Abstract
The metabolism of arachidonic acid (AA) to epoxyeicosatrienoic acids (EETs) is thought to be mediated primarily by the cytochromes P450 (P450s) from the 2 family (2C9, 2C19, 2D6, and 2J2). In contrast, P450s of the 4 family are primarily involved in omega oxidation of AA (4A11 and 4A22). The ability to determine enantioselective formation of the regioisomeric EETs is important in order to establish their potential biological activities and to asses which P450 isoforms are involved in their formation. It has been extremely difficult to analyze individual EET enantiomers in biological fluids because they are present in only trace amounts and they are extremely difficult to separate from each other. In addition, the deuterium-labeled internal standards that are commonly used for stable isotope dilution liquid chromatography/mass spectrometry (LC/MS) analyses have different LC retention times when compared with the corresponding protium forms. Therefore, quantification by LC/MS-based methodology can be compromised by differential suppression of ionization of the closely eluting isomers. We report the preparation of [13C20]-EET analog internal standards and the use of a validated high-sensitivity chiral LC/electron capture atmospheric pressure chemical ionization (ECAPCI)-MS method for the trace analysis of endogenous EETs as their pentafluorobenzyl (PFB) ester derivatives. The assay was then used to show the exquisite enantioselectivity of P4502C19-, P4502D6-, P4501A1-, and P4501B1-mediated conversion of AA into EETs and to quantify the enantioselective formation of EETs produced by AA metabolism in a mouse epithelial hepatoma (Hepa) cell line.
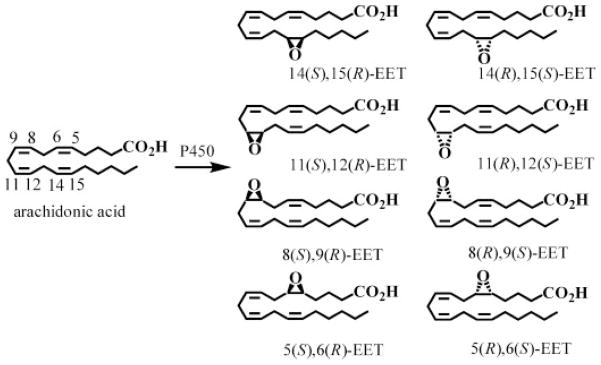
P450s are membrane-bound hemoproteins that are involved in the metabolism of drugs, carcinogens, steroids, and arachidonic acid (AA).1,2 Details of the individual genes, sequences, and basic catalytic mechanisms are now well established.3 P450s can catalyze three different types of AA oxidation. First, they can induce bis-allylic oxidation of AA to produce 7-, 10-, and 13-hydroxyeicosatetraenoic acids (HETEs) or lipoxygenase-like products such as 11-, 12-, and 15-HETEs.4 Second, P450s primarily of the 4 family can perform conventional hydroxylation reactions on the ω-terminus of AA to produce 16-, 17-, 18-, 19-, and 20-HETEs.5 Third, they can epoxidize AA at each cis-olefin to produce four EET regioisomers (5,6-EET, 8,9-EET, 11,12-EET, 14,15-EET) each of which can be formed as an enantiomeric pair (Fig. 1).6–8 The 5,6-EET regioisomer is rapidly converted into the corresponding lactone, due to the proximity of the terminal carboxylic group and the 5,6-epoxide.9 However, the other EETs are relatively stable until they are metabolized either by cytosolic epoxide hydrolases10,11 to dihydroxyeicosatrienoic acids or by glutathione S-transferases to glutathione adducts.12 The regioselectivity and enantioselectivity of EET formation is P450 isoform specific and is thought to involve primarily P450s from the 2 family in humans (2C8, 2C19, 2D6, and 2J2).13–15 Endogenous EETs16–19 are normally found esterified at the sn-2 position of cellular glycerophospholipids but can be readily released by basic hydrolysis.19,20 The EETs have potent vasodilator19,21,22 and anti-inflammatory activity.23–26 In addition, they can prevent apoptosis27–31 and, depending on their chirality and regiochemistry, they can also inhibit the platelet aggregation.13,32
Figure 1.
Biosynthesis of epoxyeicosatrienoic acids (EETs) by cytochrome P450 isoforms.
Determination of the enantioselectivity that occurs in the formation of regioisomeric EETs is important in order to establish their potential biological activities and to assess which P450 isoforms are involved in their formation. There are a number of reports describing the enantiomeric separation of the EET regioisomers using chiral columns with normal- or reversed-phase chromatography.20,33,34 However, it has been extremely difficult to determine the enantioselectivity of EET formation in cell and tissue samples because of the problems in analyzing trace amounts of these potent biologically active substances.20,33,35 Typically, this has required initial preparative chiral high-performance liquid chromatography (HPLC) separations followed by the analysis of each of the individual isomers using stable isotope dilution gas chromatography/electron capture negative chemical ionization mass spectrometry (MS).34,36 There is a diverse array of chiral HPLC stationary phases available that are derived from silica matrices coated with carbamates or benzyl esters. Some of them are suitable for the enantiomeric separation of one regioisomer but not for others. Chiralcel OJ was found to resolve all regioisomers into enantiomeric pairs, but different mobile phase flow rates were required for optimal separation of the various regioisomers.37 Previous studies have reported the analysis of EETs by LC/MS38 but the methods did not employ internal standards for the individual enantiomers37 and long chromatographic run times were required for the chiral separations.35,39 Over the last decade, we have developed highly specific and sensitive methodology for the analysis of eicosanoids based on the use of stable isotope dilution normal-phase chiral liquid chromatography/electron capture atmospheric pressure chemical ionization mass spectrometry (LC/ECAPCI-MS).40–43 This methodology has now been applied to determine the enantioselectivity of AA-mediated EETs formation by different P450 isoforms and to quantify AA-mediated chiral EETs formation in the mouse epithelial hepatoma Hepa cell line.
