Abstract
Cytotoxic T lymphocytes (CTL) are critical for lentivirus control including EIAV. Since CTL from most EIAV carrier horses recognize Gag epitope clusters (EC), the hypothesis that carrier horses would have high functional avidity CTL to optimal epitopes in Gag EC was tested. Twenty-two optimal EC epitopes were identified; two in EC1, six in EC2, and seven each in EC3 and 4. However, only five of nine horses had high functional avidity CTL (≤11 nM) recognizing six epitopes in EC; four in relatively conserved EC3; and one each in EC1 and 2. Horses with high functional avidity CTL had significantly more days since the last clinical episode than horses with low avidity CTL, and this was not explained by analyzing duration of infection. Furthermore, there was a significant inverse correlation between the CTL functional avidity of the nine horses and the days since the last clinical episode. Gag CTL epitope escape variants were found in three horses, but only one of these was recognized by high functional avidity CTL. Thus, not all carrier horses had high functional avidity CTL to Gag EC, but those that did had longer periods without disease episodes.
Keywords: CTL, Epitope cluster, EIAV, Lentivirus, Functional avidity, Gag, Matrix, Capsid
Introduction
EIAV, a lentivirus, causes persistent infection in horses that is initially characterized by recurrent viremia with fever, thrombocytopenia, and anemia (Sellon et al., 1994). Disease episodes are associated with viral antigenic variants as defined by neutralizing antibody (Hussain et al., 1987; Kono et al., 1973, 1976) and recently by CTL (Mealey et al., 2003). However, in most infected horses, disease episodes are eventually controlled within the first 6 to 12 months of infection, and a lifelong inapparent carrier stage follows (Coggins, 1984). Immune responses to EIAV are critical for termination of the initial viremia, as shown by comparison of viremia between normal and severe combined immunodeficient (SCID) foals which lack functional T and B lymphocytes (Mealey et al., 2001; Perryman et al., 1988). Based on accumulated data, the protective immune responses for the control of EIAV viremia include neutralizing antibody and CTL (Hammond et al., 1997, 2000; Mealey et al., 2001, 2003; Perryman et al., 1988; Zhang et al., 1998).
In HIV-1 and SIV infections, CTL responses correlate with virus clearance (Kuroda et al., 1999; Ogg et al., 1998). Depletion of CD8+ cells in SIV-infected macaques demonstrate that these cells are required for SIV control (Schmitz et al., 1999). Several experiments indicate that CTL responses and not neutralizing antibody resolve early episodes of viremia and clinical disease in EIAV-infected horses (Carpenter et al., 1987; McGuire et al., 1994; Mealey et al., 2003). Evaluation of CTL responses to HIV-1 (Novitsky et al., 2002) and EIAV (Chung et al., 2004; Zhang et al., 1998) demonstrates that Gag proteins are recognized by CTL from most infected individuals. HIV-1 subtype C isolates have low amino acid diversity in Gag CA protein, and infected patients have an immunodominant CTL response to this protein (Novitsky et al., 2002). EIAV Gag MA and CA proteins are the most abundant and conserved viral proteins (Montelaro et al., 1993) and are also the most frequently recognized viral proteins by CTL from longtime inapparent carrier horses infected with EIAVWSU5 (McGuire et al., 2000). CTL from EIAV carrier horses with diverse MHC class I alleles recognize epitope clusters (EC) in Gag MA and CA proteins (Chung et al., 2004). Three of four EC are recognized by CTL from >50% of horses with diverse MHC class I alleles (Chung et al., 2004). Two of these EC are from relatively conserved regions of Gag MA and CA proteins. CTL with high functional avidity to such conserved EC could be important in controlling viral load in EIAV carrier horses.
During the acute phase of SIV infection, epitopes recognized by CTL with high functional avidity escaped, and it was hypothesized that such CTL were more effective than CTL that did not select epitope change (O’Connor et al., 2002). The functional and structural constraints on particular viral regions may also influence the rate of CTL escape because mutations in those regions may result in replication defective variants (Matano et al., 2004). In EIAV-infected horses, results suggest that the epitope specificity of high and moderate functional avidity CTL is an important determinant for disease outcome (Mealey et al., 2003). Therefore, the functional avidity of CTL from EIAV carrier horses to optimal epitopes in relatively conserved Gag EC was evaluated.
