Abstract
Embryonic stem cells (ESCs) are defined by two essential features - pluripotency and self-renewal - whose balance requires the concerted action of signal transduction pathways, transcription factor networks, and epigenetic regulators. Recent findings have implicated the NuRD chromatin remodeling complex in the sophisticated choreography of ESC regulatory pathways.
Embryonic stem cells (ESCs) face a special regulatory challenge essential to their unique biological properties. These cells must maintain the capacity to self renew, while having the potential, upon receiving appropriate physiologic cues, to differentiate along diverse lineages. These properties imply an ability to maintain expression of genes integral to the pluripotency program in such a manner that they can be rapidly silenced upon receiving a signal to differentiate. Likewise, genes involved in embryonic development must be silent, with the potential for rapid activation.
The chromatin status of ESCs reflects their unique physiologic needs. ESCs have a significant number of genes that are characterized by very specific and unique histone and DNA modifications, and are enriched for essential genes in early embryonic development and developmental regulatory transcription factors (Meissner, 2010). The promoters of these genes are characterized by the simultaneous presence, under certain growth conditions, of histone modifications with seemingly opposite functions. Individual nucleosomes at these promoters exhibit trimethylation of two different residues of histone H3: lysine 4 (H3K4me3) and lysine 27 (H3K27me3). While H3K4me3 is enriched at promoters of actively transcribing genes or genes with the potential for active transcription, H3K27me3 is associated with stable gene silencing. Loci at which these two histone marks are juxtaposed on the same nucleosome are referred to as bivalent (Meissner, 2010). Bivalent genes are generally transcriptionally silent in ESCs, but are poised for rapid activation in the appropriate lineages during embryonic development. In addition, these bivalent promoters are characterized by the presence of a newly-described form of DNA modification, 5-hydroxymethyl cytosine. How the enzymes that deposit these chromatin marks interact with the core pluripotency network to regulate the balance between self-renewal and differentiation remains a central puzzle in ESC biology. A recent set of papers (Reynolds, 2012a; Whyte et al., 2012; Yildirim et al., 2011) has provided fresh insights into mechanisms underlying the regulation of the pluripotency program and its response to developmental cues, indicating a central role for the nucleosome remodeling and deacetylase (NuRD) complex in this complicated regulatory process.
The NuRD chromatin remodeling complex was defined biochemically more than a decade ago (Ramirez and Hagman, 2009). At the time of its discovery, this complex was unique in that it possessed two distinct enzymatic functions directed at chromatin-dependent gene regulation – histone deacetylase function from the HDAC1 and HDAC2 subunits and ATP-dependent chromatin remodeling, a property of the Mi-2α/β subunits. Further, NuRD contains a member of the methyl-CpG binding domain family of proteins, MBD3, although mammalian MBD3 lacks the capacity to bind methylated DNA substrates with high affinity. At the time of its discovery more than a decade ago, prevailing models predicted that this complex was recruited to promoters through direct interactions with DNA sequence-specific transcriptional repressors where its enzymatic functions – histone deacetylation and chromatin remodeling – were integral to the process of stable gene silencing (Ramirez and Hagman, 2009). This classic, static model for NuRD function is now called into question by a very recent set of papers describing a much more dynamic role for NuRD in maintenance of gene activity in ESCs (Reynolds, 2012a; Whyte et al., 2012; Yildirim et al., 2011).
NuRD and polycomb group genes antagonize LIF/Stat3 signaling in ESC differentiation
Elegant genetic analysis by Hendrich and colleagues defined an essential role for MBD3 in integrity of the NuRD complex in murine ESCs (MBD3 null ESCs fail to form functional NuRD complex) and, surprisingly, demonstrated that deletion of MBD3 and disruption of the NuRD complex did not significantly impact ESC self-renewal (Kaji et al., 2007). However, MBD3 deletion led to defects in morphological changes, down-regulation of ESC markers, and up-regulation of lineage markers upon LIF-withdrawal or during embryoid body formation, indicating that the NuRD complex is required for normal differentiation. In vivo, MBD3 null ESCs fail to make significant contributions to chimeric embryos and can block the ability of wild-type morulae to form functional embryos.