EXPERIMENTAL
Materials
( ± )-8,9-Epoxy-(5Z,11Z,14Z)-eicosatrienoic acid [( ± )8,9-EET], ( ± )-11,12-epoxy-(5Z,8Z,14Z)-eicosatrienoic acid [( ± )11,12-EET], and ( ± )-14,15-epoxy-(5Z,8Z,11Z)-eicosatrienoic acid [( ± )14,15-EET] were purchased from Biomol International (Plymouth Meeting, PA, USA). Arachidonic acid (AA) was purchased from Cayman Chemical Co. (Ann Arbor, MI, USA). [13C]-Labeled AA for the [13C]-labeled EETs synthesis was purchased from Spectra Stable Isotopes (currently Cambridge Isotope Laboratories, Andover, MA, USA). Diisopropylethylamine (DIPEA), 2,3,4,5,6-pentafluorobenzyl bromide (PFB-Br), 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), nicotinamide adenine dinucleotide phosphate-oxidase (NADPH), ethylenediaminetetraacetic acid (EDTA), 3-chloroperoxybenzoic acid (MCPBA), sodium thiosulfate, acetonitrile, methylene chloride and diethyl ether were purchased from Sigma-Aldrich (St. Louis, MO, USA). HPLC-grade hexane, isopropanol and ethanol were obtained from Fisher Scientific Co. (Fair Lawn, NJ, USA). ACS-grade ethanol was obtained from Pharmco (Brookfield, CT, USA). Gases were supplied by BOC Gases (Lebanon, NJ, USA). The Chiralpak AD-H column was obtained from Chiral Technologies (West Chester, PA, USA). Microsomes of recombinant human or rat P450 expressed in insect cells (hP4501A1, hP4501B1, hP4502D6, hP4502C19 and rP4501A1, rP4501B1) were purchased from Gentest (Woburn, MA, USA). Mouse epithelial hepatoma Hepa-1c1c7 cells (CRL-2026TM) were from the American Type Culture Collection (ATCC, Manassas, VA, USA). α-Minimum essential medium (MEM), fetal bovine serum (FBS), penicillin and streptomycin were from Invitrogen (Carlsbad, CA, USA).
Synthesis of [13C20] EETs
[13C]-AA (2 mg, 6.1 mmol) was epoxidized by adding a solution of 8 equivalents of MCPBA (8.6 mg, 50 mmol) in 1 mL of methylene chloride. The reaction mixture was stirred for 2 h at room temperature.44 Excess reagent was quenched with aqueous sodium thiosulfate (10% v/v, 1 mL). The EETs were extracted three times with 4 mL of diethyl ether. The combined organic phases were washed with aqueous sodium hydrogen carbonate (10% v/v), water and dried over anhydrous sodium sulfate. The filtrate was evaporated to dryness under a nitrogen stream. After re-suspension in 100 μL of hexanes, the reaction mixture was filtered through a small flash silica gel column under atmospheric pressure and 4 mL of hexane was added to the column to elute all the regioisomeric EETs from the silica gel. The purified [13C20]-EETs were diluted in acetonitrile, divided into 1 mL aliquot stock solutions and stored at −80°C until ready to use.
Preparation of EET-PFBs
A racemic mixture of each EET regioisomer (50 ng, 0.156 nmol) in methylene chloride (100 μL) was treated with 100 μL of DIPEA in methylene chloride (1:19, v/v) followed by 100 μL of PFB-Br in methylene chloride (1:9, v/v) and the solution was shaken at room temperature for 30 min. The solution was evaporated to dryness under a nitrogen stream at room temperature, and re-dissolved in 500 μL of hexane/ethanol (97:3, v/v) for analysis.
LC
Normal-phase chiral chromatography for LC/MS experiments was performed using a Waters Alliance 2690 HPLC system (Waters Corp., Milford, MA, USA). Gradient elution was performed in the linear mode. A Chiralpak AD-H column (250 × 4.6 mm i.d., 5 μm; Daicel Chemical Industries, Ltd., Tokyo, Japan) was employed at a flow rate of 1 mL/min. Solvent A was hexanes and solvent B was 2-propanol/hexanes (6:4, v/v). Isocratic elution was used with 1.5% B for 12 min and then a linear gradient to 100% A for 10 min for washing the column. The separation was performed at 30°C. Always, after PFB derivatization, the samples were re-suspended in 100 μL of hexanes/2-propanol (95/5, v/v) and were maintained at 4°C in the autosampler tray; injections of 20 μL were made.
ECAPCI-MS
Mass spectrometry was conducted on a Thermo Finnigan TSQ Quantum Ultra AM mass spectrometer (Thermo Finnigan, San Jose, CA, USA) equipped with an APCI source in the EC negative ion mode. The TSQ Quantum operating conditions were as follows: vaporizer temperature, 450°C; heated capillary temperature, 250°C; corona discharge needle, set at 30 μA. The sheath gas (nitrogen) and auxiliary gas (nitrogen) pressures were 25 psi and 5 (arbitrary units), respectively. Collision-induced dissociation (CID) was performed using argon as the collision gas at 1.5 mTorr in the second (rf-only) quadrupole. For full-scan and multiple reaction monitoring (MRM) analyses, unit resolution was maintained for both precursor and product ions. MS analysis was conducted by infusing a solution in hexanes of 10 ng/μL of each compound as its PFB derivative infused at 5 μL/min. The following MRM transitions were monitored: ( ± )8,9-EET-PFB, m/z 319 →155 (collision energy 12 eV), ( ± )11,12-EET-PFB, m/z 319 →208 (collision energy 14 eV), ( ± )14,15-EET-PFB, m/z 319 →219 (collision energy 12 eV). The internal standards were: ( ± )[13C20]-8,9-EET-PFB m/z 339 →163 (collision energy 12 eV), ( ± )[13C20]-11,12-EET-PFB m/z 339 →220 (collision energy 14 eV), ( ± )[13C20]-14,15-EET-PFB m/z 339 →233 (collision energy 12 eV).
Preparation of standards and QC solutions
For calibration curves of the EETs, the three racemic stereoisomers of the authentic EETs (0, 0.002, 0.004, 0.1, 0.2, 0.4, 0.8, 1.6 ng of each) together with 10 ng of each [13C20]-EET were spiked into 3 mL buffer solution. The standard solutions underwent the same sample preparation and analytical procedures as the samples. Calibration curves were calculated with a linear regression analysis of the peak area ratios of authentic standard against the appropriate [13C20]-internal standard (Fig. 2). Amounts of EET enantiomers were calculated by interpolation from the relevant calibration curve. Quality control (QC) samples were prepared by spiking a 3 mL buffer solution with a certain amount of authentic standard (0.02, 0.04, 0.2 and 0.8 ng of each stereoisomer, 3×) and internal standards (10 ng of [13C20]-EETs). The QC samples underwent the same sample preparation and analytical procedures as the study samples.
Figure 2.
Calibration curves: (A) 8(S),9(R)-EET-PFB; (B) 8(R),9(S)-EET-PFB; (C) 11(S),12(R)-EET-PFB; (D) 11(R),12(S)-EET-PFB; (E) 14(R),15(S)-EET-PFB; and (F) 14(S),15(R)-EET-PFB.
