Abstract
ETS is a family of transcriptional regulators with functions in most biological processes. Dysregulated ETS factor function leads to altered expression of multiple genes that play critical roles in many of the processes required for cancer progression. While the Ets family gene, PDEF (prostate derived ETS factor), is expressed in epithelial tissues including prostate, breast, and colon, PDEF protein expression has been found to be reduced or lost during prostate and breast cancer progression. The goal of this study was to examine the expression and biologic impact of altered PDEF expression in colon cancer. PDEF mRNA and protein are not detectable in several colon cancer derived cell lines. Re-expression of PDEF in colon cancer cells inhibits growth and migration. Growth affects are due to altered cellular proliferation, indicated by increased altered cell population in G1 and S phases of the cell cycle, as well as increased apoptosis. Relevant to its modulation of growth and migration phenotypes, PDEF expression resulted in altered expression of genes with established roles in cell cycle, motility and invasion. Furthermore, chromatin immunoprecipitation studies show that p21 and urokinase plasminogen activator (uPA) are direct PDEF transcriptional targets. While non-tumor colon epithelium expresses PDEF mRNA and protein, the majority of tumors showed decreased mRNA and/or protein expression. In human tumor tissue samples, PDEF expression was inversely correlated with the expression levels of uPA. Collectively, the data support the model that PDEF is a negative regulator of tumor progression by modulating the expression of growth and migration promoting genes.
Keywords: PDEF, Ets, Transcription factors, Colon cancer, cell growth, migration
Introduction
ETS proteins are cellular homologs of v-ets, one of the two transduced genes present in the avian E26 transforming retrovirus. Ets family members have been found in species ranging from C. elegans to humans and are a family of transcription factors defined by a conserved DNA-binding domain of ~85 amino acids [Seth and Watson, 2005; Turner et al., 2007a]. The Ets domain recognizes a core sequence of 5’-GGAA-3’ present in regulatory elements of downstream target genes. Over 200 Ets target genes were previously described [Sementchenko and Watson, 2000] and currently there are over 500 genes with functional Ets responsive sites (Watson, unpublished). Ets transcription factors control the expression of target genes that have critical roles in cell proliferation, differentiation, apoptosis, and oncogenesis. Elevated expression of several ETS genes (e.g., ETS1) has been observed in most human cancers, including prostate, breast and colon cancer [Seth and Watson, 2005].
Since the majority of cancers are of epithelial origin, an especially relevant subset of ETS factors are those that maintain a restricted pattern of expression limited to tissues with high epithelial cell content, regulating cellular proliferation and differentiation [Feldman et al., 2003b]. Among these is Prostate Derived ETS Factor (PDEF), originally isolated from normal prostate tissue, with expression demonstrated in additional epithelial tissues, including ovary, breast and colon [Feldman et al., 2003a; Ghadersohi et al., 2008; Oettgen et al., 2000]. Collective studies from our and other laboratories have shown that PDEF is a negative regulator of cell growth, migration and invasion of breast, ovarian and prostate cancer cells [Feldman et al., 2003a; Ghadersohi et al., 2008; Gu et al., 2007; Turner et al., 2008; Turner et al., 2007b].
Colon cancer is the second leading cause of cancer related deaths in the U.S. [Jemal et al., 2008]. Over 108,000 new cases of colorectal cancer will be diagnosed in 2008 with a mortality rate estimated at 46% [Jemal et al., 2008]. Normal colon epithelium undergoes continual renewal [Barker et al., 2008]. Stem cell precursors from the base of the crypts migrate to the luminal surface to replenish the epithelium. During this migration, these proliferating cells must undergo cell cycle arrest, lineage specific differentiation, and eventually apoptosis and luminal extrusion. Thus, a balance between cell proliferation and cell apoptosis is established in the normal colon. Colon cancer may develop if this normal balance is shifted towards increased proliferation together with decreased apoptosis. Numerous studies of genetic abnormalities associated with the progression of colon cancer have pinpointed several important regulators of cellular homeostasis, including transcription factors, are dysregulated and ultimately contribute to the acquisition of the more aggressive phenotype [van den Brink and Offerhaus, 2007].
We report here that several colon cancer cell lines do not express PDEF mRNA or protein. Since PDEF expression is found in epithelial tissues and has regulatory roles for cell growth, migration and invasion, we evaluated the impact of PDEF expression on phenotypes of colon cancer cells. Re-expression of PDEF into colon cancer cells results in an inhibition of cellular growth and migration. Relevant to PDEF modulation of growth and migration, chromatin immunoprecipitation studies show that p21 and uPA are direct PDEF transcriptional targets. While PDEF is expressed in epithelium of non-tumor colon tissue, PDEF mRNA and protein are reduced in the majority of tumor samples examined. Interestingly, PDEF is a negative regulator of urokinase plasminogen activator (uPA) and matrix metalloproteinase 7 (MMP7) expression, and there is a statistically significant inverse correlation between PDEF and uPA expression in colon cancer clinical specimens.
MATERIALS AND METHODS
Cell Culture
Colon cancer-derived cell lines DLD-1, CaCo-2, LoVo-2, SW620, HCT116, and HT-29 were obtained from American Type Culture Collection (ATCC) and propagated according to ATCC recommendations.
Adenoviral Infection
The construction of PDEF expressing adenovirus has been previously described [Feldman et al., 2003a; Turner et al., 2008; Turner et al., 2007b]. Cells were infected (5–10 MOI) in normal growth medium with either control virus expressing GFP (Ad-GFP), or virus expressing PDEF/GFP from a bi-cistronic promoter (Ad-PDEF). Infected cells were then incubated as normal for 14 or 36 hours. Under these conditions greater than 95% of the cells were infected as assessed by GFP expression.
PDEF Gene Knockdown
The 20-nucleotide GGCCGCTTCATTAGGTGGCT ‘loop’ located at the position 1211–1241 of the PDEF gene (GeneBank accession no: NM_012391) was used for synthesis of the shRNA duplex. The Duplex was cloned in the pSupressor vector (Imgenex, San Diego, CA). Stable clones were selected in RPMI containing 400 µg/ml G418 (Invitrogen, Carlsbad, CA, USA).
Patients and Tumor Specimens
Human colon cancer paraffin blocks were obtained from the Hollings Cancer Center Tumor Bank, Medical University of South Carolina for immunohistochemical studies. Frozen colon tumor and patient matched non-tumor tissue was obtained from the Cooperative Human Tissue Network (CHTN, NCI) and used for the preparation of RNA and protein for expression analyses.
Immunohistochemistry
Formalin-fixed and paraffin-embedded tissue sections were deparaffinized, rehydrated, and endogenous peroxidase activity was blocked using 3% H2O2. Antigen retrieval was done by heating in a microwave oven for 5 min on high power in 10 mM citrate, pH 6.0. Sections were washed and nonspecific binding was blocked with 10% horse serum in Tris-buffered saline (TBS; 50 mM Tris-HCl, 0.9% NaCl, pH 8.0) for 20 min, then incubated overnight at 4°C with a PDEF-specific primary antibody [Feldman et al., 2003a; Turner et al., 2007b] at a 1:200 dilution in the blocking solution. Overnight incubation at 4°C was followed by three 10-min washes in TBS. Slides were then incubated in Immpress™ anti-rabbit (Vector Laboratories, Burlingame, CA) for 45 min at room temperature and developed using DAB substrate (Sigma Chemicals, St. Louis, MO). Slides were counterstained with hematoxylin.