One possible mechanism by which the NuRD complex can regulate differentiation is through the proper silencing of pluripotency genes. In this issue of Cell Stem Cell, Reynolds et al (2012a) demonstrated using genetically manipulated ESCs that loss of MBD3 leads to aberrant upregulation of genes integral to the pluripotency network (including Zfp42, Tbx3, Klf4 and Klf5) under normal growth conditions (Figure 1A), although expression of the core pluripotency transcription factors Oct4, Sox2 and Nanog are not affected. MBD3 deletion resulted in their sustained expression upon LIF withdrawal, and knocking-down one of them, Klf4, could partially rescue the differentiation defects. Chromatin immunoprecipitation visualized by quantitative PCR demonstrated that MBD3 and Mi-2β both occupy the promoters and gene bodies of these pluripotency genes in a broad pattern stretching across several kilobases of DNA (Figure 2B). These findings are consistent with the LIF-independent ESC maintenance phenotype observed in the MBD3 null cells, as Tbx3, Klf4, and Klf5 act downstream in the LIF-Stat3 pathway (Niwa et al., 2009).
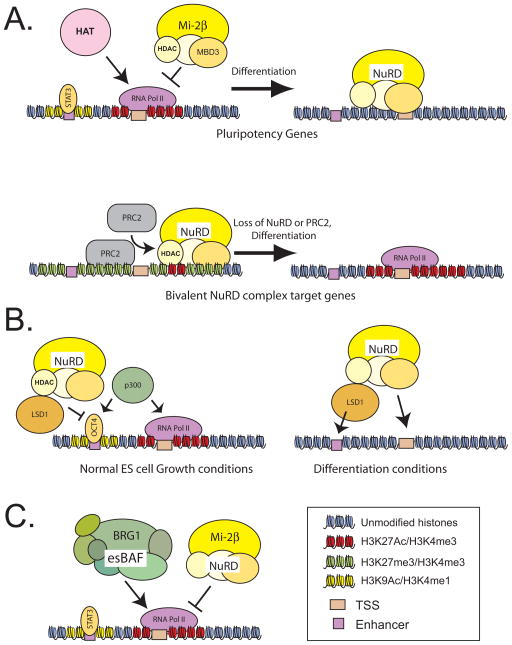
Figure 1. NuRD complex opposes histone acetyltransferases in dynamic regulation of the ESC transcriptional program.
A. A dynamic model for NuRD complex function based on the work from Hendrich and colleagues (Reynolds, 2012a; Reynolds et al., 2012b). At genes integral to the pluripotency program, histone acetyltransferase (HAT) enzymes function as chromatin regulators promoting gene activity. NuRD complex, through its histone deacetylase subunits (HDAC), opposes their function and serves to dampen expression. Balance between the activating functions of HAT enzymes and the repressive HDAC activity of NuRD fine tunes expression of these genes. At bivalent genes that are poised for activation during embryonic development, the HDAC functions of NuRD are required for stable association of PRC2, and maintenance of H3K27me3.
B. In the work of (Whyte et al. (2012), NuRD complex HDAC activity acts in competition with p300 or other HAT enzymes at active enhancers in pluripotent cells to maintain a balance of histone acetylation under normal ESC growth conditions. Under conditions favoring differentiation, NuRD deacetylates histone H3K9, making this histone a substrate for LSD1 to demethylate H3K4, a necessary step in turning off an enhancer occupied by the core pluripotency network. Loss of enhancer activity results in downregulation of transcription and, ultimately, in gene silencing.
C. Yildrim et al. (2011) depict NuRD complex in competition with the esBAF complex at highly transcribed genes, including genes downstream of LIF-Stat signaling. These two chromatin regulators act in an opposing fashion to regulate nucleosome occupancy through the ATPase subunits BRG1 (esBAF) and Mi-2β (NuRD). The balance of these activities at core promoters serves to fine tune RNA polymerase recruitment.