In this study, optimal CTL epitopes in Gag EC were mapped in order to determine the functional avidity of CTL from nine long-term EIAV carrier horses that were initially used to identify the EC. Since all the horses had been infected for ≥526 days and there had been no clinical episode for ≥167 days, the hypothesis was that carrier horses would have high avidity CTL to optimal epitopes in Gag EC. Surprisingly, the nine horses separated into two groups: five with high functional avidity CTL to optimal EC epitopes and four with low avidity CTL. Since the specificity of the entire EIAV-specific CTL responses was not mapped, it is possible that high avidity CTL were present in all the horses to epitopes in proteins other than Gag. When viral RNA could be amplified from EIAV-infected horse plasma, it was sequenced and evaluated for CTL escape variants. CTL escape variants were detected in three horses, but only one variant epitope was recognized by CTL with high functional avidity. Conserved EC3 had the most optimal epitopes recognized by high functional avidity CTL. That four carrier horses had CTL to Gag EC with low functional avidity did not support the hypothesis; however, those horses with high functional avidity CTL had significantly more days since the last clinical episode than horses with low functional avidity CTL.
Results
Mapping optimal CTL epitopes in Gag MA and CA EC
EC were previously identified using CTL from 12 EIAV-infected horses and target cells pulsed with overlapping 15–16 amino acid peptides (Chung et al., 2004). In this paper, 9 of the 12 horses were still available for use including six with unique MHC class I alleles (Table 1). To define optimal CTL epitopes, PBMC from each horse were stimulated with a pool of 15–16 amino acid peptides that were recognized by CTL from that horse (Chung et al., 2004) and evaluated with autologous EK target cells pulsed with truncated versions of the recognized peptides. Based on comparison of percent specific lysis and peptide pulsing concentrations, a total of 24 optimal epitopes were mapped in MA and CA proteins (Fig. 1). One to seven optimal epitopes were identified by CTL from each of the nine horses (Table 2). These optimal epitopes ranged in length from 8 to 12 amino acids, with 12 of 24 epitopes being 12 amino acids (Table 2).
Table 1.
MHC class I haplotypes of EIAV-infected horses used for Gag CTL epitope mapping
| Horse no. | MHC class I alleles (Chung et al., 2003) | ELA-A haplotypes |
|---|---|---|
| H593 | 113, 114 | A1a |
| H596 | 115, 116, 117, 118, 119 | A4/W11 |
| H601 | Not done | A1/W11 |
| H610 | 111, 121, 122 | A6a |
| H614 | 123, 124, 125, 126, 127 | A9a |
| H629 | Not done | A3/A5 |
| H631 | 133, 134, 135, 136 | Undeterminedb |
| A2140 | Not done | A1/W11 |
| A2147 | 7-1, 7-4 | A4a |
Only one ELA-A type was detected with the antisera used (A1–A10 and W11).
Undetermined indicates that none of the available antisera reacted with lymphocytes from this horse.
Fig. 1.

Distribution of optimal epitopes in Gag EC recognized by CTL from nine EIAV-infected horses. Horse numbers in boxes had high functional avidity CTL (≤11 nM) to the epitope.
Table 2.
