.47 ± 0.07 mM. Rates were determined at 25°C in the presence of 2.5 µM MID1-zinc buffered with 40 mM HEPES pH 8.5 and 50 mM NaCl.
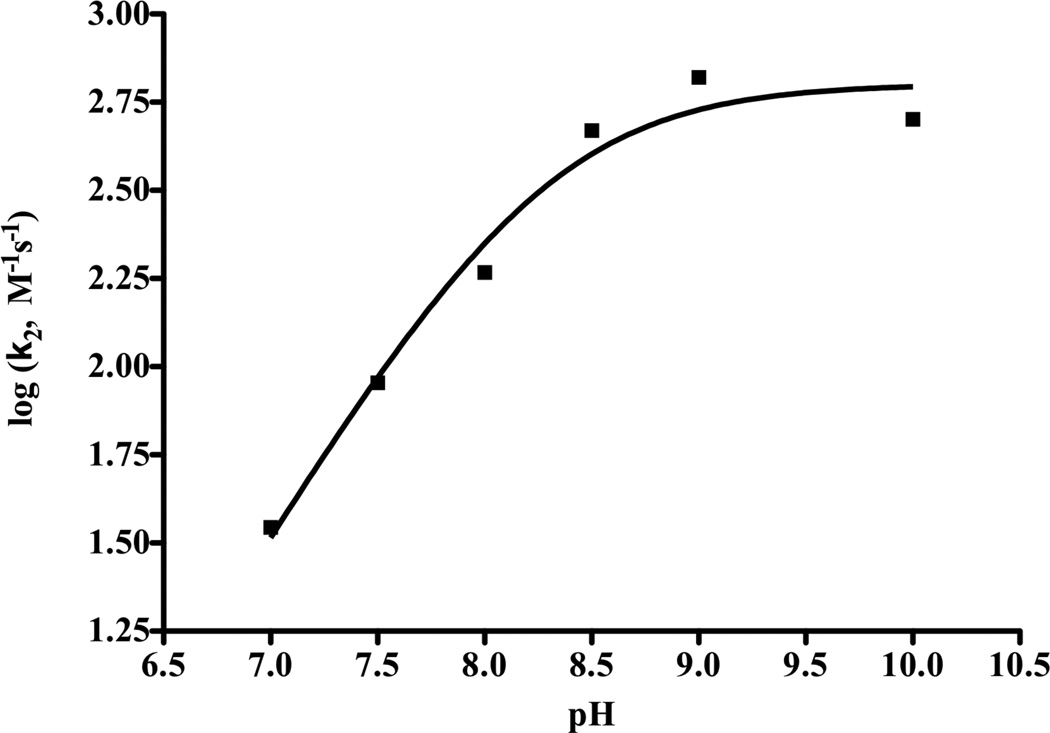
The MID1-zinc catalyzed hydrolysis of 4NPA was investigated as a function of pH. The plots of kobs versus [4NPA] determined from pH 7 – 9 showed saturation binding. At each pH, the first-order rates (s−1) were computed by converting units of Abs/second to concentration/second, then dividing by enzyme concentration (2.5 µM). Catalytic rate constants are compared to the observed uncatalyzed rate constants (s−1) in identical buffer conditions, giving values of kcat/kbuffer of up to 104 (Table 1). Also at each pH, the second-order rate constants were calculated as k2 = kcat/KM. Reactions run at pH > 9 did not show evidence of saturation, and accordingly, the k2 values for these reactions were determined as the gradients of the plots of kobs versus [4NPA]. Shown in Figure 3 is the plot of log(k2) versus pH, which when fit to Equation 2 provides a kinetic pKa of 8.2 and a maximum rate constant of kmax = 630 M−1s−1 along the high pH plateau. The high pH plateau represents the pH-independent regime, where the catalytic species (MID1-zinc-OH) is fully formed. To calculate rate acceleration (31), pH-independent rate was compared to the kneutral rate (neither acid nor base catalyzed) of 4NPA hydrolysis, 4.3 × 10−7 s−1 (32). Thus, we calculate a rate acceleration of 7 × 105 for 4NPA hydrolysis.
Table 1.
Parameters of MID1-zinc hydrolysis of 4NPA
| pH | KM (mM) | kcat (s−1) | k2 (M−1s−1) | kcat/kbuffer1 |
|---|---|---|---|---|
| 7.0 | 1.18 ± 0.10 | 0.042 ± 0.001 | 35 | 1.2×104 |
| 7.5 | 0.90 ± 0.10 | 0.081 ± 0.003 | 90 | 1.1×104 |
| 8.0 | 0.82 ± 0.16 | 0.15 ± 0.009 | 190 | 9.3×103 |
| 8.5 | 0.47 ± 0.07 | 0.22 ± 0.009 | 470 | 5.4×103 |
| 9.0 | 0.42 ± 0.10 | 0.28 ± 0.017 | 660 | 2.6×103 |
| 10.0 | no data | no data | 500 | no data |
The catalyzed rate of 4NPA hydrolysis (kcat) divided by the observed rate under identical conditions in the absence of catalyst (kbuffer). The rate acceleration (7 × 105) was determined using kcat at the high pH plateau (0.3 s−1) and the pH-neutral uncatalyzed rate constant (kneutral) at 25°C (4.3 × 10−7 s−1) (32).