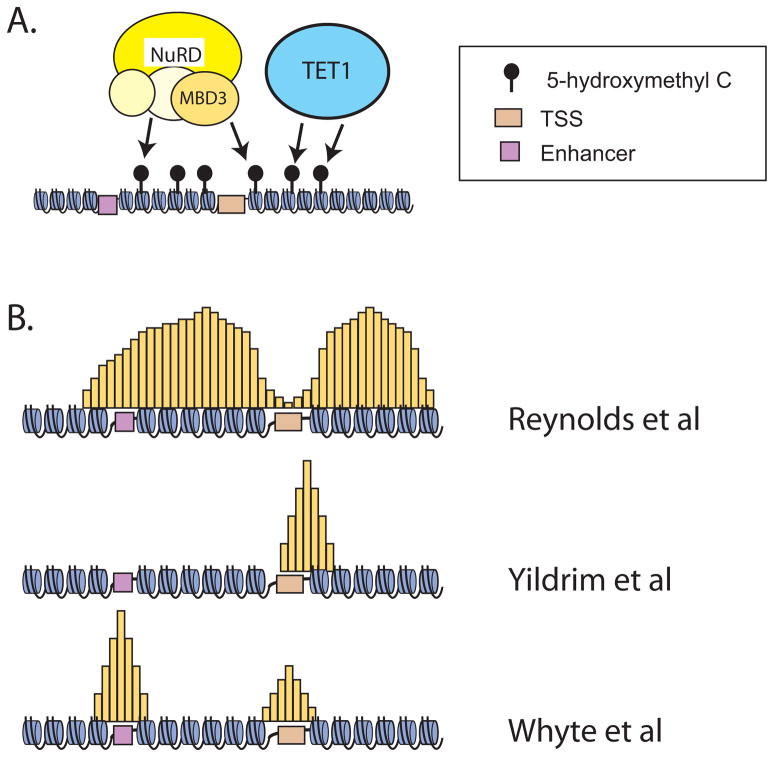
Figure 2. NuRD complex action and distribution in the ESC genome.
A. Fazzio and colleagues provide evidence suggesting that NuRD complex localizes to specific regions of the genome through direct interaction of MBD3 with 5hmC modified DNA, a mark deposited by TET1.
B. ChIP PCR (Reynolds et al., 2012a) and ChIP-Seq (Reynolds et al., 2012b) depict (yellow shaded areas) broad distribution of NuRD complex across kilobases of DNA surrounding the TSS of pluripotency genes. Note that this model depicts a gap in NuRD complex enrichment over the TSS.
ChIP-Seq (Yildrim et al., 2011) determination of focal localization of NuRD complex in ESCs (yellow shaded areas). Strongest peaks were detected slightly downstream of the TSS, little to no enrichment was noted at enhancers.
ChIP-Seq (Whyte et al., 2012) of NuRD complex in ESCs (yellow shaded areas) in focal areas of enrichment coincident with both enhancers and core promoters. Enrichment at enhancers was noted in peaks larger than those detected near the TSS.
It was previously known that the expression of Zfp42, Tbx3, Klf4, and Klf5 is heterogeneous in ESCs cultured in serum and LIF (Toyooka et al., 2008), and that there are sub-populations of cells expressing low or high levels of these proteins. In the absence of MBD3, however, the low-expressing cell population disappeared and all the cells express higher levels, suggesting that NuRD is responsible for the repression of these genes during normal ESC culture (Reynolds et al., 2012a). These findings support a model in which a balance between transcriptional activation from the LIF-Stat3 pathway and repression from the NuRD complex is in dynamic equilibrium at a subset of pluripotency genes in ESCs (Figure 1A) resulting in the observed transcriptional heterogeneity. By extension, NuRD-mediated repression of these same pluripotency genes is likely a necessary step for differentiation. It is important to note that NuRD-mediated repression does not operate to silence all pluripotency genes. For example, MBD3 deletion does not impact expression of the core pluripotency transcription factors Nanog, Oct4 and Sox2 (Reynolds et al., 2012a). Additionally, Nanog is also known to be expressed in a heterogeneous fashion in ESCs cultured in serum and LIF (Chambers et al., 2007). Its expression is controlled by an allelic regulatory mechanism independent of DNA methylation (Miyanari and Torres-Padilla, 2012).