Northern, RT-PCR and Western Blot Analysis
Tumor and non-tumor colon tissues were used for isolation of RNA and total protein. Frozen tissue samples (250 mg) were pulverized in LN2 and RNA was isolated using Trizol reagent (Invitrogen) according to the manufacturer's protocol. Total RNA (20 µg) was fractionated on 1% agarose gels containing 0.66M formaldehyde. RNA quality was assessed by ethidium bromide visualization of 28S and 18S rRNA. RNA was transferred to nylon membranes (Duralon, Stratagene, Agilent, Santa Clara, CA) in 0.1M sodium phosphate buffer (pH 6.8), UV cross-linked and hybridized for 2 hours at 65°C in QuikHyb (Stratagene). PDEF and uPA α-32P-dCTP labeled probes were prepared by random-primed synthesis using PrimeIt (Stratagene). Washed membranes were exposed to X-ray film for autoradiography. MMP7 RNA expression was examined by RT-PCR. Total RNA (2 µg) was reversed transcribed using Superscript II (single strand synthesis RT; Invitrogen). Aliquots from this reaction were subjected to PCR using primers, MMP7-F (5‘-ATCCCCCTGCATTTCAGGAA -3‘) and MMP7-R (5’-TTCCTGGCCCATCAAATGG-3’), corresponding to nucleotides 465–484 and 567–549 (GenBank No. NM_002423.3) to amplify MMP-7. The reaction mixture contained 2 mmol/L Mg2+, 0.2 mmol/L deoxynucleotide triphosphates, 1 X Taq Gold buffer, 0.88 pmol/mL Primers, and 0.02 U/µL Taq Gold (Applied Biosystems). The PCR reaction conditions were as follows: 95° for 10 min; then 34 cycles of 95°C for 30 s, 57 cycles for 45 s, 72°C for 1min, followed by 72° for 7 min. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as PCR control. GAPDH was amplified using the conditions described above for MMP-7 RT-PCR using GAPDH-F (5‘-TCCTCTGACTTCAACAGCGACA -3‘) and GAPDH-R (5‘-TTCCTCTTGTGCTCTTGCTGG -3‘)corresponding to nucleotides 949–970 and 1150–1130, respectively, of the GAPDH mRNA sequence (GenBank No. NM_002046.3). PCR products were analyzed by electrophoresis on a 1% Tris borate EDTA gel containing ethidium bromide and visualized under UV light.
Protein was extracted from pulverized tissue powders by direct lysis in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1% Triton X100, 0.1% SDS, 1% deoxycholate and protease inhibitor cocktail [Complete Protease Inhibitors, Roche]) for 15 minutes on ice. Proteins were also extracted from colon cancer cell lines. For total protein isolation, cells at 90% confluence were washed twice with ice cold PBS, and were lysed in RIPA buffer containing protease inhibitors.
Equal amounts of total protein (40 µg) were resolved by 12% SDS-PAGE and subjected to Western blot analyses using ECL system (Amersham-Pharmacia, GE Healthcare, Piscataway, NJ). Total protein lysates prepared from DLD-1 cells left untreated or infected with Ad-GFP or Ad-PDEF were examined for protein expression using antibodies against PDEF (described above), p21, PARP (Santa Cruz), cdk2, cyclin A, cyclin E, Rb (PharMingen, BD Biosciences, San Diego, CA), and caspase -3 and β-actin (Sigma).
Cell Growth Assay
CaCo-2, LoVo, SW680, and DLD-1 colon cancer cells were infected with Ad-GFP or Ad-PDEF for 14 hrs in 2% serum. The virus was then removed and media containing 10% serum was added. Cells were seeded at 2,500 to 5,000 cells per well in 96-well plates. The number of viable cells were quantified at the indicated time points, using the MTT (3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide thiazole blue) assay, performed in quadruplicate, according to the manufacturer’s instructions (Sigma). Optical density (OD) was measured at wavelength of 550 nm with reference wavelength of 690 nm.
Flow Cytometry
Cells were harvested 36 hours after adenoviral infection, fixed and dehydrated in 70% ethanol at −20°C for at least 24 hrs. Cells were washed twice in PBS containing 10% serum and resuspended in 500 ul of a solution containing 0.5 mg/ml propidium iodide and 1 mg/ml RNase A (Sigma) and analyzed using flow cytometry.
Cell viability assays
Cell counts and viability measurements were performed using the Countess optics and image automated cell counter (Invitrogen) together with the standard trypan blue dead cell staining technique. Cells were mixed with trypan blue (50/50) and loaded into a Countess cell counting chamber slide and cell images were acquired from the sample. The image analysis software was used to automatically analyze the acquired cell images to give a cell count and viability value using the trypan blue stain. Data was plotted as the number of viable and dead cells per time point for each treatment.
Migration Assays
Cell migration experiments were carried out using 8-µm pore size migration chambers (Falcon, Becton Dickinson, Franklin Lakes, NJ) pre-coated at 4°C overnight with fibronectin (Becton Dickinson) at a concentration of 5 µg per square centimeter in PBS. The following day, the fibronectin solution was aspirated and the migration chambers were rinsed once with water and allowed to air dry prior to the migration experiment. Cells at 80% confluence were trypsinized, harvested, and counted. For each condition, cells were seeded at 25,000 cells/well in 500 µL serum free media. Medium (750 µL) containing 10% serum was used as a chemoattractant in the lower chamber. Cells were allowed to migrate for eight hours at 37°C in the presence of 5% CO2. Cells that did not migrate were removed by wiping the top of the membrane with a cotton swab and the migrating cells were fixed and stained with Diff-Quik per the manufacturer’s protocol (Dade Behring, Siemens, Deerfield, IL). Migrating cells in 10 high power fields in each chamber were counted and the mean cell number was calculated. Each experiment was conducted in triplicate and repeated three times.
Chromatin immunoprecipitation (ChIP)
Chromatin was prepared and immunoprecipitation performed using HT-29 cells (express endogenous PDEF) in a two-step cross-linking protocol, as previously described [Nowak et al., 2005]. Chromatin was fragmented into 500−1000bp fragments by sonicating the cells eight times for 10s at level three in an ethanol ice bath using a Virtis Virsonic 475 sonicator (Gardiner, NY). Soluble chromatin was quantified (absorbance at 260nM) and 10 absorbance units were incubated with 2µg of PDEF rabbit polyclonal antibody or IgG alone overnight at 4°C with rotation. Collection, washing and reverse cross-linking of immune complexes was as previously described [Nowak et al., 2005].
Promoter enrichment was assessed using real time PCR analysis using primers spanning previously identified uPA [D'Orazio et al., 1997] and p21 [Funaoka et al., 1997] ETS binding sites (EBS). One microliter of a 1 in 5 dilution of DNA was used in each real-time reaction, which was conducted using a LightCycler (Roche Diagnostics, Basel, Switzerland) with the Platinum SYBER Green qPCR SuperMix (Invitrogen), as per the manufacturers’ instructions. Primers were used at a concentration of 250 nM (uPA primers: F- 5’ ATTTGTGAGGCC CATGGTTG 3’; R- 5’ AAACCGCTGCTCCCACATT 3’; p21 primers: F - 5’ TTCCACC TTTCACCATTCCCC 3’, R - 5’ TCTCCTGTCTCCTACCATCC 3’) and the cycling conditions were as follows: preincubation, 50°C for 10 min, 95°C for 2 min, followed by 40–50 cycles of denaturation at 95°C; annealing at 58°C (uPA) or 55°C (p21) and extension at 72°C, all for 20s, with a single data acquisition at the end of each extension. Melting curve and data analysis was carried out as per the manufacturer’s recommendations using the accompanying software (Roche Diagnostics).
Statistical Analysis
Statistical analyses were performed using the Student’s t-test for paired data. p <0.05 was considered significant. Clinical correlation was examined by Chi-squared test using SSPS 11.0 software package.