Figure 3.
Enzymatic hydrolysis of 4NPA by MID1-zinc becomes pH-independent at high pH. NLLSQ fitting of log(k2) versus pH gives a pKa of 8.2 ± 0.1 and a kmax of 630 ± 90 M−1s−1.
A number of control experiments were performed to support our assertion that MID1-zinc is responsible for the observed catalysis. First, MID1-zinc was subjected to size-exclusion chromatography and the collected fractions were tested for catalytic activity against 4NPA (Figure S5). The catalytic activity of the collected fractions co-eluted with a UV-visible peak at 280 nm corresponding to the expected molecular weight for MID1-zinc (11 kDa), supporting that MID1 is the catalytic entity.
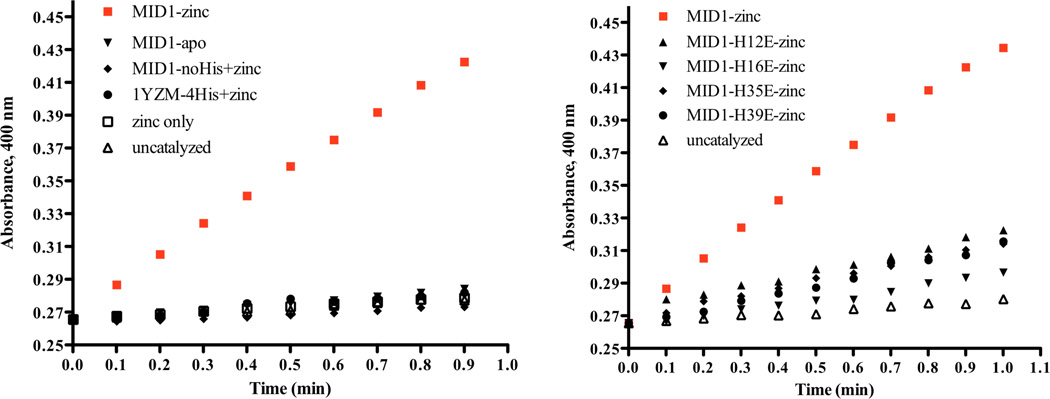
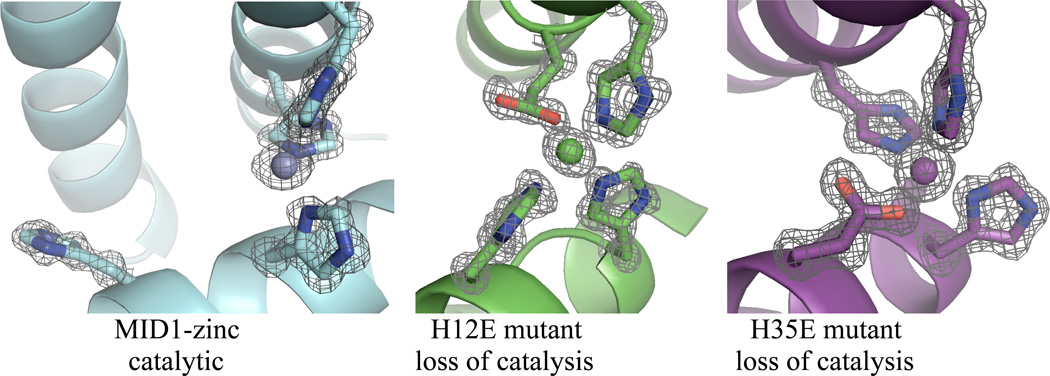
Secondly, to confirm that the zinc site at the designed protein interface was responsible for 4NPA hydrolysis rather than another unforeseen mechanism, several MID1 mutants were prepared and tested for catalytic activity (Figure 4). We tested MID1 in the absence of zinc (MID1-apo), a version of MID1 where each histidine was mutated to alanine (MID1-noHis), the wild-type scaffold with each MID1 histidine included (1YZM-4His), and free zinc. None of these variants showed activity significantly above background levels, indicating that the interface pocket and zinc-binding site are both required for catalysis (Figure 4a). Furthermore, four different single point mutants (H12E, H16E, H35E, H39E) were hypothesized to either complete four-ligand coordination of zinc or inactivate zinc by a zinc-carboxylate interaction. By solving crystal structures, we observed that the open coordination site and cleft in MID1-zinc (Figure 5a) is closed by the H12E mutation (Figure 5b) as well as the H35E mutation (Figure 5c) (22). Loss of activity in each of these histidine-to-glutamate mutants (Figure 4b) structurally supports the proposed structural mechanism of a protein interface cleft containing 3-histidine-coordinated zinc with an open zinc coordination site. Lastly, our data suggests that the MID1-zinc homodimer, which is slightly asymmetric (22), has only one active site: only one tartrate molecule was observed in the crystal structure (Figure S1), dimer is fully formed at 5 µM protein and 2.5 µM zinc (Figure S3a), and rates of catalysis do not differ when zinc is present in 1:2 molar ratio instead of 1:1 molar ratio (Figure S6).