In another very recent study from the same group, an important relationship was established between NuRD and another chromatin modifying complex, PRC2 (Reynolds et al., 2012b). PRC2, polycomb repressive complex 2, is responsible for the repressive histone mark H3K27me3 which is deposited on chromatin by its subunit EZH2 (Meissner, 2010). NuRD was shown to occupy genomic loci corresponding to genes marked by H3K4 trimethylation as well as bivalent genes in ESCs, and may thereby modulate actively transcribed genes as well as those marked by bivalent chromatin for eventual expression at a later point in development. MBD3 deletion led to an increase in H3K27 acetylation and a loss of H3K27 trimethylation at the bivalent NuRD target genes, suggesting a concerted action between NuRD (deacetylating lysine 27) and polycomb complexes – specifically PRC2 which methylates lysine 27 - at these sites (Figure 1A). Indeed, MBD3 deletion or HDAC inhibition impaired the binding of PRC2 components to some of the NuRD target genes. On the contrary, deletion of PRC2 components did not affect binding of the NuRD complex to its target genes. Thus, recruitment of PRC2 at NuRD target loci is dependent on the presence and activity of NuRD. Consistently, deletion or knockdown of MBD3 or components of the PRC complexes induced overlapping gene expression changes, including the up-regulation of several genes involved in embryonic development (Reynolds et al., 2012b).
Taken together, the above findings supported the following model for NuRD’s function in ESC differentiation (Figure 1A): MBD3/NuRD deacetylates histone lysine residues (including H3K27) at genes involved in embryonic development or pluripotency. At some genes (those downstream of LIF or of the core pluripotency network), the continued action of histone acetyltransferase (HAT) enzymes rapidly resets acetylation state, resulting in an equilibrium favoring active transcription. At other genes (genes involved in embryonic development), the hypoacetylated state at lysine 27 resulting from NuRD-dependent deacetylation is permissive for PRC2 recruitment, H3K27 trimethylation, and transcriptional repression. Thus, NuRD-mediated deacetylation serves as a brake to fine-tune the expression of pluripotency genes, and provides a mechanism for maintenance of genes involved in embryonic development in a poised or silent state. In agreement with this model, PRC2 has been shown to play important roles in ESC differentiation. Loss of PRC2 function in ESCs results in mis-regulation of pluripotency and developmental genes and defects in differentiation (Table 1).
Table 1.
| Complex | Gene | ES phenotype | Mouse phenotype | Reference |
|---|---|---|---|---|
| NuRD | MBD3 | Mbd3 null ESCs are viable and can initiate differentiation in embryoid bodies or chimeric embryos, but cannot completely silence pluripotency genes or commit to developmental lineages | Mbd3 null mice die around midgestation. Null embryos have defective epiblast expansion and extraembryonic tissue development starting at E5.5 | (Kaji et al., 2006) (Kaji et al., 2007) |
| PRC2 | Eed | Eed null ESCs are viable and can differentiate in embryoid bodies. They can contribute to chimeras (up to E9.5) | Eed null mice are embryonic lethal by E9.5. Null embryos exhibit growth defects and primitive streak formation and/or organization | (Chamberlain et al., 2008) (Leeb et al., 2010) |
| Ezh2 | N/A | Ezh2 null mice arre embryonic lethal before E8.5. Null embryos can initiate but cannot complete gastrulation. | (O'Carroll et al., 2001) | |
| Suz12 | Suz12 null cells are viable but have impaired differentiation capacity. | Suz12 null mice are embryonic lethal between E8.5 to 10.5. Null embryos show gastrulation defects that induce a developmental block around E7.5. | (Pasini et al., 2007) | |
| Tet1 | Tet1 | Tet1 null ESCs have reduced 5hmC level and subtle changes in gene expression. They can self-renew and are pluripotent with skewed differentiation toward trophectoderm in vitro. | Tet1 null mice are viable, fertile, and grossly normal, though some have a slightly smaller body size at birth. | (Dawlaty et al., 2011) |
| esBAF | Brg1 | Brg1-null ESCs show defects in self-renewal and pluripotency. | Brg1 null mice are embryonic lethal at the periimplantation stage | (Bultman et al., 2000) (Ho et al., 2011) |
| LSD1 | LSD1 | Lsd1 null ESCs have growth defects, cannot form embryoid bodies, and cannot properly silence ESC genes during differentiation. | Lsd1 null mice are embryonic lethal before E7.5 with arrested embryonic development at or before E5.5. The egg cylinder failed to elongate and undergo gastrulation. | (Wang et al., 2007) (Wang et al., 2009a) (Whyte et al., 2012) |