RESULTS
PDEF expression is lost in colon cancer-derived cell lines
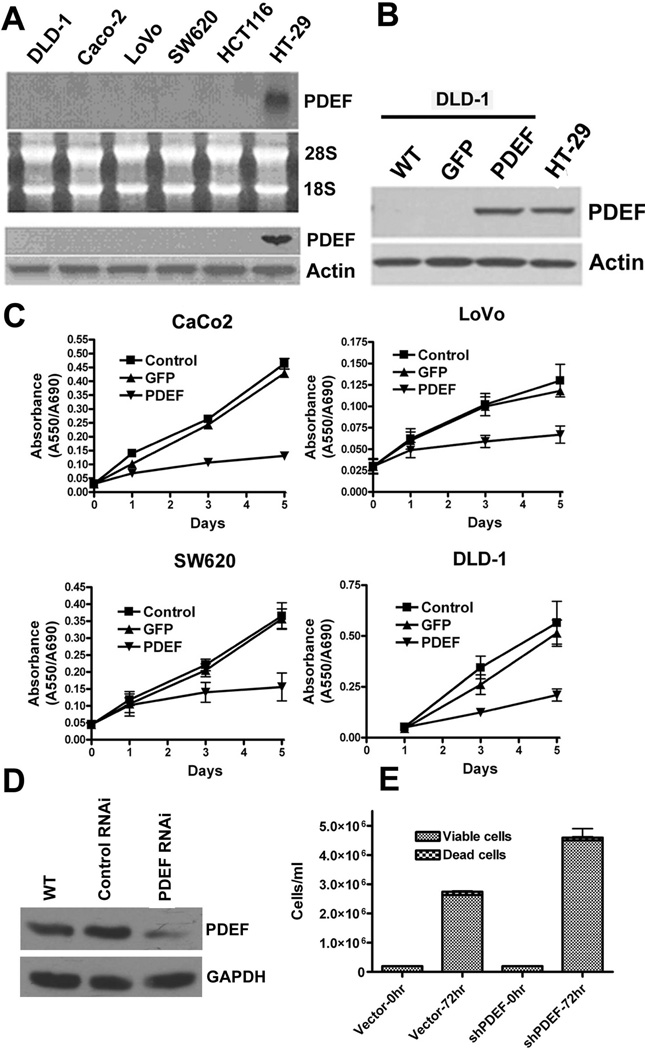
We next examined several colon cancer derived cell lines for the expression of PDEF (Figure 1A). While PDEF mRNA and protein are not detected in each of the invasive colon cancer cell Lines (DLD-1, CaCo-2, LoVo-2, SW620, HCT116), PDEF mRNA and protein expression are retained in the differentiated noninvasive cell line HT-29.
Figure 1. PDEF is lost in invasive colon cancer cell lines and re-expression of PDEF leads to growth changes in colon cancer cells.
(A) Northern and Western blot analyses of RNA and protein prepared from the indicated colon cancer cell lines. Ethidium bromide staining of 28S and 18S rRNA or actin levels are provided as a controls. (B) Western blot analysis of PDEF expression level after infection of DLD-1 colon cells and endogenous expression level observed in HT-29 cells. Actin levels provided as a control. (C) CaCo-2, LoVo-2, SW680 and DLD-1 cells were untreated (control), or infected with either Ad-GFP or Ad-PDEF and cellular growth was monitored by MTT assay for 5 days. (D) Western blot analysis of PDEF protein expression in HT-29 colon cancer cells and in HT-29 expressing vector (RNAi control) and short hairpin RNA (shRNA) PDEF. Actin levels provided as a control. (E) Number of HT-29 colon cancer cells expressing vector alone or PDEF shRNA measured after 72 hours (p<0.05).
Expression of PDEF in colon cancer derived cell lines inhibits cellular growth
To determine possible biological consequences upon expression of exogenous PDEF, we infected colon cancer derived cell lines lacking endogenous PDEF with an adenovirus expressing PDEF and GFP (Ad-PDEF) or with an adenovirus expressing GFP (Ad-GFP), as a control (Figure 1B). We are able to obtain greater than 95% cell infection using this adenoviral system.
We selected four different colon cancer cell lines for growth studies. CaCo-2, LoVo-2, SW680 and DLD-1 cells were infected with either Ad-GFP or Ad-PDEF and cellular growth was monitored by MTT assay for 5 days (Figure 1C). Compared to uninfected and Ad-GFP infected controls, cells infected with Ad-PDEF showed reduction in total cell number at 3 and 5 days post infection.
We next examined the impact of inhibition of endogenous PDEF expression on cell growth. HT29 cells were transiently transfected with a shRNA vector targeting PDEF. siRNA-mediated downregulation of PDEF resulted in ~40% reduction in protein expression (Figure 1D) in HT29 colon cancer cells. Consistent with the observation that PDEF is a negative regulator of colon cancer cell growth, the reciprocal shRNA mediated knock-down of PDEF resulted in increased colon cell number (Figure 1E).
PDEF-dependent inhibition of cell growth is mediated through cell cycle changes
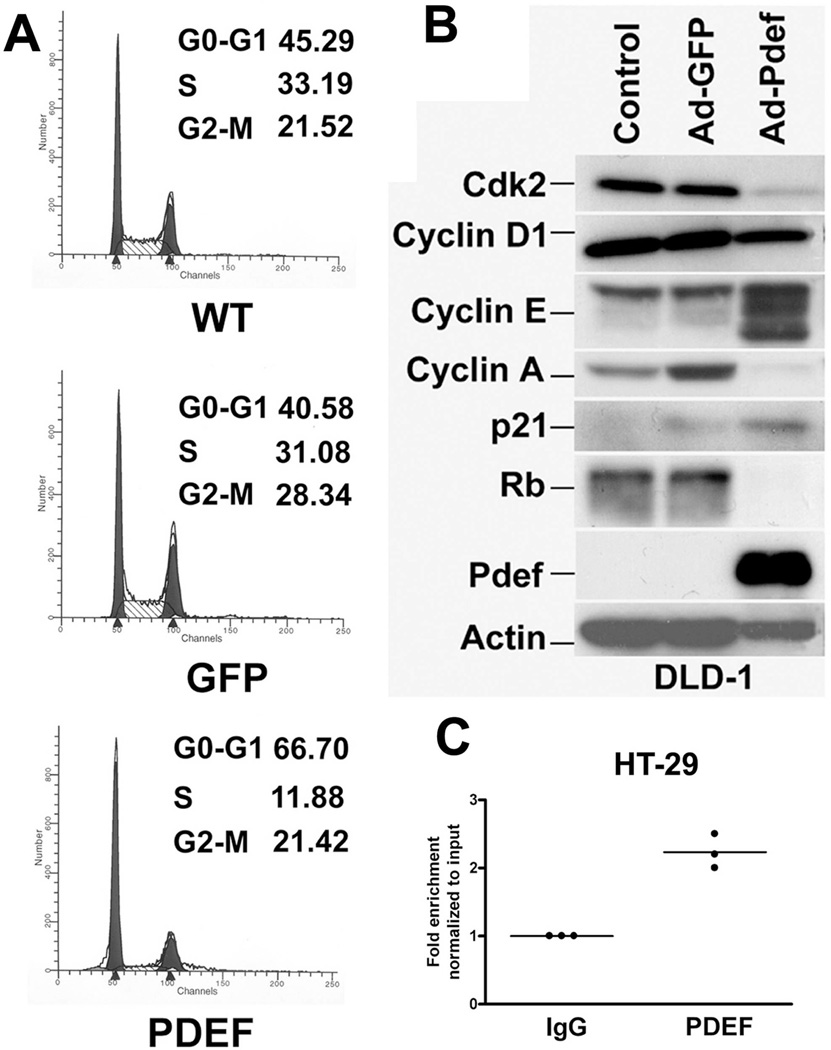
To begin to understand the molecular mechanisms for PDEF-mediated inhibition of cell growth, cell cycle analysis was performed on DLD-1 cells 36 hrs after infection with either Ad-GFP or Ad-PDEF (Figure 2A). Compared to uninfected and Ad-GFP infected cells, expression of PDEF in DLD-1 cells leads to an increase in the percentage of cells in the G0–G1 phase. There is a concomitant reduction in the percentage of cells in S phase. This suggests that expression of PDEF prolongs cell cycle progression, specifically at the G0–G1 transition.
Figure 2. PDEF expression results in altered cell cycle progression and expression of cell cycle regulatory genes.