Optimal Gag MA and CA epitopes recognized by CTL from EIAV-infected horses
| Horse no. | Optimal epitopes | Gag location (name) | Epitope cluster | Avidity (nM) |
|---|---|---|---|---|
| H593 | DCTSEEMNAF | CA 47–56 (DF10) | Non EC | >1000 |
| KSEGHPQEISKF | CA 173–184 (KF12) | EC3 | 3.7 | |
| H596 | KMGLQINNVVDG | MA 86–97 (KG12) | EC2 | >1000 |
| YPEFVDRLLSQI | CA 161–172 (YI12) | EC3 | 2.7 | |
| FVDRLLSQIKSEa | CA 164–175 (FE12) | EC3 | 9.9 | |
| IKSEGHPQEI | CA 172–181 (II10) | EC3 | 0.9 | |
| H601 | GSQKLTTGNCNW | MA 21–32 (GW12) | EC1 | 10.9 |
| H610 | YEKKTANK | MA 108–115 (YK8) | EC2 | >1000 |
| MRHLRPEDTL | CA 202–211 (ML10) | EC4 | >1000 | |
| RPEDTLEEKMYA | CA 206–217 (RA12) | EC4 | >1000 | |
| EDTLEEKMYACR | CA 208–219 (ER12) | EC4 | >1000 | |
| YACRDIGTTKQK | CA 216–227 (YK12) | EC4 | >1000 | |
| H614 | DRLLSQIKSE | CA 166–175 (DE10) | EC3 | >1000 |
| H629 | NEECRNAMRHLR | CA 195–206 (NR12) | EC4 | >1000 |
| IGTTKQKMM | CA 221–229 (IM9) | EC4 | >1000 | |
| H631 | KALKKLEKV | MA 9–17 (KV9) | EC1 | 71.3 |
| RAKYEKKTANK | MA 105–115 (RK11) | EC2 | 7.9 | |
| A2140 | GSQKLTTGNCNW | MA 21–32 (GW12) | EC1 | 3.9 |
| A2147 | RTWWAISAVK | MA 77–86 (RK10) | EC2 | >1000 |
| WWAISAVKMGL | MA 79–89 (WL11) | EC2 | >1000 | |
| WAISAVKMGLQI | MA 80–91 (WI12) | EC2 | >1000 | |
| GIKVMIGKPKAQ | CA 140–151 (GQ12) | Non EC | >1000 | |
| PEFVDRLLSQ | CA 162–171 (PQ10) | EC3 | >1000 | |
| RLLSQIKSEG | CA 167–176 (RG10) | EC3 | >1000 | |
| TLTIQNANEECRb | CA 188–199 (TR12) | EC4 | >1000 |
Peptide EFVDRLLSQIKS was also recognized by CTL from H596 with high functional avidity (10 nM) but was considered the same epitope as FVDRLLSQIKSE.
Only C-terminal eight amino acid residues of TLTIQNANEECR were in EC4 (Chung et al., 2004).
It was of interest to evaluate the number of optimal epitopes that occurred in the previously described EC (Fig. 2). EC1 contained two optimal epitopes recognized by CTL from three horses, EC2 contained six recognized by four horses, EC3 contained seven recognized by four horses, and EC4 contained seven recognized by three horses (Fig. 2 and Table 3). There were two optimal epitopes located in two non-EC regions recognized by CTL from one horse each (Table 3).
Fig. 2.
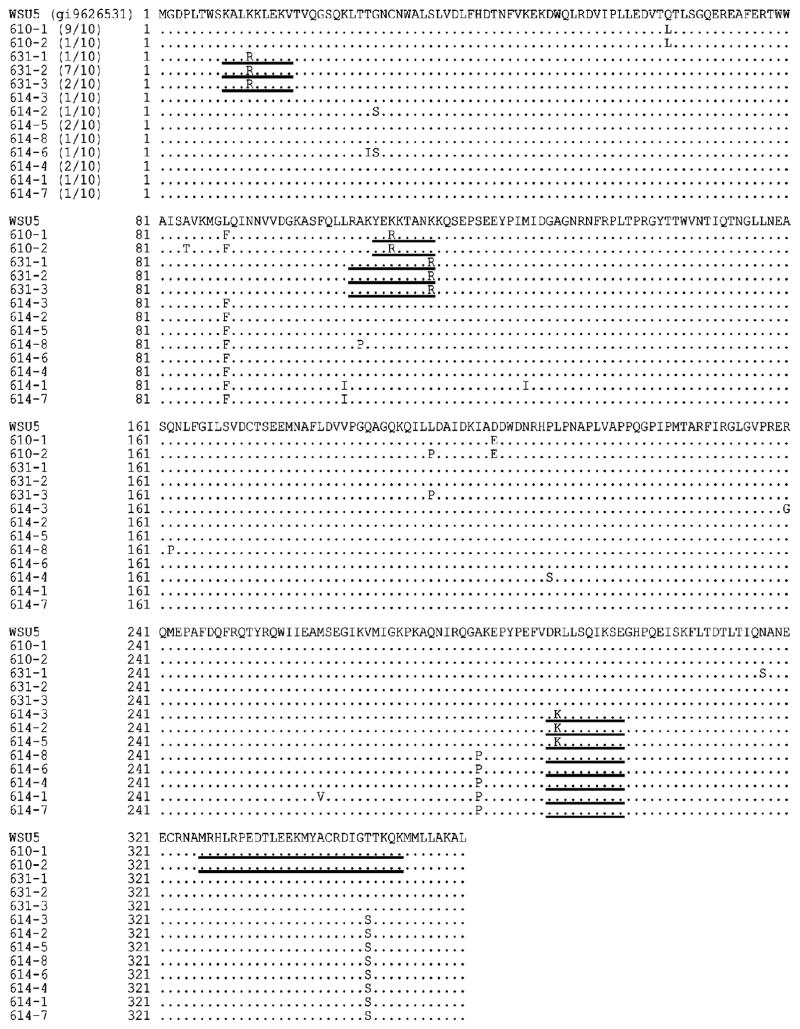
Multiple alignments of variant Gag MA and CA protein sequences identified from EIAV-infected carrier horses H610, H614, and H631. Underlined are optimal peptide sequences recognized by CTL from each horse. In parentheses, following the horse number and sequence designation are the number of clones with that sequence/total number of clones sequenced from each horse.