The genetic functional data presented by these two papers provides an attractive model to explain important aspects of the role of NuRD in the pluripotency program. It should be noted that, like all models, this one does not explain all the existing data. In particular, analysis of animals and cells genetically engineered at PRC2 genes suggests that the story may not be entirely this simple (Table 1). Animals null for the PRC2 components Suz12, Eed, and Ezh2 show developmental defects at slightly later stages than animals lacking MBD3. In these models, development fails around or after the time of gastrulation, suggesting that these animals have some capacity for differentiation. In ESCs, PRC2 deficiency can lead to a much less severe block on differentiation than loss of MBD3, as Eed or Suz12-null ESCs can still form embryoid bodies, and in some cases differentiate into three germ layers and contribute to chimeras (Table 1). These data suggest that a model with NuRD acting solely as a mediator for PRC2-induced repression does not explain all the requirements for NuRD in this process.
Maintaining pluripotency by repression of pluripotency genes
Repression of pluripotency genes is required for differentiation. Whyte et al recently reported that LSD1, a histone H3K4/K9 demethylase, is required for ESC differentiation by decommissioning, or silencing, ESC specific enhancers (Whyte et al., 2012). They found that inhibition of LSD1 in ESCs through RNA interference or pharmacologic methods prevented the normal silencing of pluripotency genes such as Nanog and Sox2 under differentiation conditions, without affecting the up-regulation of lineage markers. A genome-wide ChIP approach demonstrated that LSD1 occupies the enhancers and promoters of roughly 90% of actively transcribed and 2/3 of bivalent genes. Occupancy was noted at both promoter and enhancer regions, with peaks being higher at enhancer regions (Figure 2B). Focusing their attention on enhancer elements occupied by the core pluripotency transcription factors Oct4, Nanog and Sox2, the authors found that inhibition of LSD1 blocked the removal of H3K4me1 at LSD1 occupied enhancer regions during differentiation. Thus, LSD1 demethylates H3K4me1 at ESC-specific enhancers during differentiation to facilitate repression of pluripotency genes, defining LSD1 as essential for decommissioning enhancers. Partially consistent with these findings, another study in human ESCs also found that LSD1-bound genes are highly enriched for those with bivalent domains and those occupied by Oct4 and Nanog (Adamo et al., 2011). However, in this case LSD1 and its demethylase activity were required to maintain ESC self-renewal and suppress differentiation into meso-endodermal lineages, possibly by reducing H3K4 methylation at its target promoters. The precise reason(s) for these discrepancies remain unclear, although human and murine ESCs do represent different pluripotent stem cells and require different mechanisms for self-renewal.
Interestingly, Whyte et al showed that LSD1 physically interacts with and co-occupies enhancer regions with the NuRD complex in ESCs (of approximately 5500 genes bound by Mi-2β, 4800 are also bound by LSD1), suggesting that LSD1 and NuRD have broad functional overlap (Whyte et al., 2012). Consistent with that idea, biochemical and genomic association between LSD1 and NuRD had been previously described, albeit in a different biological context (Wang et al., 2009b). The authors proposed that NuRD complex and LSD1 function together at these enhancers to alter the local chromatin status and suppress enhancer function during the differentiation process (Figure 1B). Depletion of the core NuRD ATPase subunit, Mi2-β, resulted in defects in ESC differentiation in vitro and failure to downregulate a set of pluripotency genes in a manner very similar to loss of LSD1 function. Mechanistically, the authors invoke the enzymatic properties of LSD1 to explain its reliance on NuRD function – the enzyme has greatly reduced affinity for peptide substrates containing acetylation at lysine 9 of histone H3 (Forneris et al., 2005). Somewhat surprisingly, Whyte and colleagues fail to find significant colocalization of LSD1 with CoREST/REST proteins which have been previously tightly linked physically and functionally to LSD1 in other systems (Lee et al., 2005), including human ESCs (Adamo et al., 2011).