AA incubation with supersomes
To supersomes containing 50 pmol of the relevant P450 in 0.25 mL incubation buffer (0.05 mM Tris-HCl, pH 7.4, containing 1 mM EDTA) and 1 mM NADPH was added 5 μM AA in ethanol (95% (v/v)). The mixtures were incubated at 37°C for 30 min. All incubations were performed in duplicate. The reactions were terminated by placing the samples on ice and adding 1 mL of ice-cold ethanol. The internal standard mixture (10 μL stock solution of [13C20]-EET) was added to all samples and they were centrifuged at 10 000 rpm for 10 min. The supernatants were moved to larger glass tubes and lipids were extracted with diethyl ether (3 mL × 2) and the combined organic layers were evaporated to dryness under nitrogen. Analysis of the PFB derivatives (prepared as described above for the standards) by LC/ECAPCI-MRM-MS analysis was conducted on a 20 μL aliquot of this solution. Concentrations of EETs from the supersomes were calculated by interpolation from the calculated regression lines.
Culture of epithelial mouse Hepa-1c1c7 cells
Hepa1c1c7 cells were cultured in α-MEM with 10% FBS and 100 U/mL penicillin/streptomycin. Cells grew to 90% confluence, approximately 1 × 106 cells per plate, at 37°C and 5% CO2 and were passaged every 4 days at a 1:3 dilution.
Incubations of Hepa cells with AA
When the cells were approximately 90% confluent, the media was removed and the cells washed with phosphate-buffered saline (PBS). Fresh FBS-free media was added and the cells were treated with vehicle alone (dimethyl sulfoxide (DMSO), ethanol) or 5 nM TCDD in DMSO (95% (v/v)) and 10 μM AA in ethanol (95% (v/v)). All treatments were performed in duplicate. After completion of the treatment, the media was removed, and the cells were scraped into polypropylene Eppendorf tubes. They were centrifuged at 10 000 rpm for 10 min. The supernatants were removed, 1 mL of fresh PBS was added to the cell pellet, and the cells were re-suspended by pipetting for 10 s. The cell suspension (typically 106 cells) was transferred to a clean glass tube containing chloroform and methanol (2:1 v/v, 5 mL). The lipids were extracted by Folch extraction.45 Briefly: the samples were shaken for 15 min and centrifuged at 3000 g for 10 min. The supernatant from each tube was transferred to a new tube and washed with 1 mL of 0.9% NaCl solution. After vortex mixing and centrifugation to separate the two phases, the upper phase was removed. The steps for washing with 0.9% NaCl solution and separation were repeated and the combined lower phases were evaporated to dryness under nitrogen. Hydrolysis of the esterified lipids was performed after dissolving the dry residue in 500 μL of 80% methanolic NaOH for 30 min at 60°C under nitrogen. After completion of the hydrolysis, samples were transferred to larger glass tubes and acidified to pH 4 with 2 N HCl. Then 10 ng of the internal standard mixture was added and the lipids were extracted with diethyl ether (3 mL × 2). The combined organic layers were evaporated to dryness under nitrogen. Analysis of the PFB derivatives (prepared as described above for the standards) by LC/ECAPCI-MRM-MS analysis was conducted on a 20 μL aliquot of this 100 μL solution. Concentrations of EETs from the cells were calculated by interpolation from the calculated regressions lines.
Data analysis
All data analysis was performed using Xcalibur software (version 2.0 SR2; Thermo Analytical) from raw mass spectral data. Calibration curves were plotted using a linear regression with weighting index of 1/x. Concentrations of EETs in validation samples were determined from the calibration line, and used to calculate the accuracy and precision of the method within the study.
RESULTS
MS analysis of EETs
Each PFB derivative of the three EET regioisomers was analyzed under ECAPCI conditions. All of the EETs exhibited an intense precursor ion at m/z 319 corresponding to the loss of the PFB moiety in the source of the mass spectrometer.40 The product spectra of the [M–PFB]− ion for each of the regioisomers were almost identical with those reported by Bernstrom et al. from CID of the fast atom bombardment-generated carboxylate anions46 and Nakamura et al. from CID of the same carboxylate anions generated by electrospray ionization in a tandem quadrupole mass spectrometer.47 The EET-PFB derivatives had common product ions formed by loss of water at m/z 301, at m/z 275 (-COO) and at m/z 257 (-(H2O + COO)). The MS and MS/MS spectra for the R,S isomer and S,R isomers were identical. The most intense product ion from the CID and MS/MS analysis of 8,9-EET-PFB [M–PFB]− was observed at m/z 155 (Fig. 3(A)). This corresponded to the breaking of the C8–C9 bond and C9–O. CID and MS/MS analysis of 11,12-EET-PFB [M–PFB]− showed a characteristic odd-electron ion at m/z 208, corresponding to the cleavage of the C12–C13 bond (Fig. 3(B)). The product ions at m/z 167 and 179 corresponded to the breaking of the C10–C11 bond with an additional proton and the breaking of C11–C12 and C11–O, respectively. CID and MS/MS analysis of 14,15-EET-PFB [M–PFB]− revealed a product ion at m/z 219 resulting from the cleavage of the C14–C15 bond and C15–O (Fig. 3(C)) and a product ion at m/z 175 corresponding to the loss of water from the m/z 219 ion. The mechanisms of formation of all the EET-derived product ions were assumed to be similar to those described by Murphy et al.48
Figure 3.
Product ion spectra of EETs: (A) CID of 8,9-EET-PFB after PFB loss in the source; (B) CID of 11,12-EET-PFB after PFB loss in the source; and (C) CID of 14,15-EET-PFB after PFB loss in the ion source of the mass spectrometer.