(A) Cell cycle analysis of untreated DLD-1 cells or cells treated with Ad-GFP or Ad-PDEF and harvested 36 hours after infection. (B) Western blot analysis of total protein isolated from cells infected with Ad-GFP or Ad-PDEF using antibodies against the indicated cell cycle proteins. Blots were re-hybridized using antibodies against PDEF and β-Actin. Control lanes represent uninfected cells. (C) PDEF is bound to the p21 expression in vivo. Chromatin immuno-precipitation (CHIP) analysis using chromatin prepared from the HT-29 cell line. Q-PCR analysis using primers for the p21 promoter and chromatin immunoprecipitation with a PDEF-specific antibody, compared to IgG control antibody.
PDEF-dependent inhibition of cell growth is mediated through changes in cell cycle regulatory proteins
To better characterize the G1 arrest in DLD-1 cells, cell lysates prepared from infected cells were examined for expression of G1 cell cycle regulatory proteins by Western blot (Figure 2B). The protein levels of cyclin dependent kinase 2 (cdk2), cyclin A and Rb were decreased in DLD-1 cells infected with Ad-PDEF compared to controls. In contrast, the expression of cyclin E and the cyclin dependent kinase inhibitor, p21 (p21WAF1/CIP1) were increased. The elevation in p21 and reduced cyclin A/cdk2 could explain the observed G1 arrest in DLD-1 cells.
We selected p21 to test whether PDEF may directly regulate its expression at the transcriptional level. Chromatin was prepared from the HT-29 cell line which expresses endogenous PDEF. Chromatin immunoprecipitation (CHIP) and RT-QPCR analysis demonstrate that the p21 promoter region corresponding to that previously shown to have functional Ets binding sites (EBS) [Funaoka et al., 1997] was enriched following immunoprecipitation with a PDEF-specific antibody, compared to IgG control antibody. Thus, PDEF occupies functional EBS present in the p21 promoter in vivo and thus, that p21 is a direct PDEF target (Figure 2C).
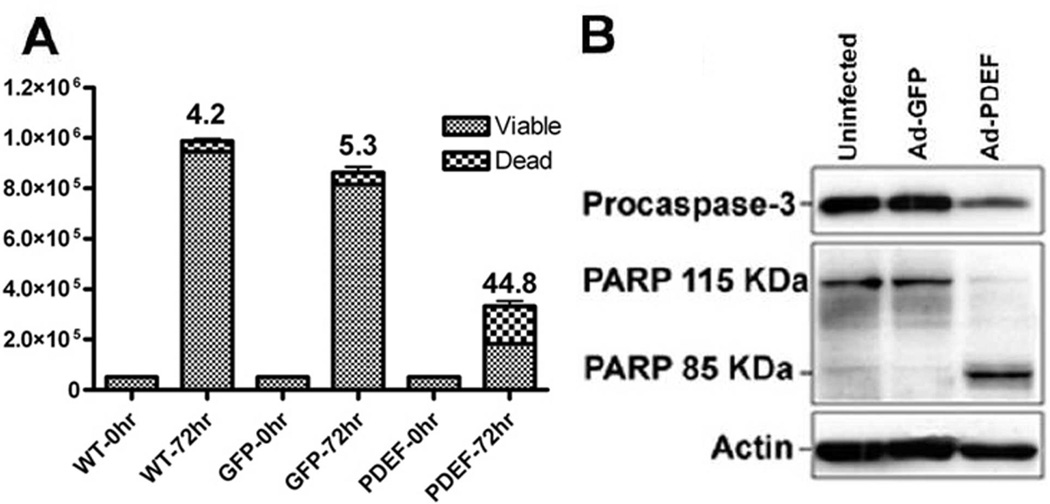
Expression of PDEF in DLD-1 cells results in an induction of apoptosis
During our studies with DLD-1 cells, we noted a sub-population of cells were easily dislodged from the plate after PDEF expression. To determine whether increased apoptosis also contributes to PDEF-mediated changes in cellular growth, we determined the impact on viable cell number by trypan blue staining. Cells infected with Ad-PDEF exhibit 48% apoptotic cells by day 3 after infection (Figure 3A). These results support the model that PDEF causes cellular death in DLD-1 cells, possibly by activating apoptotic machinery. Caspase activation is a major component during the execution phase of apoptosis. Proteolytic cleavage of the prodomain of caspase-3 activates this effector caspase to degrade a variety of apoptotic substrates. Infection of DLD-1 cells with Ad-PDEF results in activation of caspase-3 as seen by a decrease in the level of procaspase-3 (Figure 3B). This is in contrast to uninfected cells and cells infected with Ad-GFP. Caspase-3 cleaves at the signature motif (DXXD), first characterized in the poly ADP-ribose polymerase (PARP) [Cohen, 1997]. We performed western blot analyses on lysates prepared from infected cells to examine the cleavage of PARP in response to PDEF expression. Compared to controls, cells infected with Ad-PDEF exhibit the characteristic cleaved 85-kDa fragment (Figure 3B).
Figure 3. PDEF expression increases apoptosis.
(A) Quantitation of trypan blue cell viability assays of parental (uninfected) DLD-1 cells, or cells infected with Ad-GFP or Ad-PDEF. The columns represent the average values for apoptotic index, the percentage of apoptotic vs. total cell number. Results are statistically significant with a p-value <0.05. (B) Western blot analysis of procaspase 3 and PARP expression levels in DLD-1 cells infected with Ad-GFP or Ad-PDEF. Actin levels provided as a control.
PDEF expression levels alter cell migration
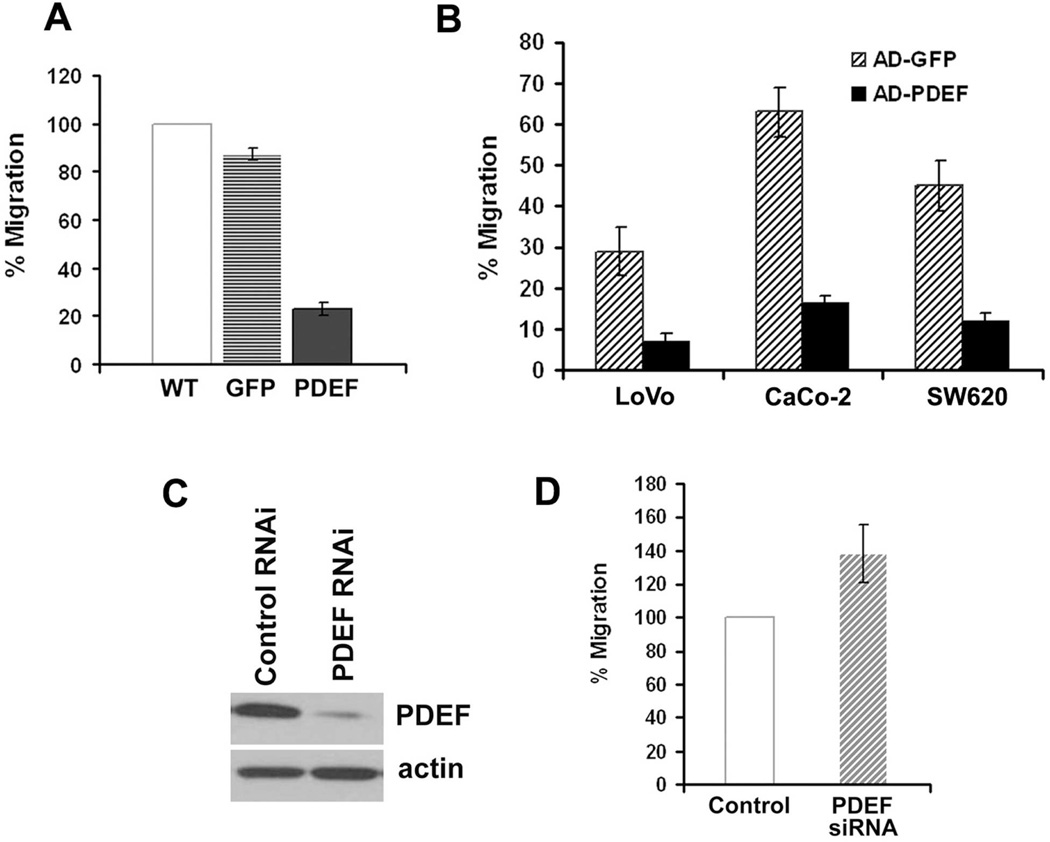
The effect of PDEF re-expression on chemokinetic migration (movement towards a stimulant) was examined in transwell migration assays using serum as a chemoattractant. Compared to control cells, adenoviral mediated expression of PDEF in DLD-1 (Figure 4A), LoVo, CaCo-2, or SW620 (Figure 4B) cells reduced the number of cells able to migrate across fibronectin-coated membranes by 60–75%.