A common theme emerges from the above studies of NuRD, PRC2, and LSD1: dynamic equilibria between various histone modification enzymes results in a plastic regulatory state under the control of the core pluripotency transcription factor network as well as the signaling pathways known to contribute to ESC pluripotency. Action of the core pluripotency transcription factors and signaling pathways opposes the repressive functions of NuRD and LSD1, ensuring continued expression. LIF withdrawal (or other physiologic cues) results in loss of positive signals, upon which the repressive functions of NuRD and LSD1 dominate, permitting normal differentiation (Figure 1A and B). In the absence of these critical repressive factors, proper fine-tuning is lost at many genes and the ability to down-regulate the pluripotency network is severely compromised. This delicate balance of opposing chromatin modification functions constitutes a feasible mechanism to ensure developmental plasticity while maintaining the ability to self renew.
Integration of the activating and repressive signals
The LIF-Stat3 pathway constitutes a positive signal balancing the negative regulation of pluripotency genes by the NuRD and PRC2 complexes. Crabtree and colleagues recently demonstrated that LIF-Stat3 signaling in ESCs is partly dependent on Brg1 (Ho et al., 2011), the ATPase subunit of a specialized chromatin remodeling complex in ESCs termed esBAF. Brg1 is required for maintaining the binding of Stat3 and preventing PRC2-mediated H3K27me3 at some Stat3 target sites. LIF withdrawal and Brg1 deletion induced similar gene expression changes which could be rescued through deletion of the PRC2 component Suz12. These results suggested that Brg1 helps to propagate the LIF/Stat3 signaling axis in part through blocking the action of PRC2 – and thus acting in a yin/yang fashion with NuRD.
More recently, Fazzio and colleagues found that MBD3 silencing by RNAi led to gene expression changes that are largely opposite to those caused by Brg1 silencing, and double knockdown of Brg1 and MBD3 resulted in a more wild-type expression profile (Yildirim et al., 2011). Furthermore, MBD3 and Brg1 interact with each other biochemically and co-occupy many gene promoter regions where they antagonistically control nucleosome occupancy and regulate recruitment of RNA polymerase II. Surprisingly, normal localization of MBD3 was completely lost in cells depleted for Brg1, suggesting some physical or functional requirement for esBAF function in NuRD localization. Together, these results indicated that the repressive signals from the NuRD and PRC2 complexes and the activation signals from LIF-Stat3 and Brg1, function in opposition to fine-tune the expression of genes required for ESC self-renewal (Figure 1C). Analyses of genetically manipulated mice (Table 1) also indicate that loss of either MBD3 or Brg1 result in early failure of development at the periimplantation stage.
Indeed, the interplay between LIF-Stat3 and NuRD contributed to the heterogeneous expression of pluripotency genes such as Zfp42, Tbx3, Klf4, and Klf5 in ESCs as described by Hendrich and colleagues. As mentioned above, these genes are normally expressed at low or high levels in sub-populations of ESCs grown in serum and LIF medium. LIF withdrawal led to the collapse of the sub-populations into one low-expressing population, while MBD3 deletion generated one high-expressing population. Interestingly, NuRD protein levels and its occupancy at target gene promoters do not change between sub-populations of ESCs that express high or low levels of the pluripotency gene Zfp42. On the contrary, Stat3 activity was found to be higher in Zfp42-high cells, suggesting that LIF-Stat3, but not NuRD, is the rate limiting factor for pluripotency gene expression. Taken together, these data suggest that LIF-dependent self renewal results in part from the action of Brg1 in opposing the repressive functions of NuRD and PRC2 at Stat3 target loci.