Analysis of EET-PFB derivatives by chiral phase LC/ECAPCI-MRM-MS
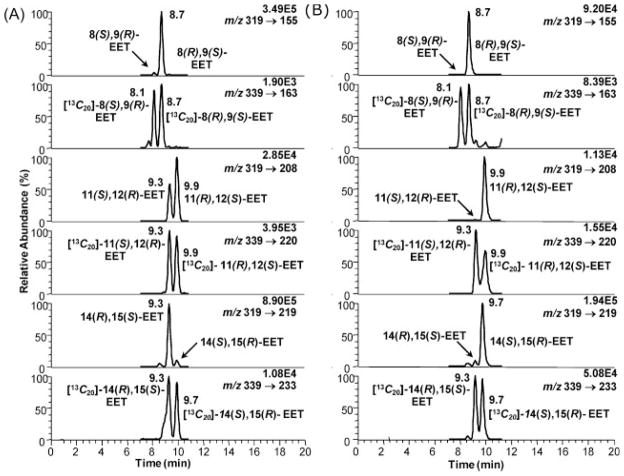
Specific product ions were selected for each EET-PFB derivative. For 11,12-EET, the product ion at m/z 208 had to be picked over the more intense ion at m/z 167 because 8,9-EET also exhibited a ion fragment at m/z 167, which showed peaks in the channel for 11,12-EET. For quantification of the EETs the [13C20]-EET mixture was added as the internal standard (Fig. 4). The following specific MRM transitions were monitored for the EETs: m/z 319 [M–PFB]− to m/z 155 for 8,9-EET, m/z 319 [M–PFB]− to m/z 208 for 11,12-EET and m/z 319 [M–PFB]− to m/z 219 for 14,15-EET. For the internal standards the following transitions were monitored: m/z 339 [M–PFB]− to m/z 163 for [13C20]-8,9-EET, m/z 339 [M–PFB]− to m/z 220 for [13C20]-11,12-EET and m/z 339 [M–PFB]− to m/z 233 for [13C20]-14,15-EET. The chiral separation of the EET-PFB derivatives was performed on the Chiralpak AD-H column, an amylose tris(3,5-dimethyl-phenyl carbamate) chiral stationary phase. The separation of the EETs enantiomers required a non-polar phase. The best separation was archived with 0.9% 2-propanol in hexanes. Addition of more polar (ethanol, methanol) or less polar (butanol) solvents to hexanes did not separate all the regioisomers into their corresponding enantiomers in the same chromatographic run. The retention times were 7.9 min for 8(S),9(R)-EET-PFB and 8.4 min for 8(R),9(S)-EET-PFB; 9.0 min for 11(S),12(R)-EET-PFB and 9.6 min for 11(R),12(S)-EET-PFB; 8.9 min for 14(R),15(S)-EET-PFB and 9.6 min for 14(S),15(R)-EET-PFB. The [13C20]-EETs had the exact same retention times as their corresponding unlabeled EETs (Fig. 4).
Figure 4.
LC/MRM-MS chromatograms of EET-PFB standards and corresponding [13C]-labeled internal standards.
The authentic EET standards were commercially available as a racemic mixture. LC/MS analysis of 14,15-EET-PFB revealed that interference from MRM transitions m/z 319 to 155 (8,9-EET-PFB) and m/z 319 to 208 (11,12-EET-PFB) was <0.5% (0.5% and 0.3%, respectively; Fig. 5). Thus, although m/z 155 and 208 were minor product ions present in the MS/MS spectrum of 14,15-EET-PFB, they did not cause significant interference in the LC/MRM-MS analysis of 8,9- and 11–12-EET enantiomers. It is known42 that different polar modifiers of the mobile phase will change the elution order. Due to lack of the pure enantiomers of each EET, the elution order was established by comparison of the most abundant enantiomer formed by different supersome isoforms with the enantiomer reported in the literature. For P4502C8, an enzyme with major epoxygenase activity, it was reported49 that the 11(R),12(S)-EET was produced with 82% enantiomeric excess (ee). In our experiments, we found the later peak eluting in the 11,12-EET channel to be the most abundant (data not shown), so the first eluting peak was from 11(S),12(R)-EET and the later eluting peak arose from 11(R),12(S)-EET (Fig. 4). This assignment was confirmed by the order of elution of the products of P4502C9-mediated AA metabolism. The reported ee for 11(S),12(R)-EET was 69% and we obtained almost exactly the same value for P4502C9-mediated AA metabolism to 11,12-EET (data not shown). The order of elution of the 14,15-EET-PFB derivatives was established by a similar rationale; the first eluting peak was from 14(R),15(S)-EET and the second one was from 14(S),15(R)-EET (Fig. 4). Previous studies of supersome-mediated 8,9-EET formation do not provide data for unequivocal identification of the elution order for the individual EET-PFB enantiomers. However, Wei et al.37 reported that on the Chiralcel OJ column, 8(R),9(S)-EET and 8(S),9(R)-EET were separated by 7 min. Each enantiomer was collected after Chiralcel OJ separation, mixed with the appropriate [13C20]-internal standards and analyzed after PFB derivatization using the AD-H column. This made it possible to assign the first eluting enantiomer as 8(S),9(R)-EET and the later eluting enantiomer as 8(R),9(S)-EET (Fig. 6). Therefore, the order of elution of the EET PFB-derivatives on the AD-H column was the opposite of that reported for the underivatized EETs on the OJ column.37
Figure 5.
LC/MRM-MS chromatograms of 14,15-EET-PFB standard injected alone. Intensities in the MRM channels for 8,9-EET-PFB and 11,12-EET-PFB were <0.5%.
Figure 6.
LC/MRM-MS chromatograms of synthetic pure 8(R),9(S)-EET-PFB and 8(S),9(R)-EET-PFB standards and their corresponding 13C-labeled internal standards for establishing the order of elution of the each enantiomer.
Sensitivity and linearity
To determine the limit of detection (LOD), a serial dilution of each racemic EET was prepared (0.02 to 80 ng/mL). The LOD determined at a S/N (signal/noise) ratio of 3:1 for ± )8,9-EET-PFB, ( ± )11,12-EET-PFB and ( ± )14,15-EET-PFB were 800, 100 and 100 fg on-column, respectively. Calibration curves were prepared in the range from 1 to 40 ng/mL (Fig. 2). Samples were stored in hexanes/2-propanol (95:5, v/v). Calibration curves for 8(S),9(R)-EET (y =0.408× – 0.0001; r2= 0.996), 8(R),9(S)-EET (y =0.4863× – 0.0002; r2 =1), 11(S),12(R)-EET (y =0.2283× – 0.0004; r2 =0.999), 11(R),12(S)-EET (y = 0.2247× – 0.0002; r2 =0.999), 14(R),15(S)-EET (y =0.2143× –0.0014; r2 =0.998) and 14(S),9(R)-EET (y=0.2227× – 0.0002; r2 =0.998) were fitted to a linear regression with a 1/x weighting.
Assay validation
Validations were performed on five replicate samples at the lower limit of quantification (LLOQ) of 0.02 ng, lower quality control (LQC) of 0.04 ng, middle quality control (MQC) of 0.2 ng, and high quality control (HQC) of 0.8 ng. Precision was between 2.0% and 14.7% and accuracy was between 91.8% and 107.8% for each individual enantiomer (Table 1).
Table 1.