Table 3.
Number of CTL epitopes in EC and non-EC regions and the number (in parentheses) recognized by CTL with high functional avidity (≤11)
Non-EC indicates epitopes in regions outside EC1–4 (Chung et al., 2004).
One of these two epitopes was recognized by CTL from two horses sharing an ELA-A haplotype.
This number includes one epitope recognized by CTL from horse H593 and three epitopes recognized by CTL from H596.
Functional avidity of CTL recognizing optimal epitopes
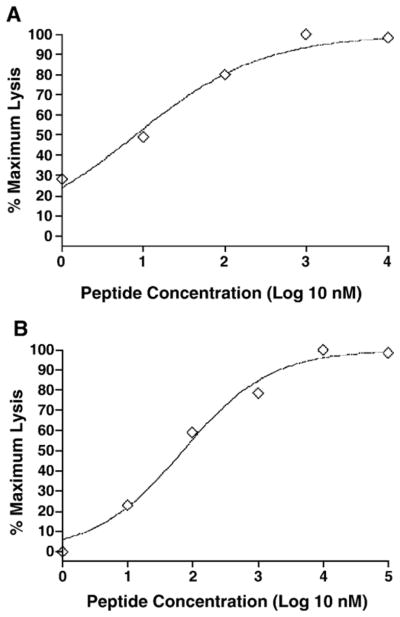
To determine the functional avidity, PBMC from each horse were stimulated with a pool of 15–16 amino acid peptides (final concentration of each peptide 103 nM) recognized by CTL from each horse. They were tested for recognition of EK cell targets pulsed with 100 to 104 nM of truncated versions of each peptide recognized by CTL from each horse. Representative data of a high functional avidity CTL and data from the moderate functional avidity CTL are in Fig. 3. Only five of nine carrier horses had CTL with high functional avidity (≤11 nM peptide, Table 2), whereas the other four horses had CTL with low functional avidity (>1000 nM peptide, Table 2). The CTL with the highest functional avidity (0.9 nM) were from horse H596 which recognized autologous EK cell targets pulsed with an optimal ten amino acid CA epitope (IKSEGHPQEI) (Table 2). Two other optimal epitopes were recognized by high avidity CTL from horse H596 (2.7 and 9.9 nM, Table 2). High functional avidity CTL from four other horses (H593, H631, A2140, and H601) recognized one optimal epitope each. Horse H631 also had CTL that recognized an optimal epitope with moderate functional avidity (71.3 nM). Among the six unique epitopes recognized by high avidity CTL (≤11 nM), four were 12 amino acids long, and two were 11 and 10 amino acids long (Table 2).
Fig. 3.
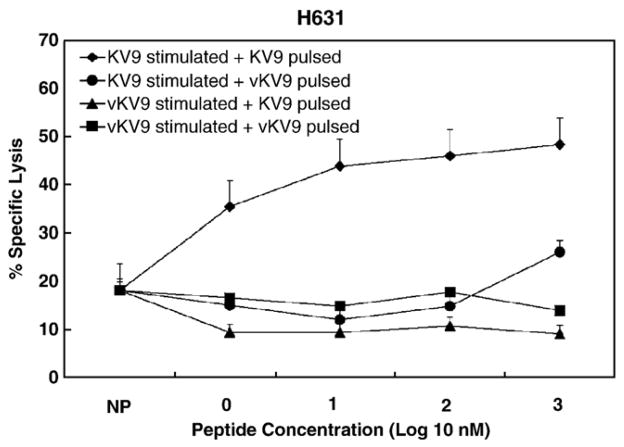
Representative data used for determining CTL functional avidity. Panel A contains data from high avidity H631 CTL (EC50 = 7.9) stimulated with peptide RK11 and evaluated on EK cell targets pulsed with different concentrations of RK11. Panel B contains data from moderate avidity H631 CTL (EC50 = 71.3) stimulated with peptide KV9 and evaluated on EK cell targets pulsed with different concentrations of KV9.