NuRD recruitment to target sites in the genome
A critical question unaddressed to this point is how are promoters/enhancers selected for local enrichment of NuRD, LSD1, or other chromatin modification enzymes? Considerable evidence suggests sequence dependent recruitment of polycomb complexes in flies, and similar recruitment sequences may also exist in mammals (Woo et al., 2010). Crabtree and colleagues suggested transcription factor dependent recruitment of Brg1 by Stat3 (Ho et al., 2011). What about NuRD? Is the complex uniquely recruited to focal regions by specific interactions or does it act in a more broad sense across large regions of the genome? An interesting analysis by Yildirim et al indicated that the genomic binding patterns of MBD3 as measured by ChIP-Seq strongly overlapped with that of Tet1 (Figure 2A), an enzyme that catalyzes the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), and MBD3 was completely delocalized in Tet1 knockdown cells (Yildirim et al., 2011). In a biochemical assay, MBD3/NuRD complex could bind to both unmodified and hydroxymethylated DNA substrates, suggesting that 5hmC may recruit MBD3/NuRD to 5hmC-marked genes.
While attractive at first glance, it is unclear how consistent this model may be with the existing literature. First, the biological data appear, at best, unresolved. The role of Tet1 or 5hmC in ESCs remains controversial. A role for either in self-renewal has not been fully established, and Tet1 deletion or silencing by RNAi has not been associated, to date, with defects in differentiation (reviewed in (Wu and Zhang, 2011)). Second, recent biophysical analysis by Xiaodong Cheng and colleagues indicates that MBD3 binds double stranded DNA with a micromolar dissociation constant and has no significant difference in affinity when comparing unmodified to 5hmC modified substrates of identical sequence (Hashimoto et al., 2012). Finally, Hendrich and colleagues have elegantly demonstrated genetic complementation of MBD3 KO ESCs using the MBD3b isoform – a splice variant that disrupts the primary sequence (and likely destroys normal structure) of the methyl CpG binding domain (Reynolds et al., 2012a; Reynolds et al., 2012b). In a similar vein, the genetics and biochemistry of Tet1 and NuRD seem contradictory. Yildrim et al report that depletion of Tet1 in ESCs results in a complete loss of NuRD localization at all promoters examined, despite normal levels of MBD3 protein in these cells. This finding is difficult to understand given the recent report that Tet1 null animals survive to birth and appear normal (Dawlaty et al., 2011). Thus, elucidation of mechanisms for recruitment of NuRD to promoters/enhancers may not have reached the point of consensus.
Is it really all so simple?
Collectively, the recent papers reviewed here have added considerable depth to the literature linking NuRD complex to the ESC transcriptional program. These papers collectively support a model wherein a complicated balance between positive and negative forces dictates transcriptional output at genes integral to self-renewal and development. Despite being performed with different goals and using diverse methods, these experiments display considerable convergence in outcomes and conclusions.
In stark contrast to the big picture conclusions, there are striking areas in which these papers simply do not agree on matters of critical importance. Principal among them is the precise localization pattern of NuRD in ESCs. Reynolds and colleagues describe localization of Mi-2β and MBD3 in broad regions of several kilobases containing promoters and gene bodies (Figure 2B). It is evident in their data that NuRD complex peaks both upstream and downstream of a pronounced dip at the transcription start site (TSS). In contrast, both the Yildrim and Whyte papers depict focal accumulation at core promoters. Fazzio and colleagues provide further definition, stipulating the presence of a prominent peak of NuRD immediately downstream from the TSS. These authors posit that MBD3 localizes primarily to promoters with little association seen at enhancers (Figure 2B). This contrasts with the study by Whyte and colleagues who describe NuRD localization at roughly 2500 known enhancers (Figure 2B). How do we reconcile these differences? All three groups report results with compromised signal to noise ratios and/or inexhaustive coverage. Reynolds et al (2012a, 2012b) and Yildrim (2011) report similar results for multiple antibodies targeting MBD3 and/or Mi-2β, making it unlikely that the differences observed result from use of vastly different reagents. In the absence of any glaringly obvious explanations, we are left with the unsatisfying probability that the problem could be technical. Chromatin regulators are notoriously difficult to ChIP productively (Ram et al., 2011). These complexes are generally large, their proximity to DNA may be an issue (standard formaldehyde cross linking has an effective radius of only 1.9 Å), their chemistry may impair effective cross linking (formaldehyde cross linking occurs through lysine residues), and their residence time on chromatin is generally unknown. Alternatively, given the large datasets involved, each group may have chosen to focus on one particular aspect of the data. Analysis of each dataset in an identical manner by independent analysts may provide additional insight into the differences/commonalities between the actual data. Whatever the reasons, technical or otherwise, underlying the differences in results reported by these three groups, the issue of precisely where NuRD complex is distributed relative to genes will require additional work.