Figure 4.
Enzymatic hydrolysis of 4NPA by MID1-zinc requires three-histidine-coordinated zinc with an open zinc coordination site. Shown are plots of Abs400 nm versus time for the hydrolysis of 1 mM 4NPA at pH 8.0 and 25°C in the presence of 2.5 µM of the indicated protein. MID1-zinc catalyses 4NPA hydrolysis (red squares), while the following variants are not catalytic: a) MID1-apo (without metal), MID1 with all four histidines mutated to alanine (MID1-noHis) with zinc, the wild-type scaffold (1YZM) with the four histidine mutations (1YZM-4His) with zinc, and free zinc. b) Four histidine-to-glutamate point mutants lead to significant decrease in activity. Note: relative slopes of catalyzed v. uncatalyzed timecourses do not indicate the magnitude of rate acceleration because catalyzed hydrolysis must be divided by catalyst concentration for comparison of kcat to kuncat (Equation S1). Thus, the reported 10,000-fold increase in rate is not apparent in the slopes of these raw data plots.
Figure 5.
Crystallographic evidence for the catalytic mechanism. A) The MID1-zinc crystal structure (PDB code 3V1C) reveals a cleft and open zinc coordination site. B) The H12E mutation (PDB code 3V1E) and C) the H35E point mutation (PDB code 3V1F) close the cleft and complete the four-coordination of zinc, and these mutants demonstrate loss of catalytic activity.
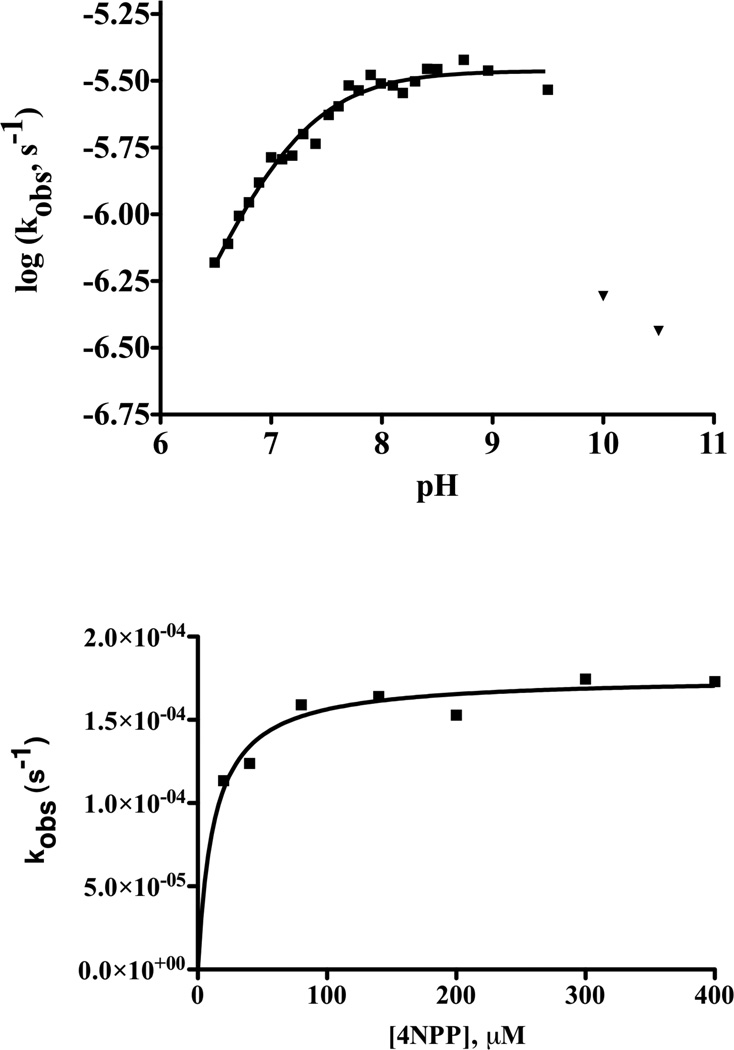
In addition to 4NPA, MID1-zinc also catalyses the hydrolysis of 4-nitrophenyl phosphate (4NPP). 4NPP is intrinsically less reactive than 4NPA by several orders of magnitude, and reactions catalyzed by MID1-zinc were performed to measure phosphoester hydrolysis. The rate increases from pH 6.5 to 8.0 but becomes pH-independent from pH 8.0 to 9.5 (Figure 6a). At pH 10.0 and higher, the catalyst is degraded as observed by a very steep drop-off in rate. The kinetic parameters of MID1-zinc for 4NPP hydrolysis were determined as the following: kcat = 2 × 10−4 s−1, KM = 12 µM, and kcat/KM = 14 M−1s−1, rate acceleration = 1 × 104 (Figure 6b). The 4NPP KM is 50-times lower than 4NPA, likely due to a favorable electrostatic interaction between the negatively-charged phophspate group in 4NPP and the positively-charged active site zinc.
Figure 6.