Figure 4. PDEF expression levels alter cell migration.
The effect of PDEF expression on the migration of colon cancer cells was measured by transwell assay. (A) DLD-1 cells were untreated or infected with Ad-GFP or Ad-PDEF and migration measured by transwell assays. Data is representative of three experiments performed in triplicate. (B) LoVo, CaCo-2, and SW620 cells were infected with Ad-GFP or Ad-PDEF. Cell migration was measured by transwell migration assay. (C) Western blot analysis of PDEF protein expression in HT-29 colon cancer cells and in HT-29 expressing short hairpin RNA (shRNA) PDEF and control vectors. Actin levels provided as a control. (D) Cell migration of HT-29 colon cancer cells and of HT-29 cells expressing PDEF shRNA. Each of the measured differences were statistically significant with a p-value <0.05 by students t-test.
We next wanted to determine whether inhibition of endogenous PDEF expression level would result in an increase in cell migration. siRNA-mediated downregulation of PDEF resulted in >90% reduction in mRNA (data not shown) and protein expression (Figure 4C) in HT29 colon cancer cells. Consistent with the data obtained from our gain of function studies that show that PDEF is a negative regulator of colon cancer cell migration, the reciprocal shRNA mediated knock-down of PDEF resulted in increased colon cell migration (Figure 4D).
PDEF protein expression is lost in colon cancer
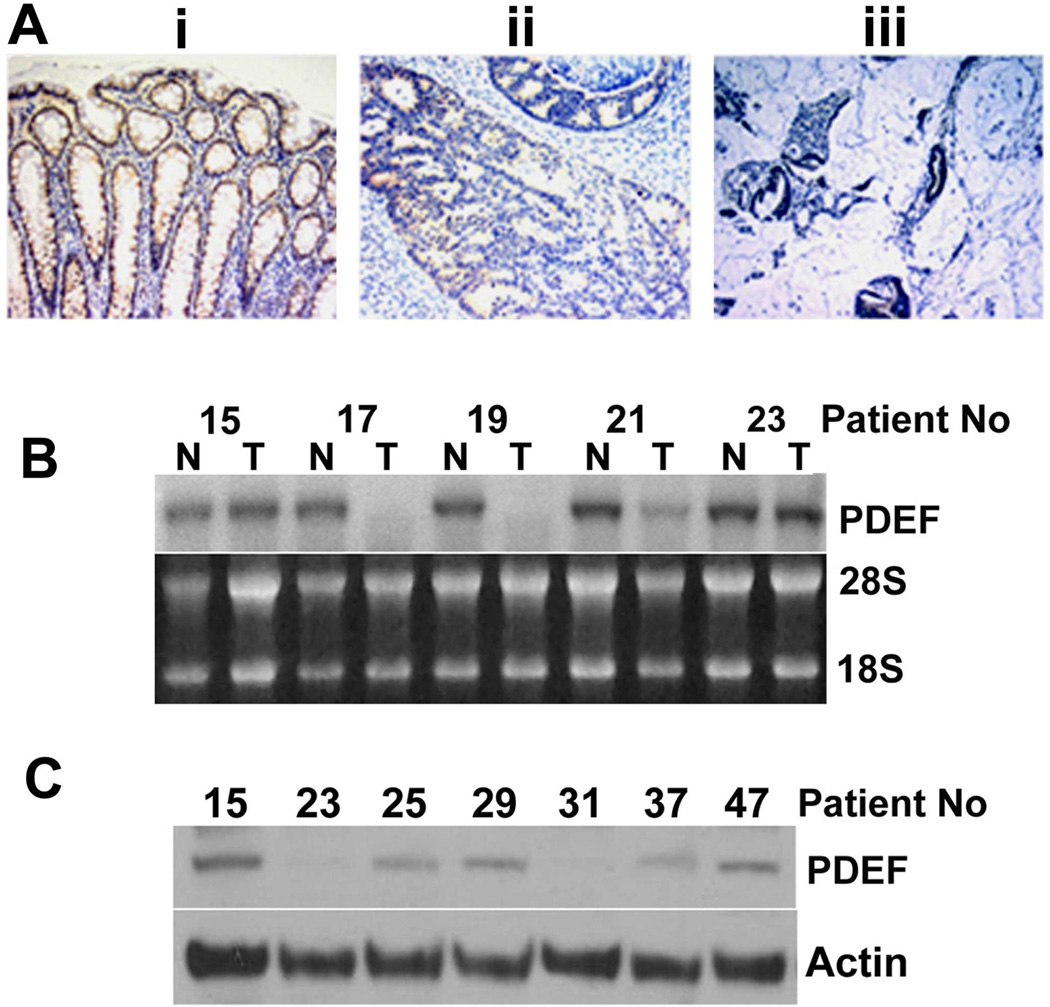
Human PDEF mRNA is expressed at high levels primarily in tissues with high epithelial content, including colon, prostate, and breast [Feldman et al., 2003a]. Several reports have indicated that modulation of PDEF expression occurs during carcinogenesis [Mitas et al., 2002; Nozawa et al., 2000]. Immunohistochemical analysis was used to examine PDEF expression and distribution in paraffin embedded human colon cancer specimens (Figure 5). PDEF was expressed predominately in the nuclei of non-tumor tissues (Figure 5A, panel i), while PDEF protein was lost in the tumor areas in 5 of the 6 cases examined (Figure 5A, panels ii and iii).
Figure 5. PDEF expression is reduced in colon cancer tissue.
(A) Immunohistochemical analysis of PDEF expression in colon tissue: (i) non-tumor, (ii) tumor and (iii) invasive mucinous portion of tumor. Panels are from the same section. (Magnification, X400). Data representative of 5 of 6 cases. (B) Northern blot analysis for PDEF mRNA in a series of matched tumor (T) and non-tumor (N) tissues from the same patient (indicated by numbers). Data representative of 25 matched tumor/non-tumor samples. Ethidium bromide staining of 28S and 18S rRNA provided as control. (C) Western blot analysis of PDEF protein in 7 tumors that retain PDEF mRNA expression. Actin levels provided as a control.
PDEF mRNA and protein are lost in human colon cancer tissue
To examine possible mechanisms for loss of PDEF protein expression in colon cancer, we evaluated PDEF mRNA expression in 25 matched pairs of non-tumor/tumor colon tissues (subset shown in Figure 5B). These matched specimens consisted of tissues obtained from areas of colon adenocarcinoma and a corresponding non-tumor area of colon that was removed from a distal resection from a single patient. Thirteen out of 25 (52%) colon adenocarcinoma samples show loss of the single 1.9 kb PDEF mRNA transcript compared to corresponding non-tumor colon tissue. PDEF mRNA is reduced in 5/25 (20%), while expression retained in 7/25 (28%). Out of the 7 samples expressing PDEF mRNA, only 5 had PDEF protein expression (Figure 5C). Thus, in this set of samples, the majority show correlated loss or reduced expression of both PDEF mRNA and protein. In a small set of samples, PDEF protein is lost even in the presence of mRNA.