High functional avidity CTL recognition of optimal epitopes in EC
EC1–4 were evaluated to determine the number of optimal epitopes recognized by high functional avidity CTL (≤11 nM). EC1 contained one optimal epitope recognized by high functional avidity CTL from horses A2140 and H601, and EC2 contained one optimal epitope recognized by high avidity CTL from horse H631 (Table 3, in parentheses). High functional avidity CTL from horse H593 recognized one EC3 epitope, whereas high avidity CTL from H596 recognized three EC3 epitopes. Neither EC4 nor the non-EC regions had epitopes recognized by CTL with high functional avidity (Table 3). Therefore, conserved EC3 (Chung et al., 2004) had the most optimal epitopes recognized by high functional avidity CTL.
Amino acid sequence variation in Gag MA and CA optimal CTL epitopes in plasma virus from carrier horses
EIAV Gag RNA amplification from plasma by RT-PCR was attempted in all but successful in only three of the nine EIAV carrier horses during this study. DNA sequences of ten clones from each amplified product were evaluated to determine if variation occurred in optimal CTL epitopes. Two types of variant Gag sequences from horse H610 were found among the ten clones (Fig. 2). All ten clones had one amino acid change in MA epitope YK8 recognized by horse H610 CTL, whereas there were no changes in the four overlapping CA epitopes (Fig. 2). The ten clones from horse H631 plasma had three variant Gag sequences; however, all ten had one amino acid change in each of the two MA epitopes (KV9 and RK11) recognized by horse H631 CTL (Fig. 2). The ten clones from horse H614 plasma contained eight variant Gag sequences, and three of these sequences representing a total of four clones had a single amino acid change in epitope DE10 which was the only epitope recognized by CTL from H614 (Fig. 2). The other six clones had no amino acid changes in epitope DE10 (Fig. 2).
Surprisingly, all of the amino acid changes identified in optimal epitopes were clearly conservative changing lysine (K) to arginine (R) or vice versa (Table 4). The K in position three of epitope YK8, recognized by CTL from horse H610, was replaced with R in all 10 Gag sequences (Table 4). The R in position two of CTL epitope DE10 was changed to K in only four of ten clones from horse H614 plasma (Table 4). The K in position four of CTL epitope KV9 and the K in C-terminal position 11 of CTL epitope RK11 were both changed to R in all 10 clones from horse H631 plasma (Table 4).
Table 4.
Gag MA and CA CTL epitope variant in virus sequences from carrier horse plasma
| Horse no. | No. of sequences with epitope variant/total | Epitopes and location | Epitope variant and amino acid change |
|---|---|---|---|
| H610 | 10/10 | YK8 (YEKKTANK), MA 108–115 | vYK8 (3K → R) |
| H614 | 4/10 | DE10 (DRLLSQIKSE), CA 166–175 | vDE10 (2R → K) |
| H631 | 10/10 | KV9 (KALKKLEKV), MA 9–17 | vKV9 (4K → R) |
| H631 | 10/10 | RK11 (RAKYEKKTANK), MA 105–115 | vRK11 (11K → R) |
One conservative amino acid change in three different Gag epitopes resulted in CTL escape
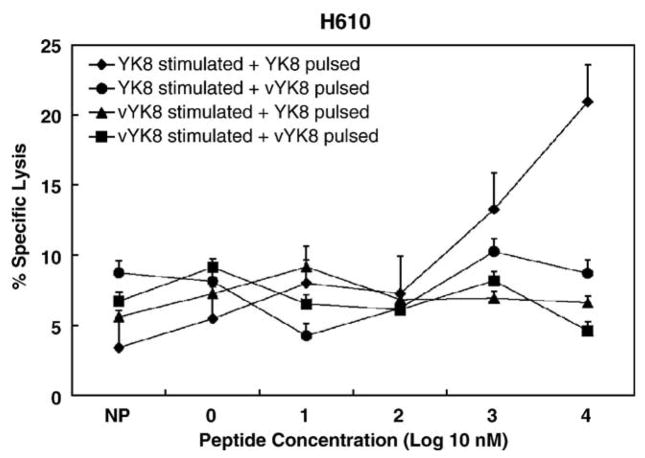
To determine whether the amino acid changes in Gag epitopes recognized by CTL resulted in escape variants, PBMC were stimulated with wild-type or variant peptides and evaluated using EK cell targets pulsed with different concentrations of these peptides. Horse H610 CTL stimulated with peptide YK8 significantly lysed EK cells pulsed with YK8, but not EK cells pulsed with variant peptide vYK8 (Fig. 4). Furthermore, CTL stimulated with vYK8 did not significantly lyse EK cells pulsed with either YK8 or vYK8 peptide. Therefore, the K → R change at position 3 in YK8 resulted in escape from horse H610 CTL, even though these CTL had low functional avidity (>1000 nM, Table 2).