MID1-zinc catalyses 4NPP hydrolysis. a) pH rate profile for the MID1-zinc catalyzed hydrolysis of 4-nitrophenyl phosphate determined by monitoring product formation at 400 nm. The data from pH 6.5 to 9.5 (squares) were fit to Equation 2, giving a kcat = (3.5 ± 0.09) × 10−6 s−1 and a pKa = 7.1 ± 0.03. The data obtained at pH > 9.5 (inverted triangles) indicate a slope of < −1 suggesting catalyst instability at high pH. The high pH points were excluded from the NLLSQ fit of the data. Reaction conditions were: 2.5 µM MID1-zinc, 40 µM 4NPP, 40 mM buffer (HEPES for pH 6.5 – 8.5, or potassium hydrogen carbonate for pH > 8.5), 50 mM NaCl, and 37°C. b) Plot of kobs versus [4NPP] for MID1-zinc catalyzed hydrolysis at pH 8.5 and 37°C. A fit of the data to a standard binding equation gives kcat = (1.8 ± 0.05) × 10−4 s−1 and KM = (12 ± 2) µM.
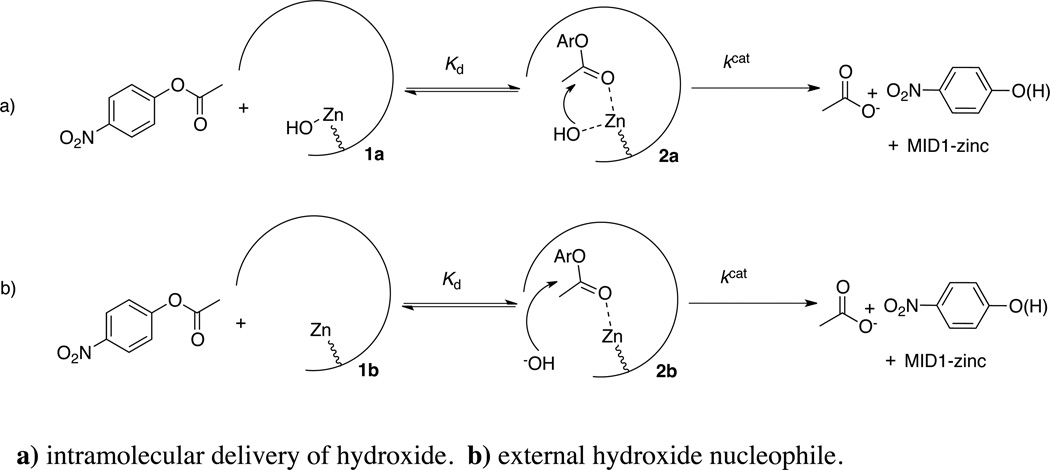
Given the minimalist characteristics of this enzyme, it is useful to consider the MID1-zinc hydrolytic mechanism. Shown in Scheme 1 is a mechanism for the catalytic hydrolysis of 4NPA consistent with the available data. In the first mechanism, upon formation of the Michaelis complex, there is intramolecular delivery of a zinc-hydroxide nucleophile (33, 34). A kinetically equivalent process posits that upon Michaelis complex formation, nucleophilic attack occurs by an external hydroxide.
Scheme 1.
Proposed reaction mechanisms for the MID1-zinc catalyzed hydrolysis of 4NPA
To discern which mechanism is more likely, we performed a limited Brønsted analysis using three substrates to investigate accumulation of charge on the aryloxide leaving group in the transition state. The Brønsted equilibrium coefficient (Beq) for hydrolysis of aryl acetates is estimated to be −1.7 (35). In the uncatalyzed mechanism of aryl acetate cleavage in aqueous conditions where hydroxide addition is the rate-limiting step, the Brønsted coefficient (β) is −0.45 (32), corresponding to a Leffler parameter α = β/βeq = 25%. In our limited Brønsted analysis plotting log(kcat) versus pKa, we observe a Brønsted coefficient of βLG = −1.2 ± 0.07 (Figure S7), corresponding to a Leffler parameter α = βLG/βeq = 70%, indicating that bond cleavage in the enzymatic transition state has progressed ~3-times further than that of the uncatalyzed transition state. Thus, while hydroxide addition is the rate-limiting step in the uncatalyzed mechanism, the large negative βLG indicates that bond cleavage and breakdown of the tetrahedral intermediate is rate-limiting in the enzymatic reaction (Figure S8). This Brønsted analysis suggests that the mechanism of carboxyester hydrolysis by MID1-zinc likely proceeds as depicted in Scheme 1a rather than Scheme 1b: in order for the tetrahedral intermediate to preferentially partition backwards such that aryloxy bond cleavage is rate limiting, the zinc ion must directly interact with the hydroxide to promote the rapid dissociation of hydroxide from the tetrhadetral intermediate.