PDEF loss alters the expression of genes associated with aggressive colon cancer
Since PDEF is a transcription factor, it is likely that its loss in tumors would alter the expression of many genes and that this altered transcriptome would contribute to cancer progression. Based on our microarray studies ([Turner et al., 2008], and data not shown), we selected two candidate PDEF responsive genes which have been reported to be associated with invasive behavior of colon cancer: uPA and MMP-7.
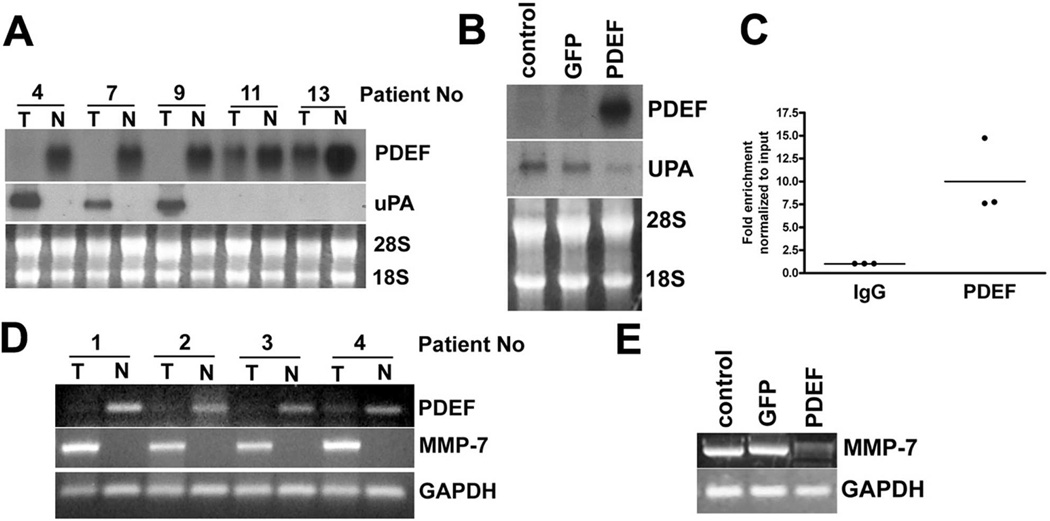
uPA is up-regulated and associated with poor outcome in patients with several cancers, including colon cancer [Zlobec et al., 2008]. Northern blot analysis was used to quantify PDEF and uPA mRNA expression in RNA prepared from tumor tissue and matched adjacent non-tumor tissues. There is a statistically significant correlation between PDEF loss and upregulation of uPA (Figure 6A and Table 1). We also found that re-expression of PDEF in DLD-1 cells decreased uPA mRNA expression in these cells (Figure 6B). To test the possibility that PDEF may directly regulate uPA expression at the transcriptional level, we performed chromatin immunoprecipitation (CHIP) analysis. RT-QPCR analysis indicated that uPA promoter fragments were enriched following immunoprecipitation with a PDEF-specific antibody, compared to IgG control antibody. The uPA promoter has previously been shown to have functional Ets binding sites [D'Orazio et al., 1997] and the current data supports the model that these sites in uPA are bound by PDEF in vivo and thus, that uPA is a direct PDEF target (Figure 6C).
Figure 6. PDEF loss alters the expression of the cancer associated genes, uPA and MMP-7.
(A) Northern blot analysis of RNA prepared from tumor tissue and matched adjacent non-tumor tissues. 28S and 18S RNA provided as control for loading and RNA integrity. Data is representative of 25 tumor and non-tumor pairs examined. (B) Northern blot analysis of RNA prepared from parental DLD-1 cells and DLD-1 cells infected with Ad-GFP or Ad-PDEF. Ethidium bromide staining of 28S and 18S provided as a control. (C) Chromatin immuno-precipitation (CHIP) analysis using chromatin prepared from the HT-29 cell line. Q-PCR analysis using primers for the uPA promoter and chromatin immunoprecipitation with a PDEF-specific antibody, compared to IgG control antibody. (D) RT-PCR analysis MMP-7 mRNA in RNA prepared from tumor tissue and matched adjacent non-tumor tissues. GAPDH RT-PCR provided as a control. Data representative of 25 matched samples. (E) RT-PCR analyses of MMP-7 mRNA in RNA prepared from uninfected DLD-1 cells and cells infected with Ad-GFP or Ad-PDEF. GAPDH provided as control.
Table 1. Correlation between PDEF expression and Disease parameters.
PDEF expression or loss in patient samples was compared to uPA and MMP7 expression, node status and tumor differentiation using the χ2 test. A statistically significant correlation is found between PDEF loss and uPA expression. Although, PDEF re-expression down-regulated MMP-7 mRNA levels in DLD-1 cells, there is not a statistically significant correlation between MMP-7 expression in tumor tissues and PDEF loss. No statistical significant correlation between PDEF loss and lymph node involvement or tumor differentiation.
| PDEF Expression | |||||
|---|---|---|---|---|---|
| Lost | Expressed | X2 | P value | ||
| NEG | 5 | 4 | 6.6 | 0.01 | |
| uPA | POS | 15 | 1 | ||
| NEG | 7 | 2 | 0.043 | 0.8 | |
| MMP-7 | POS | 13 | 3 | ||
| NEG | 9 | 3 | 0.157 | 0.6 | |
| Node Status | POS | 9 | 2 | ||
| Well | 2 | 2 | 2.8 | 0.2 | |
| Tumor | Moderate | 9 | 1 | ||
| Differentiation | Poor | 9 | 2 | ||
MMP-7 has been correlated with growth, proliferation and metastatic properties of colon cancer [Roeb et al., 2004]. RT-PCR analysis of RNA prepared from tumor tissue and matched adjacent non-tumor tissues indicates MMP-7 mRNA levels are elevated in the majority of tumors with lost or reduced PDEF expression, although this correlation did not reach statistical significance (Figure 6D and Table 1). PDEF re-expression resulted in a reduction of MMP-7 mRNA levels in DLD-1 cells (Figure 6E).
Discussion
PDEF expression is not detected in several colon cancer derived cell lines. In this manuscript, we examined the impact of PDEF expression on cell growth and migration. Consistent with the phenotype observed upon PDEF expression in breast cancer cells [Feldman et al., 2003a], PDEF expression in DLD-1 colon cancer cells results in a prolonged G1, concomitant with reduction in Rb, cdk2, and cyclin A levels and increased p21 and cyclin E [Feldman et al., 2003a]. PDEF may directly regulate expression of genes involved in cell cycle progression as has been shown for other ETS factors. It has been previously shown that Cyclin D1, Rb and p21 are each Ets response genes [Sementchenko and Watson, 2000]. Reduced ETS2 levels inhibit proliferation and induce apoptosis of prostate cancer cells, and is associated with reduced levels of cyclin D1 and the anti-apoptotic protein bcl-x(L) [Carbone et al., 2004]. Fli1 mediated repression of Rb during erythropoiesis occurs through an upstream ETS consensus site [Tamir et al., 1999]. PEA3 is able to activate the promoter of p21 independently of p53 [Funaoka et al., 1997]. Prolonged activation of MAPK signaling in primary hepatocytes results in an Ets2 mediated increase in p21 expression [Park et al., 2000], while ETS2 is indispensable for p21 activity in chondrocytes [Beier et al., 1999]. While further experiments should delineate whether PDEF mediated G1 arrest is acting through an indirect or direct regulation of cell cycle protein expression, the ChIP studies presented here support the model that at least p21 is a direct transcriptional target of PDEF in colon cells.