Precision and accuracy of EETs analyses (n =5)
| Analyte | Parameter | LLOQ (ng) | LQC (ng) | MQC (ng) | HQC (ng) |
|---|---|---|---|---|---|
| 0.02 | 0.04 | 0.2 | 0.8 | ||
| 8(R),9(S)-EET | Mean (ng) | 0.02 | 0.04 | 0.203 | 0.86 |
| Precision (%) | 10.3 | 11.3 | 3.3 | 5.6 | |
| Accuracy (%) | 99.3 | 100.5 | 101.6 | 100.7 | |
| 8(S),9(R)-EET | Mean (ng) | 0.02 | 0.04 | 0.205 | 0.839 |
| Precision (%) | 14.8 | 12.1 | 7.3 | 8.9 | |
| Accuracy (%) | 98.2 | 100.4 | 102.7 | 104.8 | |
| 11(R),12(S)-EET | Mean (ng) | 0.021 | 0.038 | 0.208 | 0.863 |
| Precision (%) | 7.8 | 9.3 | 7.6 | 3.5 | |
| Accuracy (%) | 103.5 | 95.9 | 103.9 | 107.8 | |
| 11(S),12(R)-EET | Mean (ng) | 0.018 | 0.04 | 0.213 | 0.848 |
| Precision (%) | 8.8 | 6.6 | 6.4 | 2.7 | |
| Accuracy (%) | 91.8 | 99.7 | 106.3 | 106.1 | |
| 14(R),15(S)-EET | Mean (ng) | 0.021 | 0.04 | 0.220 | 0.860 |
| Precision (%) | 7.2 | 9.6 | 4.1 | 3.0 | |
| Accuracy (%) | 107.2 | 101.2 | 110.2 | 107.5 | |
| 14(S),15(R)-EET | Mean (ng) | 0.02 | 0.042 | 0.198 | 0.790 |
| Precision (%) | 14.7 | 2.0 | 2.4 | 6.8 | |
| Accuracy (%) | 101.7 | 105.6 | 99.1 | 98.8 | |
Analysis of EET enantiomers in supersomes treated with AA
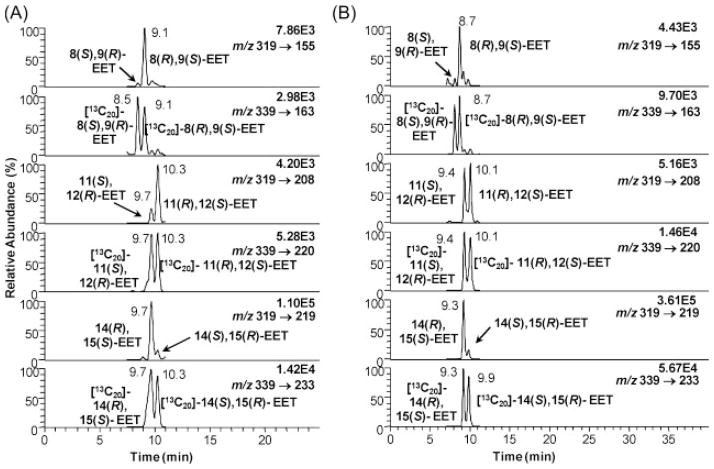
Recombinant supersomes containing appropriate P450s were treated with 5 μM AA for 30 min after adding the NADPH. The control experiments were the exact same amounts minus the NADPH. There was a striking difference between hP4502C19 and hP4502D6 (Fig. 7, Table 2). 14(R),15(S)-EET was formed in 96% ee in hP4502C19, whereas 14(S),15(R)-EET was formed in 96% ee in hP4502D6. hP4502C19 and 2D6 were also very enantioselective for 8(R),9(S)-EET formation (Figs. 7(A) and 7(B), Table 2). However, hP4502C19-mediated formation of 11,12-EET was almost racemic (Fig. 7(A)), whereas hP4502D6 was highly enantioselective for the formation of 11(R),12(S)-EET (Fig. 7(B)). hP4501A1 and rP4501A1 gave a remarkably similar pattern of EET metabolites (Fig. 8). Enantioselective formation of 8(R),9(S)-EET (Fig. 8) was similar to hP4502D6 and 2C19 (Fig. 7), whereas enantioselective formation of 14(R),15(S)-EET (Fig. 8) and racemic 11,12-EET (Fig. 8) was similar to hP4502C19 (Fig. 7(A)) and contrasted with the enantioselective formation of 14(S),15(R)-EET and 11(R),12(S)-EET by hP4502D6 (Fig. 7(B)). rP4501B1 formed 8(R),9(S)-EET and racemic 11,12-EET in a similar manner to P4502C19 but preferentially formed 14(S),15(R)-EET in a manner analogous to hP4502D6 (Table 2).
Figure 7.
Enantioselective biosynthesis of EETs by P450 family 2 isoforms: (A) hP4502C19 and (B) hP4502D6.
Table 2.
Enantiomeric composition of each EET regioisomer formed through metabolism of AA by recombinant P450-containing supersomes
| 8(S),9(R)-EET (%) | 8(R),9(S)-EET (%) | 11(S),12(R)-EET (%) | 11(R),12(S)-EET (%) | 14(R),15(S)-EET (%) | 14(S),15(R)-EET (%) | |
|---|---|---|---|---|---|---|
| hP4501A1 | 2 | 98 | 13 | 87 | 90 | 10 |
| hP4501B1 | 4 | 96 | 47 | 53 | 92 | 8 |
| rP4501A1 | 2 | 98 | 49 | 51 | 90 | 10 |
| rP4501B1 | 2 | 98 | 50 | 50 | 25 | 75 |
| hP4502C19 | 2 | 98 | 40 | 60 | 96 | 4 |
| hP4502D6 | 0 | 100 | 1 | 99 | 4 | 96 |
Figure 8.
Enantioselective biosynthesis of EETs by P450 family 1 isoforms:(A) hP4501A1 and (B) rP4501A1.