DISCUSSION
Protein engineering is a valuable approach to uncover minimal requirements for protein structure/function. MID1-zinc is a minimalist engineered protein – it was derived from a 5 kDa helical hairpin monomer, and the zinc coordination features no second-shell interactions (hydrogen bonds to backside histidine nitrogens). It offers a compelling example for the evolutionary benefits of metal coordination in protein structure/function: requiring only a limited number of mutations, zinc binding confers significant stability (Tm increase of 24°C), homodimer binding affinity (>100-fold increase), and catalytic function (hydrolytic rate acceleration of 105). The gains in stability and homodimer affinity were contrived goals of this computational zinc-based design, though the discovery of carboxyester and phosphoester hydrolysis was serendipitous. Although the catalytic activity was serendipitous, this actually strengthens our assertion of the evolutionary plausibility of a primordial enzyme featuring a zinc-mediated homodimeric interface; the arrangment of a zinc binding site at a homodimeric protein interface embodies what we consider to be a minimalist evolutionary path to a cleft containing a catalytic motif. First, clefts are larger and more common at protein interfaces than monomer surfaces. Secondly, the easiest way to form a primordial interface is through a symmetric homodimer – symmetry is energetically favorable in primordial complexes (36), and co-evolution of two genes for a heterodimer is not required. Third and fourthly, metal binding sites effectively mediate both protein interactions and catalytic function.
Previously reported activities by artificial enzymes allow us to investigate a correlation between minimialist active site properties and catalytic rates. First we investigate zinc-containing artificial hydrolases to assess the impact of the binding cleft (Table 2). Macrocyclic amine Zn(II) complexes contain an activated zinc but no binding cleft, and these catalysts have weak activity for 4NPA (second-order rate constants <1 M−1s−1). Rates significantly improve when these catalysts are used in apolar solvents, simulating the apolarity of an active site cleft (37). A zinc hydrolase was created by building a Zn(His)3O zinc binding site at the center of a 3-helix coiled-coil trimer – in our assessment, the zinc within the trimer is not very accessible to a substrate (the catalytic form is likely to be partially dissociated), and this artificial enzyme has a kcat of 0.04 s−1 and a kcat/KM of 23 M−1s−1. MID1-zinc has a shallow but easily accessible cleft, and the pH-independent values are kcat = 0.3 s−1 and kcat/KM = 630 M−1s−1. The kcat/KM of MID1-zinc surpasses by 66-fold the base-catalyzed rate of 9.5 M−1s−1 (32), which is further evidence (in addition to saturable kinetics) for an actual substrate binding event rather than simply providing a hydroxide nucleophile. The naturally occurring enzyme carbonic anhydrase II contains a Zn(His)3O site and features a buried tunnel-like active site, and hydrolysis of 4NPA, a nonnatural substrate, proceeds with a kcat of 53 s−1 and kcat/KM of 2,550 M−1s−1. The difference in rates is likely explained by more than just the structure of the cleft – for example, the three active-site histidines in CAII each have a backside hydrogen bond (second shell interaction), and other sidechains in the active site cleft may also play a functional role in mediated binding/catalysis/release. Nevertheless, in primordial zinc-based enzyme models, we make an overall observation that cleft formation is a major determinant of catalytic rates.
Table 2.
Comparison of rates of 4NPA hydrolysis by artificial esterases.
| name | reference | molecule type | kcat (s−1) | KM (mM) | k2 (M−1s−1) |
|---|---|---|---|---|---|
| Zinc esterases (4NPA) | |||||
| MID1-zinc | (22) | helix-turn-helix dimer | 0.30 | 0.42 | 630 |
| Macrocyclic amine Zn(II) complexes | (33, 49, 50) | small molecule | -- | -- | < 1 |
| [Hg(II)]s[Zn(II)(H2O/OH−)]N(TRIL9CL23H)3n+ | (20) | trimeric peptide | 0.04 | 1.7 | 23 |
| Carbonic anhydrase II | (51) | natural protein, unnatural substrate |
53 | 21 | 2,550 |
| Metal-free esterases (4NPA) | |||||
| −OH | (32) | hydroxide ion | -- | -- | 9.5 |
| KO-42 | (26) | helix-turn-helix dimer | -- | -- | 0.29 |
| JNIIRO | (38) | peptide | -- | -- | 0.06 |
| S-824 | (25) | unselected catalytic antibody |
0.0054 | 3 | 2 |
| ECH13 | a | protein monomer | 0.018 | 0.057 | 320 |
| 43C9 | (28) | selected catalytic antibody |
25 | 0.05 | 470,000 |
| PDZ2 | (30) | thioredoxin redesign | 0.0005 | 0.17 | 3 |
Richter, Baker, personal communication
Comparing MID1-zinc to metal-free artificial hydrolases provides only a crude persepective on the catalytic power of zinc given the unique features of each enzyme. However, given that MID1-zinc is an unoptimized primitive active site, we can use MID1-zinc as a metric for sophistication in other examples of artificial esterases (Table 2). The catalytic peptides KO-42 (26) and JNIIRO (38) do not show saturable kinetics and have slow second-order rate constants (0.29 M−1s−1 and 0.056 M−1s−1, respectively). A catalytic antibody S-824 from an unselected library (kcat = 0.005 s−1) (25) and a computationally designed metal-free esterase ECH13 (kcat = 0.018 s−1, Richter et al., Baker lab, personal communication) do show saturable kinetics but are only moderately active. The 43C9 catalytic antibody (39) hydrolyses 4NPA with an impressive kcat = 25 s−1 and kcat/KM = 4.7 × 105 M−1s−1; however, as with many catalytic antibodies, 43C9 experiences product inhibition, Ki = 1 µM (28).