Summary and Perspectives
Since its initial biochemical discovery, the NuRD complex has been predicted to contribute to stable gene silencing through the enzymatic actions of histone deacetylation and chromatin remodeling. While useful at the time of its inception, it has become increasingly clear that such a model cannot explain all the data. Of particular note, beautiful genetic studies by Georgopoulos and colleagues have provided clear in vivo evidence for roles for NuRD complex in gene activation as well as repression in lymphocytes (Zhang et al., 2012). The current set of new manuscripts carry this concept further. Collectively, they depict genes integral to self-renewal and genes critical to early embryonic development as regulatory targets of the NuRD complex. In all cases (Figure 1), normal homeostasis results from a delicate balance between the repressive functions of NuRD (along with its functional partners PRC2 and/or LSD1) in opposition to activating functions of transcription factors (Stat3 or Oct4), histone acetyltransferases (p300), and other chromatin remodeling complexes (esBAF). The model is simple, and greatly advances our concept of how ESCs maintain the exquisite plasticity of gene expression necessary to support development while retaining the ability to self renew.
Like all models, this one does not explain all the data and will ultimately be proven wrong in at least some aspects. It remains unclear the reason why specifics regarding localization of NuRD differ so much in the various studies. Likewise, the mode of recruitment of NuRD to genes – whether it is ultimately agreed that it is focal, broadly distributed, or the pattern depends on which gene is analyzed - remains poorly understood. Furthermore, the mechanisms by which cells overcome the combination of NuRD and PRC2 at genes integral to early embryonic development is not understood in any degree of detail. Regardless of the limitations, this new model prompts a reexamination of how NuRD functions and will undoubtedly inspire a new generation of studies that will further dissect the interplay of transcription factors, signaling pathways, and chromatin regulators in the unique biology of ESCs.
Acknowledgments
The authors express gratitude to Dr. Raja Jothi for many useful discussions during the course of prerparation of this manuscript. This manuscript was substantially improved by critical comments derived from the peer review process at Cell Stem Cell. Work in the authors’ laboratories is supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, NIH (ES102745 to G.H. and ES101965 to P.A.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamo A, Sese B, Boue S, Castano J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13:652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Dawlaty MM, Ganz K, Powell BE, Hu YC, Markoulaki S, Cheng AW, Gao Q, Kim J, Choi SW, Page DC, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F, Binda C, Vanoni MA, Battaglioli E, Mattevi A. Human histone demethylase LSD1 reads the histone code. J Biol Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13:903–913. doi: 10.1038/ncb2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kaji K, Nichols J, Hendrich B. Mbd3, a component of the NuRD co-repressor complex, is required for development of pluripotent cells. Development. 2007;134:1123–1132. doi: 10.1242/dev.02802. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Torres-Padilla ME. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J, Hagman J. The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics. 2009;4:532–536. doi: 10.4161/epi.4.8.10108. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, O'Shaughnessy A, Mosaku O, Signolet J, Brennecke P, Kalkan T, Costello I, Humphreys P, Mansfield W, Nakagawa K, Stroubolis J, Behrens A, Bertone P, Hendrich B. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012a;XXXXX:1–13. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Salmon-Divon M, Dvinge H, Hynes-Allen A, Balasooriya G, Leaford D, Behrens A, Bertone P, Hendrich B. NuRD-mediated deacetylation of H3K27 facilitates recruitment of Polycomb Repressive Complex 2 to direct gene repression. EMBO J. 2012b;31:593–605. doi: 10.1038/emboj.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009a;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009b;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482:221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jackson AF, Naito T, Dose M, Seavitt J, Liu F, Heller EJ, Kashiwagi M, Yoshida T, Gounari F, et al. Harnessing of the nucleosome-remodeling-deacetylase complex controls lymphocyte development and prevents leukemogenesis. Nat Immunol. 2012;13:86–94. doi: 10.1038/ni.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]