Quantification of EETs from AA-treated Hepa cells
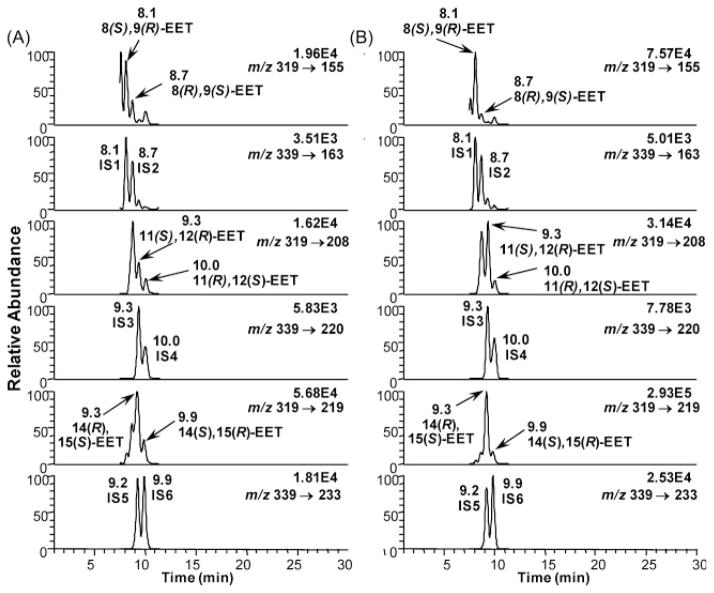
Hepa cells were treated with 10 μM AA before or after pre-treatment with 5 nM TCDD. The total amount of EETs (esterified and free) was determined by LC/ECAPCI-MRM-MS. Enantioselective formation of 8(S),9(R)-EET, 11(S),12(R)-EET, and 14(R),15(S)-EET was observed (Fig. 9). From the chromatograms of the Hepa cells (Fig. 9) one can appreciate the usefulness of the [13C20]-internal standards. In the channel corresponding to 8,9-EET (m/z 319 to 155) (Fig. 9(A)), there were two intense peaks, that could have been easily mistaken for both enantiomers of the 8,9-EET, if the corresponding [13C20]-internal standards had not shown that only one of the intense peaks was indeed the 8(S),9(R)-EET. The other enantiomer appeared as a much smaller peak, corresponding to the enantioselective formation of 8(R),9(S)-EET. The [13C20]-internal standards proved even more useful for the correct assignment of 11,12-EET enantiomers (Fig. 9(A)). In the channel m/z 319 to 208 there were three peaks, and the most intense one did not correspond to any of the peaks of the internal standard, so the two lower intensity peaks were assigned as 11(S),12(R)-EET and 11(R),12(S)-EET, respectively. In the case of the 14,15-EET enantiomers, the [13C20]-internal standards showed the enantiomeric formation of 14(R),15(S)-EET (Fig. 9(A)). In Fig. 9(B), the enantiomer assignments were easier since increased amounts were formed after TCDD induction, but using the labeled internal standards, it was possible to unequivocally quantify the 14(R),15(S)-EET. When the cells were treated with 10 μM AA for 1 h the amounts of 8(S),9(R)-EET and 8(R),9(S)-EET were 0.53 ± 0.71 ng and 0.26 ± 0.01 ng/106 cells (Fig. 10(A)). The amounts of 11(S),12(R)-EET and 11(R),12(S)-EET were 0.25 ± 0.09 ng and 0.2 ± 0.05 ng/106 cells (Fig. 10(B)). The amounts of 14(R),15(S)-EET and 14(S),15(R)-EET were 1.25 ± 0.09 ng and 0.2 ± 0.02 ng/106 cells (Fig. 10(C)). After 4 h treatment, the amounts of 8(S),9(R)-EET and 8(R),9(S)-EET increased to 0.95 ± 0.08 ng and 0.45 ± 0.09 ng/106 cells. The amounts of 11(S),12(R)-EET and 11(R),12(S)-EET were 0.5 ± 0.06 ng and 0.4 ± 0.06 ng/106 cells. The amounts of 14(R),15(S)-EET and 14(S),15(R)-EET were 2.5 ± 0.14 ng and 0.4 ± 0.1 ng/106 cells.
Figure 9.
LC/MRM-MS chromatograms of EET-PFB in Hepa cells and their corresponding 13C-labeled internal standards: (A) 1 h treatment with 10 μM AA without TCDD induction and (B) 1 h treatment with 10 μM AA after 6 h TCDD (5 nM) induction. Abbreviations: IS, internal standard; IS1, [13C20]-8(S),9(R)-EET-PFB; IS2, [13C20]-8(R),9(S)-EET-PFB; IS3, [13C20]-11(S)129(R)-EET-PFB; IS4, [13C20]-11(R),12(S)-EET-PFB; IS5, [13C20]-14(R),15(S)-EET-PFB; IS6, [13C20]-14(S),15(R)-EET-PFB.
Figure 10.
EET formation in Hepa cells treated with 10 μM of arachidonic acid with or without 5 nM TCDD induction.
TCDD pre-treatment of the Hepa cells also resulted in enantioselective formation of the EETs. The 1 h treatment with AA gave: 8(S),9(R)-EET and 8(R),9(S)-EET 1.5 ± 0.17 ng and 0.3 ± 0.04 ng/106 cells. The amounts of 11(S),12(R)-EET and 11(R),12(S)-EET were 0.9 ± 0.05 ng and 0.2 ± 0.03 ng/106 cells. The amounts of 14(R),15(S)-EET and 14(S),15(R)-EET were 3.2 ± 0.07 ng and 0.5 ± 0.3 ng/106 cells. The 4 h treatment with AA gave: 8(S),9(R)-EET and 8(R),9(S)-EET 1.5 ± 0.17 ng and 0.3 ± 0.04 ng/106 cells. The amounts of 11(S),12(R)-EET and 11(R),12(S)-EET were 1 ± 0.02 ng and 0.4 ± 0.06 ng/106 cells. The amounts of 14(R),15(S)-EET and 14(S),15(R)-EET were 4.9 ± 0.02 ng and 0.6 ± 0.08 ng/106 cells. Therefore, the predominant isomer with or without TCDD induction was 14(R),15(S)-EET (Fig. 10(C)). The second most abundant regioisomer was 8,9-EET (Fig. 10(A)) with the 8(S),9(R)-EET enantiomer being formed preferentially. The amounts of 11,12-EETs (Fig. 10(B)) were almost eight times lower than 14,15-EET (Fig. 10(C)). TCDD alone did not induce significant amounts of EET formation (Fig. 10).
DISCUSSION
There was a striking difference in the enantioselectivity of 14,15-EET formation between P450 2C19 and P450 2D6. 14(R),15(S)-EET was formed with a high ee by P4502C19, whereas 14(S),15(R)-EET was the predominant enantiomer formed from P4502D6 (Fig. 7). As expected, hP4501A1 and rP4501A1 had similar enantioselectivity, converting AA primarily into the 14(R),15(S)-EET (Fig. 8). P450-mediated metabolism of AA by mouse Hepa cells also resulted in the formation of the EETs with high regioselectivity for 14(R),15(S)-EET (Figs. 9 and 10). Hepa cells constitutively express CYP1A1 and CYP1B1,50 and so a predominance of the 14(R),15(S)-EET would have been predicted from the supersome studies (Fig. 8, Table 2). Up-regulation of these P450s would also be expected to increase the amounts of EETs that are formed from AA.