To compare rates of 4NPP hydrolysis, a recent work describes a large supramolecular cyclen complex containing eight zinc and two copper ions that mimicks a protein nanostructure – this complex is less efficient than MID1-zinc in hydrolysis of 4NPP: kcat = 1.5 × 10−5 s−1, KM = 470 µM, kcat/KM = 0.03 M−1s−1 (40). Additionally, a histidine-containing pseudopeptide hydrolyses 4NPP with a similar rate constant, kcat = 2.1 × 10−5 s−1 (41). A natural and highly proficient (often diffusion-limited) enzyme, alkaline phosphatase, is much more efficient than MID1-zinc in hydrolysis of 4NPP: kcat = 40 s−1, KM = 7 µM, kcat/KM = 5 × 106 M−1s−1 (40).
The above same-substrate comparisons with 4NPA and 4NPP used kcat and kcat/KM as the metric for comparison. The catalytic rate acceleration, defined as kcat/kneutral, has been instrumental in measuring the power of a large number of enzymes that catalyze a variety of reactions (42, 43). The pH-neutral uncatalyzed hydrolysis (31) of 4NPA has a reported kneutral of 4.3 × 10−7 s−1 at 25°C (32), so we calculate a MID1-zinc rate acceleration of 7 × 105. A 105 rate acceleration is better or comparable to previously reported de novo enzymes (7, 8, 10, 11, 16, 18, 26, 27, 30, 38, 44–46) and many catalytic antibodies (47). However, MID1-zinc achieves only a modest fraction of the rate accelerations observed in natural hydrolases (31, 48).
In conclusion, a metal-mediated protein interface has a high probability of having catalytic function given the requirements of a cleft and a catalytic motif, which is supported by our serendipitous discovery of hydrolysis by our computationally designed zinc-mediated protein homodimer. This recipe for catalysis may have been a critical route for evolution of natural enzymes and may provide an effective strategy for rational engineering of new catalytic activities.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Richard Wolfenden for his expertise in determining rate accelerations and for his comments and perspectives on this work. We thank Dr. Greg Young of the UNC Biomolecular NMR Lab for his contributions.
ABBREVIATIONS
- 4NPA
4-nitrophenyl acetate
- 4NPP
4-nitrophenyl phosphate
- MID1-zinc
metal interface design 1 with zinc
Footnotes
This work was funded by the NIH grant GM073960 and by the National Science Foundation graduate research fellowship (2009070950 to B.D.).
Supporting Information Available. Crystal structure of MID1-zinc co-crystallized with tartrate, absorbance spectrum of 4NPA hydrolysis, gel filtration fractions tested for catalytic activity, binding affinity of zinc for MID1 monitored by fluorescence polarization. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Scalley-Kim M, Baker D. Characterization of the folding energy landscapes of computer generated proteins suggests high folding free energy barriers and cooperativity may be consequences of natural selection. J Mol Biol. 2004;338:573–583. doi: 10.1016/j.jmb.2004.02.055. [DOI] [PubMed] [Google Scholar]
- 2.Grishin NV, Phillips MA. The subunit interfaces of oligomeric enzymes are conserved to a similar extent to the overall protein sequences. Protein Sci. 1994;3:2455–2458. doi: 10.1002/pro.5560031231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali MH, Imperiali B. Protein oligomerization: how and why. Bioorg Med Chem. 2005;13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 4.Sonavane S, Chakrabarti P. Cavities and atomic packing in protein structures and interfaces. PLoS Comput Biol. 2008;4:e1000188. doi: 10.1371/journal.pcbi.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao M, Skolnick J. The distribution of ligand-binding pockets around protein-protein interfaces suggests a general mechanism for pocket formation. Proc Natl Acad Sci U S A. 2012;109:3784–3789. doi: 10.1073/pnas.1117768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Rothlisberger D, Baker D. New algorithms and an in silico benchmark for computational enzyme design. Protein Science. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, 3rd, Hilvert D, Houk KN, Stoddard BL, Baker D. De novo computational design of retro-aldol enzymes. Science. 2008;319:1387–1391. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 9.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St Clair JL, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khare SD, Kipnis Y, Greisen PJ, Takeuchi R, Ashani Y, Goldsmith M, Song Y, Gallaher JL, Silman I, Leader H, Sussman JL, Stoddard BL, Tawfik DS, Baker D. Computational redesign of a mononuclear zinc metalloenzyme for organophosphate hydrolysis. Nat Chem Biol. 2012;8:294–300. doi: 10.1038/nchembio.