Expression of PDEF in DLD-1 cells also resulted in significant cell death. ETS factors have also been implicated in the regulation of apoptosis [Sementchenko and Watson, 2000]. ETS1 is able to activate the promoter of PARP in an in vitro luciferase assay [Soldatenkov et al., 1999]. Maximal activation of the rat caspase 3 promoter requires an intact ETS consensus sequence [Liu et al., 2002]. Examination of protein lysates from PDEF infected cells also revealed increased activation of the effector caspase-3. This results in the downstream cleavage of the apoptotic substrate PARP and subsequent increases in cellular apoptosis. Recently, it has been shown that shRNA mediated silencing of PDEF expression resulted in the upregulation of survivin expression in MCF-7 cells, as well as increased in vitro and in vivo cell growth and resistance to drug-induced apoptosis, consistent with our findings [Ghadersohi et al., 2007].
Consistent with previous studies in breast and prostate cancer [Feldman et al., 2003a; Gu et al., 2007; Turner et al., 2007b], PDEF is a negative regulator of colon cancer cell migration. The reduced expression of uPA and MMP-7 observed upon PDEF expression may contribute to the inhibition of migration. In addition to the established role for these genes in cell migration and invasion, their reduced expression could also contribute to growth inhibition and apoptosis. Pro-proliferative effects are the result of protease-mediated release of sequestered growth factors and cytokines [Cheng et al., 2007]. MMP-7 proteolysis of the insulin-like growth factor binding proteins promotes colon cancer cell survival via IGF-1 [Nakamura et al., 2005]. While previous studies have defined ETS regulatory pathways that activate transcription of uPA and MMP-7 genes, our studies demonstrate that uPA is a direct PDEF target gene and that binding to the uPA promoter in vivo results in reduced transcription. In addition to defining uPA as a direct PDEF target gene, our previous studies have demonstrated that exogenous expression of uPA on a heterologous promoter is able to restore the migratory phenotype of breast cancer cells in the presence of exogenous PDEF [Turner et al., 2008], supporting the model that uPA is a critical target associated with PDEF-mediated negative regulation of cellular migration.
We also demonstrate that PDEF is expressed in non-tumor human colon tissue. Intestinal mucosa is covered with an epithelial lining that turns over continuously throughout life. Thus, the precise regulation of cell proliferation, differentiation, and migration is crucial for maintenance of an intact epithelial layer. PDEF is one of the subset of Ets factors that have a restricted pattern of expression limited to tissues with high epithelial content. These Epithelial-specific Ets (Ese) Factors include Ese1 (also designated Ert/Jen/Elf3/Esx), Ese2 (Elf5), Ese3 (Ehf), and PDEF (Pse). Epithelial specific Ets factors have been demonstrated to regulate genes involved in epithelial cell differentiation [Feldman et al., 2003b]. Ese1, Ese3 and PDEF are expressed in colon epithelium. Knockout studies of Ese1 have also demonstrated that Ese factors may be important for proper maturation of intestinal epithelium [Ng et al., 2002]. Mice lacking Ese1 have differentiation defects in both absorptive enterocytes and goblet cells. These studies highlight the potential of ETS transcription factors to control events leading to cell cycle arrest and subsequent terminal differentiation.
Colon cancer may develop if this balance is shifted towards increased proliferation together with decreased apoptosis. Genetic abnormalities associated with the progression of colon cancer have pinpointed several key mutational events. We have previously described a model where conversion of tumor suppressor ETS factors towards oncogenic ETS factors occurs during cancer progression [Turner and Watson, 2008]. ETS1 and ETS2 are upregulated in colon adenocarcinomas [Ito et al., 2002] and their pro-proliferative and pro-invasive activities place them among the oncogenic ETS factors. PDEF likely plays a role in regulating processes related to proliferation, differentiation, or apoptosis in the colon. Dysregulation of these processes occurs in cancer whereby cells can assume a more undifferentiated state and become resistant to apoptosis. In support of the model of PDEF as a tumor suppressor ETS factor, we have shown that PDEF mRNA and protein are lost or significantly reduced in the majority of colon cancer tissue specimens compared to non-tumor tissue. This data is consistent with the findings of loss of PDEF protein expression in prostate [Nozawa et al., 2000], breast [Feldman et al., 2003a] and ovarian [Ghadersohi et al., 2008] cancer tissues. Interestingly, PDEF loss in colon cancer is predominantly regulated at the transcriptional level, unlike the posttranscriptional regulation described in prostate [Nozawa et al., 2000] and breast [Feldman et al., 2003a; Findlay et al., 2008] cancer. The possibility that the PDEF promoter may become methylated in colon cancer remains to be examined.
We find an inverse correlation between PDEF protein expression and uPA and MMP-7 mRNA expression in colon cancer clinical samples. Elevated expression of uPA [Zlobec et al., 2008] and MMP-7 [Roeb et al., 2004] are a characteristic of colon cancer and are correlated with significantly poorer prognosis. The functional significance of uPA in colon cancer tumor growth and progression has been demonstrated in mice. Tumor formation in APC/Mmin+ mice was reduced in a uPA null background [Ploplis et al., 2007]. MMP-7 expression has been shown to enhance metastatic potential of human colon cancer cells in mice [Kioi et al., 2003]
In summary, PDEF is an epithelial tissue specific ETS transcription factor that is down-regulated during colon carcinogenesis. Recent studies have demonstrated an inverse correlation between PDEF expression in human breast [Ghadersohi et al., 2007] and ovarian [Ghadersohi et al., 2008] tumor specimens and cancer patient survivability. Cancer death is due in large part to metastases, leading to one of the more interesting challenges to understand cellular changes that promote progression to invasive cancer. The studies reported here and these clinical studies further support the role of PDEF as a negative regulator of progression to invasive and metastatic cancer. These observations also support the notion that PDEF has a significant role in maintaining proper homeostatic balance in the colon epithelium.
Acknowledgments
We thank Drs. Victoria J. Findlay and Patricia M. Watson for critical review of this manuscript. This work was supported in part by a grant from the National Institutes of Health (P01CA78582). The authors also acknowledge support from Margaret Romano (HCC Tissue Bank) and the Flow Cytometry & Cell Sorting Shared Resource of the Hollings Cancer Center. This shared resource is supported in part by a Cancer Center Support Grant (P30 CA 138313). Tissue samples were also provided by the Cooperative Human Tissue Network which is funded by the National Cancer Institute.