TCDD is a polycyclic aromatic hydrocarbon that binds to the aryl hydrocarbon receptor (AhR), translocates into the nucleus, and up-regulates P4501A1 and 1B1 expression.51,52 Therefore, TCDD pre-treatment of the Hepa cells resulted in formation of almost twice the amount of EETs when compared with non-induced cells at both 1 h and 4 h (Fig. 10). Furthermore, the enantioselectivity of 14,15-EET formation was preserved after TCDD induction (Fig. 10(C)), which provided additional confirmation that rodent P4501A1 and 1B1 were responsible for its formation. Surprisingly, 8(S),9(R)-EET was the major AA-derived 8,9-EET in both the non-induced and TCDD-induced Hepa cells (Fig. 10(A)). None of the P450s that were tested produced significant quantities of this enantiomer (Table 2), which has been shown previously to be a major metabolite of the rat cortex.53 This suggests that there is another P450 in the mouse Hepa cell line, which is responsible for the formation of 8(S),9(R)-EET. Interestingly, the 8(S),9(R)-EET enantiomer has potent vasoactive properties and undergoes COX-mediated metabolism to a potent mitogen for mesangial cells.54 The low abundance of the 8(R),9(S)-EET in the TCDD-induced cells at 1 h and 4 h (Fig. 10(A)), a significant product of both rP4501A1 and 1B1 (Fig. 8, Table 2), suggests that preferential hydrolysis of this EET enantiomer11 could have occurred as a result of TCDD treatment. Recent studies have suggested that TCDD-mediated up-regulation of antioxidant genes such as epoxide hydrolase could occur through an interaction between the Ahr and Nrf-2.55
11,12-EET, a minor product of AA metabolism of P4501A1 and 1B1 in the supersomes (Fig. 8, Table 2), was also the least abundant product in the Hepa cell incubations (Fig. 10(B)). The expected racemic 11,12-EET was observed in the non-induced cells, whereas TCDD induction caused an apparent selective induction of 11(S),12(R)-EET formation (Fig. 10(B)). However, this could have been due to selective hydrolysis of the 11(R),12(S)-EET isomer as suggested above for 8(R),9(S)-EET. Taken together, these data suggest that P4501A1- and 1B1-mediated AA metabolism can produce significant quantities of EETs in addition to the widely recognized P450s of the 2 family. Finally, it is evident that the chiral LC/ECAPCI-MS method described above will provide an excellent basis for the quantification of enantiomeric EETs in urine and tissue samples.
CONCLUSIONS
We have developed a stable isotope dilution chiral phase LC/MRM-MS assay for all six enantiomers corresponding to the three stable regioisomeric EETs. The use of [13C20]-internal standards for each of the EET enantiomers overcame the problems encountered with deuterium-labeled standards, which separated from the corresponding protium forms. The new method made it possible to assess the enantioselectivity of AA-derived EETs from individual P450s and to quantify the individual enantiomers in AA-treated cultured cell lines.
Acknowledgments
We gratefully acknowledge the support of grants UO1ES016004 and P30ES0135080 from the National Institutes of Health.
References
- 1.Guengerich FP. Drug Metab Rev. 2004;36:159. doi: 10.1081/dmr-120033996. [DOI] [PubMed] [Google Scholar]
- 2.Capdevila JH, Falck JR, Imig JD. Kidney Int. 2007;72:683. doi: 10.1038/sj.ki.5002394. [DOI] [PubMed] [Google Scholar]
- 3.Guengerich FP. Chem Res Toxicol. 2008;21:70. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 4.Bylund J, Kunz T, Valmsen K, Oliw EH. J Pharmacol Exp Ther. 1998;284:51. [PubMed] [Google Scholar]
- 5.Hsu MH, Savas U, Griffin KJ, Johnson EF. Drug Metab Rev. 2007;39:515. doi: 10.1080/03602530701468573. [DOI] [PubMed] [Google Scholar]
- 6.Capdevila JH, Falck JR, Estabrook RW. FASEB J. 1992;6:731. doi: 10.1096/fasebj.6.2.1537463. [DOI] [PubMed] [Google Scholar]
- 7.Harder DR, Gebremedhin D, Narayanan J, Jefcoat C, Falck JR, Campbell WB, Roman R. Am J Physiol. 1994;266:H2098. doi: 10.1152/ajpheart.1994.266.5.H2098. [DOI] [PubMed] [Google Scholar]
- 8.Schwartzman ML, da Silva JL, Lin F, Nishimura M, Abraham NG. Nephron. 1996;73:652. doi: 10.1159/000189154. [DOI] [PubMed] [Google Scholar]
- 9.Fulton D, Falck JR, McGiff JC, Carroll MA, Quilley J. J Lipid Res. 1998;39:1713. [PubMed] [Google Scholar]
- 10.Chacos N, Capdevila J, Falck JR, Manna S, Martin-Wixtrom C, Gill SS, Hammock BD, Estabrook RW. Arch Biochem Biophys. 1983;223:639. doi: 10.1016/0003-9861(83)90628-8. [DOI] [PubMed] [Google Scholar]
- 11.Zeldin DC, Wei S, Falck JR, Hammock BD, Snapper JR, Capdevila JH. Arch Biochem Biophys. 1995;316:443. doi: 10.1006/abbi.1995.1059. [DOI] [PubMed] [Google Scholar]
- 12.Spearman ME, Prough RA, Estabrook RW, Falck JR, Manna S, Leibman KC, Murphy RC, Capdevila J. Arch Biochem Biophys. 1985;242:225. doi: 10.1016/0003-9861(85)90496-5. [DOI] [PubMed] [Google Scholar]
- 13.Smith HE, Jones JP, III, Kalhorn TF, Farin FM, Stapleton PL, Davis CL, Perkins JD, Blough DK, Hebert MF, Thummel KE, Totah RA. Pharmacogenet Genomics. 2008;18:943. doi: 10.1097/FPC.0b013e32830e1e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaspera R, Totah RA. Expert Opin Drug Metab Toxicol. 2009;5:771. doi: 10.1517/17425250902932923. [DOI] [PubMed] [Google Scholar]
- 15.Spector AA. J Lipid Res. 2009;50(Suppl):S52. doi: 10.1194/jlr.R800038-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capdevila JH, Wei S, Yan J, Karara A, Jacobson HR, Falck JR, Guengerich FP, DuBois RN. J Biol Chem. 1992;267:21720. [PubMed] [Google Scholar]
- 17.Karara A, Dishman E, Blair I, Falck JR, Capdevila JH. J Biol Chem. 1989;264:19822. [PubMed] [Google Scholar]