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Privett HK, Kiss G, Lee TM, Blomberg R, Chica RA, Thomas LM, Hilvert D, Houk KN, Mayo SL. Iterative approach to computational enzyme design. Proc Natl Acad Sci U S A. 2012;109:3790–3795. doi: 10.1073/pnas.1118082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 13.Lu Y, Yeung N, Lin YW, Gao YG, Zhao X, Russell BS, Lei LY, Miner KD, Robinson H. Rational design of a structural and functional nitric oxide reductase. Nature. 2009;462:1079–U1144. doi: 10.1038/nature08620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nanda V, Rosenblatt MM, Osyczka A, Kono H, Getahun Z, Dutton PL, Saven JG, DeGrado WF. De novo design of a redox-active minimal rubredoxin mimic. Journal of the American Chemical Society. 2005;127:5804–5805. doi: 10.1021/ja050553f. [DOI] [PubMed] [Google Scholar]
- 15.Nanda V, Koder RL. Designing artificial enzymes by intuition and computation. Nature Chemistry. 2010;2:15–24. doi: 10.1038/nchem.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan J, DeGrado WF. De novo design of catalytic proteins. Proc Natl Acad Sci U S A. 2004;101:11566–11570. doi: 10.1073/pnas.0404387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faiella M, Andreozzi C, de Rosales RTM, Pavone V, Maglio O, Nastri F, DeGrado WF, Lombardi A. An artificial di-iron oxo-protein with phenol oxidase activity. Nature Chemical Biology. 2009;5:882–884. doi: 10.1038/nchembio.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson DE, Wisz MS, Hellinga HW. Rational design of nascent metalloenzymes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6292–6297. doi: 10.1073/pnas.97.12.6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sigman JA, Kwok BC, Lu v. From myoglobin to heme-copper oxidase: Design and engineering of a Cu-B center into sperm whale myoglobin. Journal of the American Chemical Society. 2000;122:8192–8196. [Google Scholar]
- 20.Zastrow ML, Peacock AF, Stuckey JA, Pecoraro VL. Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat Chem. 2012;4:118–123. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salgado EN, Faraone-Mennella J, Tezcan FA. Controlling protein-protein interactions through metal coordination: assembly of a 16-helix bundle protein. J Am Chem Soc. 2007;129:13374–13375. doi: 10.1021/ja075261o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Der BS, Machius M, Miley MJ, Mills JL, Szyperski T, Kuhlman B. Metal-mediated affinity and orientation specificity in a computationally designed protein homodimer. J Am Chem Soc. 2012;134:375–385. doi: 10.1021/ja208015j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nilsson J, Baltzer L. Reactive-site design in folded-polypeptide catalysts--the leaving group pKa of reactive esters sets the stage for cooperativity in nucleophilic and general-acid catalysis. Chemistry. 2000;6:2214–2220. doi: 10.1002/1521-3765(20000616)6:12<2214::aid-chem2214>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 24.Nicoll AJ, Allemann RK. Nucleophilic and general acid catalysis at physiological pH by a designed miniature esterase. Org Biomol Chem. 2004;2:2175–2180. doi: 10.1039/B404730C. [DOI] [PubMed] [Google Scholar]
- 25.Wei Y, Hecht MH. Enzyme-like proteins from an unselected library of designed amino acid sequences. Protein Eng Des Sel. 2004;17:67–75. doi: 10.1093/protein/gzh007. [DOI] [PubMed] [Google Scholar]
- 26.Broo KS, Brive L, Ahlberg P, Baltzer L. Catalysis of hydrolysis and transesterification reactions of p-nitrophenyl esters by a designed helix-loop-helix dimer. Journal of the American Chemical Society. 1997;119:11362–11372. [Google Scholar]
- 27.Nicoll AJ, Allemann RK. Nucleophilic and general acid catalysis at physiological pH by a designed miniature esterase. Organic & Biomolecular Chemistry. 2004;2:2175–2180. doi: 10.1039/B404730C. [DOI] [PubMed] [Google Scholar]
- 28.Gibbs RA, Benkovic PA, Janda KD, Lerner RA, Benkovic SJ. Substituent Effects on an Antibody-Catalyzed Hydrolysis of Phenyl-Esters - Further Evidence for an Acyl-Antibody Intermediate. Journal of the American Chemical Society. 1992;114:3528–3534. [Google Scholar]
- 29.Baltzer L, Broo KS, Nilsson H, Nilsson J. Designed four-helix bundle catalysts--the engineering of reactive sites for hydrolysis and transesterification reactions of p-nitrophenyl esters. Bioorg Med Chem. 1999;7:83–91. doi: 10.1016/s0968-0896(98)00218-1. [DOI] [PubMed] [Google Scholar]
- 30.Bolon DN, Mayo SL. Enzyme-like proteins by computational design. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolfenden R, Yuan Y. The "Neutral" Hydrolysis of Simple Carboxylic Esters in Water and the Rate Enhancements Produced by Acetylcholinesterase and Other Carboxylic Acid Esterases. Journal of the American Chemical Society. 2011;133:13821–13823. doi: 10.1021/ja204116a. [DOI] [PubMed] [Google Scholar]