Funded by:
NIH; Grant number: P01CA78582
References
- Barker N, van de Wetering M, Clevers H. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier F, Taylor AC, LuValle P. The Raf-1/MEK/ERK pathway regulates the expression of the p21(Cip1/Waf1) gene in chondrocytes. J Biol Chem. 1999;274:30273–30279. doi: 10.1074/jbc.274.42.30273. [DOI] [PubMed] [Google Scholar]
- Carbone GM, Napoli S, Valentini A, Cavalli F, Watson DK, Catapano CV. Triplex DNA-mediated downregulation of Ets2 expression results in growth inhibition and apoptosis in human prostate cancer cells. Nucleic Acids Res. 2004;32:4358–4367. doi: 10.1093/nar/gkh744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Xie G, Raufman JP. Matrix metalloproteinase-7-catalyzed release of HB-EGF mediates deoxycholyltaurine-induced proliferation of a human colon cancer cell line. Biochem Pharmacol. 2007;73:1001–1012. doi: 10.1016/j.bcp.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orazio D, Besser D, Marksitzer R, Kunz C, Hume DA, Kiefer B, Nagamine Y. Cooperation of two PEA3/AP1 sites in uPA gene induction by TPA and FGF-2. Gene. 1997;201:179–187. doi: 10.1016/s0378-1119(97)00445-9. [DOI] [PubMed] [Google Scholar]
- Feldman RJ, Sementchenko VI, Gayed M, Fraig MM, Watson DK. PDEF expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res. 2003a;63:4626–4631. [PubMed] [Google Scholar]
- Feldman RJ, Sementchenko VI, Watson DK. The Epithelial-Specific Ets Factors Occupy a Unique Position in Defining Epithelial Proliferation, Differentiation, and Carcinogenesis. Anticancer Research. 2003b;23:2125–2132. [PubMed] [Google Scholar]
- Findlay VJ, Turner DP, Moussa O, Watson DK. MicroRNA-mediated inhibition of PDEF mRNA translation affects PDEF regulatory networks in human breast cancer. Cancer Research. 2008;68:8499–8506. doi: 10.1158/0008-5472.CAN-08-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funaoka K, Shindoh M, Yoshida K, Hanzawa M, Hida K, Nishikata S, Totsuka Y, Fujinaga K. Activation of the p21(Waf1/Cip1) promoter by the ets oncogene family transcription factor E1AF. Biochem Biophys Res Commun. 1997;236:79–82. doi: 10.1006/bbrc.1997.6909. [DOI] [PubMed] [Google Scholar]
- Ghadersohi A, Odunsi K, Zhang S, Azrak RG, Bundy BN, Manjili MH, Li F. Prostate-derived Ets transcription factor as a favorable prognostic marker in ovarian cancer patients. Int J Cancer. 2008;123:1376–1384. doi: 10.1002/ijc.23667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadersohi A, Pan D, Fayazi Z, Hicks DG, Winston JS, Li F. Prostate-derived Ets transcription factor (PDEF) downregulates survivin expression and inhibits breast cancer cell growth in vitro and xenograft tumor formation in vivo. Breast Cancer Res Treat. 2007;102:19–30. doi: 10.1007/s10549-006-9314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Zerbini LF, Otu HH, Bhasin M, Yang Q, Joseph MG, Grall F, Onatunde T, Correa RG, Libermann TA. Reduced PDEF expression increases invasion and expression of mesenchymal genes in prostate cancer cells. Cancer Res. 2007;67:4219–4226. doi: 10.1158/0008-5472.CAN-06-3689. [DOI] [PubMed] [Google Scholar]
- Ito Y, Takeda T, Okada M, Matsuura N. Expression of ets-1 and ets-2 in colonic neoplasms. Anticancer Res. 2002;22:1581–1584. [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Kioi M, Yamamoto K, Higashi S, Koshikawa N, Fujita K, Miyazaki K. Matrilysin (MMP-7) induces homotypic adhesion of human colon cancer cells and enhances their metastatic potential in nude mouse model. Oncogene. 2003;22:8662–8670. doi: 10.1038/sj.onc.1207181. [DOI] [PubMed] [Google Scholar]
- Liu W, Wang G, Yakovlev AG. Identification and functional analysis of the rat caspase-3 gene promoter. J Biol Chem. 2002;277:8273–8278. doi: 10.1074/jbc.M110768200. [DOI] [PubMed] [Google Scholar]
- Mitas M, Mikhitarian K, Hoover L, Lockett MA, Kelley L, Hill A, Gillanders WE, Cole DJ. Prostate-Specific Ets (PSE) factor: a novel marker for detection of metastatic breast cancer in axillary lymph nodes. Br J Cancer. 2002;86:899–904. doi: 10.1038/sj.bjc.6600190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Miyamoto S, Maeda H, Ishii G, Hasebe T, Chiba T, Asaka M, Ochiai A. Matrix metalloproteinase-7 degrades all insulin-like growth factor binding proteins and facilitates insulin-like growth factor bioavailability. Biochem Biophys Res Commun. 2005;333:1011–1016. doi: 10.1016/j.bbrc.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Ng AY, Waring P, Ristevski S, Wang C, Wilson T, Pritchard M, Hertzog P, Kola I. Inactivation of the transcription factor Elf3 in mice results in dysmorphogenesis and altered differentiation of intestinal epithelium. Gastroenterology. 2002;122:1455–1466. doi: 10.1053/gast.2002.32990. [DOI] [PubMed] [Google Scholar]
- Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- Nozawa M, Yomogida K, Kanno N, Nonomura N, Miki T, Okuyama A, Nishimune Y, Nozaki M. Prostate-specific transcription factor hPSE is translated only in normal prostate epithelial cells. Cancer Res. 2000;60:1348–1352. [PubMed] [Google Scholar]
- Oettgen P, Finger E, Sun Z, Akbarali Y, Thamrongsak U, Boltax J, Grall F, Dube A, Weiss A, Brown L, Quinn G, Kas K, Endress G, Kunsch C, Libermann TA. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem. 2000;275:1216–1225. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]
- Park JS, Qiao L, Gilfor D, Yang MY, Hylemon PB, Benz C, Darlington G, Firestone G, Fisher PB, Dent P. A role for both Ets and C/EBP transcription factors and mRNA stabilization in the MAPK-dependent increase in p21 (Cip-1/WAF1/mda6) protein levels in primary hepatocytes. Mol Biol Cell. 2000;11:2915–2932. doi: 10.1091/mbc.11.9.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploplis VA, Tipton H, Menchen H, Castellino FJ. A urokinase-type plasminogen activator deficiency diminishes the frequency of intestinal adenomas in ApcMin/+ mice. J Pathol. 2007;213:266–274. doi: 10.1002/path.2236. [DOI] [PubMed] [Google Scholar]
- Roeb E, Arndt M, Jansen B, Schumpelick V, Matern S. Simultaneous determination of matrix metalloproteinase (MMP)-7, MMP-1,-3, and-13 gene expression by multiplex PCR in colorectal carcinomas. Int J Colorectal Dis. 2004;19:518–524. doi: 10.1007/s00384-004-0592-6. [DOI] [PubMed] [Google Scholar]
- Sementchenko VI, Watson DK. Ets target genes: past, present and future. Oncogene. 2000;19:6533–6548. doi: 10.1038/sj.onc.1204034. [DOI] [PubMed] [Google Scholar]
- Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–2478. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Soldatenkov VA, Albor A, Patel BK, Dreszer R, Dritschilo A, Notario V. Regulation of the human poly(ADP-ribose) polymerase promoter by the ETS transcription factor. Oncogene. 1999;18:3954–3962. doi: 10.1038/sj.onc.1202778. [DOI] [PubMed] [Google Scholar]
- Tamir A, Howard J, Higgins RR, Li YJ, Berger L, Zacksenhaus E, Reis M, Ben-David Y. Fli-1, an Ets-related transcription factor, regulates erythropoietin-induced erythroid proliferation and differentiation: evidence for direct transcriptional repression of the Rb gene during differentiation. Molecular & Cellular Biology. 1999;19:4452–4464. doi: 10.1128/mcb.19.6.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DP, Findlay VJ, Kirven AD, Moussa O, Watson DK. Global Gene Expression Analysis Identifies PDEF Transcriptional Networks Regulating Cell Migration during Cancer Progression. Mol Biol Cell. 2008;19:3745–3757. doi: 10.1091/mbc.E08-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DP, Findlay VJ, Moussa O, Watson DK. Defining ETS transcription regulatory networks and their contribution to breast cancer progression. J Cell Biochem. 2007a;102:549–559. doi: 10.1002/jcb.21494. [DOI] [PubMed] [Google Scholar]
- Turner DP, Moussa O, Sauane M, Fisher PB, Watson DK. Prostate-derived ETS factor is a mediator of metastatic potential through the inhibition of migration and invasion in breast cancer. Cancer Res. 2007b;67:1618–1625. doi: 10.1158/0008-5472.CAN-06-2913. [DOI] [PubMed] [Google Scholar]
- Turner DP, Watson DK. ETS transcription factors: oncogenes and tumor suppressor genes as therapeutic targets for prostate cancer. Expert Rev Anticancer Ther. 2008;8:33–42. doi: 10.1586/14737140.8.1.33. [DOI] [PubMed] [Google Scholar]
- van den Brink GR, Offerhaus GJ. The morphogenetic code and colon cancer development. Cancer Cell. 2007;11:109–117. doi: 10.1016/j.ccr.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Zlobec I, Minoo P, Baumhoer D, Baker K, Terracciano L, Jass JR, Lugli A. Multimarker phenotype predicts adverse survival in patients with lymph node-negative colorectal cancer. Cancer. 2008;112:495–502. doi: 10.1002/cncr.23208. [DOI] [PubMed] [Google Scholar]