- 18.Karara A, Dishman E, Jacobson H, Falck JR, Capdevila JH. FEBS Lett. 1990;268:227. doi: 10.1016/0014-5793(90)81014-f. [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Moomaw CR, Tomer KB, Falck JR, Zeldin DC. J Biol Chem. 1996;271:3460. doi: 10.1074/jbc.271.7.3460. [DOI] [PubMed] [Google Scholar]
- 20.Capdevila JH, Dishman E, Karara A, Falck JR. Methods Enzymol. 1991;206:441. doi: 10.1016/0076-6879(91)06113-h. [DOI] [PubMed] [Google Scholar]
- 21.Harder DR, Campbell WB, Roman RJ. J Vasc Res. 1995;32:79. doi: 10.1159/000159080. [DOI] [PubMed] [Google Scholar]
- 22.Roman RJ. Physiol Rev. 2002;82:131. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- 23.Campbell WB. Trends Pharmacol Sci. 2000;21:125. doi: 10.1016/s0165-6147(00)01472-3. [DOI] [PubMed] [Google Scholar]
- 24.Fleming I. Trends Pharmacol Sci. 2007;28:448. doi: 10.1016/j.tips.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Node K, Huo Y, Ruan X, Yang B, Spiecker M, Ley K, Zeldin DC, Liao JK. Science. 1999;285:1276. doi: 10.1126/science.285.5431.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spector AA, Norris AW. Am J Physiol Cell Physiol. 2007;292:C996. doi: 10.1152/ajpcell.00402.2006. [DOI] [PubMed] [Google Scholar]
- 27.Davis BB, Thompson DA, Howard LL, Morisseau C, Hammock BD, Weiss RH. Proc Natl Acad Sci USA. 2002;99:2222. doi: 10.1073/pnas.261710799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nieves D, Moreno JJ. Apoptosis. 2007;12:1979. doi: 10.1007/s10495-007-0123-3. [DOI] [PubMed] [Google Scholar]
- 29.Dhanasekaran A, Gruenloh SK, Buonaccorsi JN, Zhang R, Gross GJ, Falck JR, Patel PK, Jacobs ER, Medhora M. Am J Physiol Heart Circ Physiol. 2008;294:H724. doi: 10.1152/ajpheart.00979.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C, Li G, Liao W, Wu J, Liu L, Ma D, Zhou J, Elbekai RH, Edin ML, Zeldin DC, Wang DW. J Pharmacol Exp Ther. 2009;329:908. doi: 10.1124/jpet.109.152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, El-Sikhry H, Chaudhary K, Batchu SN, Shayeganpour A, Jukar TO, Bradbury JA, Graves JP, Degraff LM, Myers PH, Rouse DC, Foley J, Nyska A, Zeldin DC, Seubert JM. Am J Physiol Heart Circ Physiol. 2009;297:H37. doi: 10.1152/ajpheart.00983.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fitzpatrick FA, Ennis MD, Baze ME, Wynalda MA, McGee JE, Liggett WF. J Biol Chem. 1986;261:15334. [PubMed] [Google Scholar]
- 33.Capdevila JH, Kishore V, Dishman E, Blair IA, Falck JR. Biochem Biophys Res Commun. 1987;146:638. doi: 10.1016/0006-291x(87)90576-6. [DOI] [PubMed] [Google Scholar]
- 34.Hammonds TD, Blair IA, Falck JR, Capdevila JH. Anal Biochem. 1989;182:300. doi: 10.1016/0003-2697(89)90598-8. [DOI] [PubMed] [Google Scholar]
- 35.Vander Noot VA, Van Rollins M. Anal Chem. 2002;74:5866. [Google Scholar]
- 36.Zhang JY, Blair IA. J Chromatogr B Biomed Appl. 1994;657:23. doi: 10.1016/0378-4347(94)80065-0. [DOI] [PubMed] [Google Scholar]
- 37.Wei S, Brittin JJ, Falck JR, Anjaiah S, Nithipatikom K, Cui L, Campbell WB, Capdevila JH. Anal Biochem. 2006;352:129. doi: 10.1016/j.ab.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Bylund J, Ericsson J, Oliw EH. Anal Biochem. 1998;265:55. doi: 10.1006/abio.1998.2897. [DOI] [PubMed] [Google Scholar]
- 39.Kiss L, Roder Y, Bier J, Weissmann N, Seeger W, Grimminger F. Anal Bioanal Chem. 2008;390:697. doi: 10.1007/s00216-007-1718-9. [DOI] [PubMed] [Google Scholar]
- 40.Singh G, Gutierrez A, Xu K, Blair IA. Anal Chem. 2000;72:3007. doi: 10.1021/ac000374a. [DOI] [PubMed] [Google Scholar]
- 41.Lee SH, Williams MV, DuBois RN, Blair IA. Rapid Commun Mass Spectrom. 2003;17:2168. doi: 10.1002/rcm.1170. [DOI] [PubMed] [Google Scholar]
- 42.Lee SH, Williams MV, Blair IA. Prostaglandins Other Lipid Mediat. 2005;77:141. doi: 10.1016/j.prostaglandins.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Lee SH, Blair IA. BMB Rep. 2009;42:401. doi: 10.5483/bmbrep.2009.42.7.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ken Lie, Jie MS, Lau MM, Lam CN, Alam MS, Metzger JO, Biermann U. Chem Phys Lipids. 2003;125:93. doi: 10.1016/s0009-3084(03)00071-9. [DOI] [PubMed] [Google Scholar]
- 45.Folch J, Lees M, Sloane Stanley GH. J Biol Chem. 1957;226:497. [PubMed] [Google Scholar]
- 46.Bernstrom K, Kayganich K, Murphy RC. Anal Biochem. 1991;198:203. doi: 10.1016/0003-2697(91)90530-7. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura T, Bratton DL, Murphy RC. J Mass Spectrom. 1997;32:888. doi: 10.1002/(SICI)1096-9888(199708)32:8<888::AID-JMS548>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 48.Murphy RC, Barkley RM, Zemski BK, Hankin J, Harrison K, Johnson C, Krank J, McAnoy A, Uhlson C, Zarini S. Anal Biochem. 2005;346:1. doi: 10.1016/j.ab.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 49.Zeldin DC, Moomaw CR, Jesse N, Tomer KB, Beetham J, Hammock BD, Wu S. Arch Biochem Biophys. 1996;330:87. doi: 10.1006/abbi.1996.0229. [DOI] [PubMed] [Google Scholar]
- 50.Eltom SE, Zhang L, Jefcoate CR. Mol Pharmacol. 1999;55:594. [PubMed] [Google Scholar]
- 51.Whitlock JP., Jr Chem Res Toxicol. 1993;6:754. doi: 10.1021/tx00036a003. [DOI] [PubMed] [Google Scholar]
- 52.Denison MS, Fisher JM, Whitlock JP., Jr Proc Natl Acad Sci USA. 1988;85:2528. doi: 10.1073/pnas.85.8.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh T, Takahashi K, Capdevila J, Karara A, Falck JR, Jacobson HR, Badr KF. Am J Physiol. 1991;261:F578. doi: 10.1152/ajprenal.1991.261.4.F578. [DOI] [PubMed] [Google Scholar]
- 54.Homma T, Zhang JY, Shimizu T, Prakash C, Blair IA, Harris RC. Biochem Biophys Res Commun. 1993;191:282. doi: 10.1006/bbrc.1993.1214. [DOI] [PubMed] [Google Scholar]
- 55.Yeager RL, Reisman SA, Aleksunes LM, Klaassen CD. Toxicol Sci. 2009;111:238. doi: 10.1093/toxsci/kfp115. [DOI] [PMC free article] [PubMed] [Google Scholar]