- 32.Jencks WP, Gilchris M. Nonlinear Structure-Reactivity Correlations. Reactivity of Nucleophilic Reagents toward Esters. Journal of the American Chemical Society. 1968;90:2622. doi: 10.1021/ja01016a040. -&. [DOI] [PubMed] [Google Scholar]
- 33.Kimura E, Shiota T, Koike T, Shiro M, Kodama M. A Zinc(Ii) Complex of 1,5,9-Triazacyclododecane ([12]Anen3) as a Model for Carbonic-Anhydrase. Journal of the American Chemical Society. 1990;112:5805–5811. [Google Scholar]
- 34.Suh JH, Son SJ, Suh MP. Kinetics of hydrolysis of phenyl acetates catalyzed by the zinc(II) complex of 1,5,9-triazacyclododecane. Evidence for attack of water or hydroxide ion at the coordinated ester. Inorganic Chemistry. 1998;37:4872–4877. doi: 10.1021/ic980205x. [DOI] [PubMed] [Google Scholar]
- 35.Williams A. Effective charge and Leffler's index as mechanistic tools for reactions in solution. Accounts of Chemical Research. 1984;17:425–430. [Google Scholar]
- 36.Andre I, Strauss CE, Kaplan DB, Bradley P, Baker D. Emergence of symmetry in homooligomeric biological assemblies. Proc Natl Acad Sci U S A. 2008;105:16148–16152. doi: 10.1073/pnas.0807576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gomez-Tagle P, Vargas-Zuniga I, Taran O, Yatsimirsky AK. Solvent effects and alkali metal ion catalysis in phosphodiester hydrolysis. J Org Chem. 2006;71:9713–9722. doi: 10.1021/jo061780i. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson J, Baltzer L. Reactive-site design in folded-polypeptide catalysts – The leaving group pK(a) of reactive esters sets the stage for cooperativity in nucleophilic and general-acid catalysis. Chemistry-a European Journal. 2000;6:2214–2220. doi: 10.1002/1521-3765(20000616)6:12<2214::aid-chem2214>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 39.Janda KD, Schloeder D, Benkovic SJ, Lerner RA. Induction of an Antibody That Catalyzes the Hydrolysis of an Amide Bond. Science. 1988;241:1188–1191. doi: 10.1126/science.3413482. [DOI] [PubMed] [Google Scholar]
- 40.Zulkefeli M, Suzuki A, Shiro M, Hisamatsu Y, Kimura E, Aoki S. Selective hydrolysis of phosphate monoester by a supramolecular phosphatase formed by the self-assembly of a bis(Zn(2+)-cyclen) complex, cyanuric acid, and copper in an aqueous solution (cyclen = 1,4,7,10-tetraazacyclododecane) Inorg Chem. 2011;50:10113–10123. doi: 10.1021/ic201072q. [DOI] [PubMed] [Google Scholar]
- 41.Ichikawa K, Tarnai M, Uddin MK, Nakata K, Sato S. Hydrolysis of natural and artificial phosphoesters using zinc model compound with a histidine-containing pseudopeptide. J Inorg Biochem. 2002;91:437–450. doi: 10.1016/s0162-0134(02)00452-x. [DOI] [PubMed] [Google Scholar]
- 42.Radzicka A, Wolfenden R. A Proficient Enzyme. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 43.Wolfenden R. Degrees of difficulty of water-consuming reactions in the absence of enzymes. Chemical Reviews. 2006;106:3379–3396. doi: 10.1021/cr050311y. [DOI] [PubMed] [Google Scholar]
- 44.Johnsson K, Allemann RK, Widmer H, Benner SA. Synthesis, structure and activity of artificial, rationally designed catalytic polypeptides. Nature. 1993;365:530–532. doi: 10.1038/365530a0. [DOI] [PubMed] [Google Scholar]
- 45.Rossi P, Tecilla P, Baltzer L, Scrimin P. De novo metallonucleases based on helix-loop-helix motifs. Chemistry-a European Journal. 2004;10:4163–4170. doi: 10.1002/chem.200400160. [DOI] [PubMed] [Google Scholar]
- 46.Korendovych IV, Kulp DW, Wu Y, Cheng H, Roder H, DeGrado WF. Design of a switchable eliminase. Proc Natl Acad Sci U S A. 2011;108:6823–6827. doi: 10.1073/pnas.1018191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao DN, Wootla B. Catalytic Antibodies: Concept and Promise. Resonance. 2007;12:6–21. [Google Scholar]
- 48.Bryant RAR, Hansen DE. Direct measurement of the uncatalyzed rate of hydrolysis of a peptide bond. Journal of the American Chemical Society. 1996;118:5498–5499. [Google Scholar]
- 49.Konig B, Subat M, Woinaroschy K, Anthofer S, Malterer B. 1,4,7,10-Tetraazacyclododecane metal complexes as potent promoters of carboxyester hydrolysis under physiological conditions. Inorganic Chemistry. 2007;46:4336–4356. doi: 10.1021/ic070101z. [DOI] [PubMed] [Google Scholar]
- 50.Koerner TB, Brown RS. The hydrolysis of an activated ester by a tris(4,5-di-n-propyl-2-imidazolyl)phosphine- Zn(2+) complex in neutral micellar medium as a model for carbonic anhydrase. Canadian Journal of Chemistry-Revue Canadienne De Chimie. 2002;80:183–191. [Google Scholar]
- 51.Verpoort Ja, Mehta S, Edsall JT. Esterase Activities of Human Carbonic Anhydrases B and C. Journal of Biological Chemistry. 1967;242:4221. -&. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.