Fig. 4.
CTL escape by vYK8 peptide from horse H610 CTL recognizing wild-type peptide YK8. The peptides used to stimulate PBMC and for pulsing EK cell targets are listed in the legend. Bars above the data points are SEs.
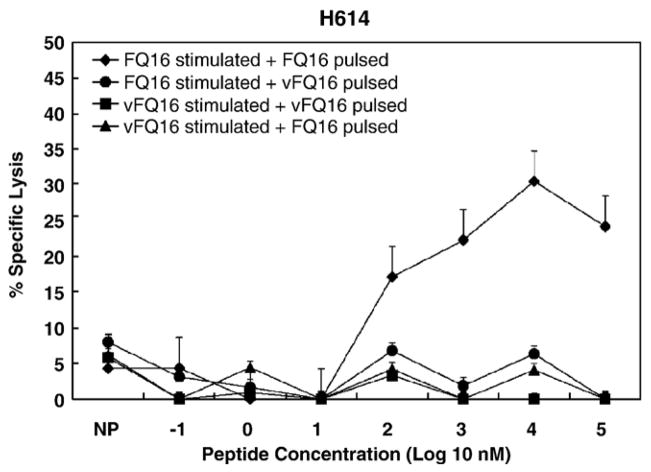
A one amino acid change occurred in wild-type FQ16 (FVDRLLSQIKSEGHPQ) peptide used for initial CTL detection with PBMC from horse H614. CTL stimulated with either FQ16 or vFQ16 failed to lyse EK targets pulsed with vFQ16, whereas CTL stimulated with FQ16 significantly lysed EK targets pulsed with FQ16, but not those pulsed with vFQ16 (Fig. 5). The optimal epitope in FQ16 was DE10 (DRLLSQIKSE), but CTL escape of this optimal epitope was not determined due to euthanasia of horse H614 following a clinical problem unrelated to EIAV infection. Since the R → K change at position four in the FQ16 peptide caused escape from horse H614 CTL, it is likely that this change in position two of optimal epitope DE10 had a similar effect.
Fig. 5.
CTL escape by vFQ16 peptide from horse H614 CTL recognizing wild-type FQ16 peptide. The peptides used to stimulate PBMC and for pulsing EK cell targets are listed in the legend. Bars above the data points are SEs.
CTL from horse H631 stimulated with optimal epitope KV9 (KALKKLEKV) recognized EK cell targets pulsed with KV9; however, there was no recognition of targets pulsed with vKV9 (4K → R) (Fig. 6). Moreover, there was no recognition of targets pulsed with either KV9 or vKV9 when the effectors were stimulated with vKV9 (Fig. 6).
Fig. 6.
CTL escape by vKV9 peptide from horses H631 CTL recognizing wild-type KV9 peptide. The peptides used to stimulate PBMC and for pulsing EK cell targets are listed in the legend. Bars above the data points are SEs.
Thus, a conservative amino acid change in three different Gag epitopes resulted in CTL escape in three different EIAV-infected horses. For horse H614, this resulted in loss of Gag recognition by CTL because only a single Gag epitope was recognized; however, only four of ten clones sequenced had the variant epitope (Table 4). Horse H610 had plasma virus with variation in one epitope that resulted in CTL escape; however, CTL responses to Gag were maintained in this horse because other Gag epitopes that did not change were still recognized. Plasma virus variants from horse H631 had changes in both optimal epitopes recognized in Gag protein; KV9 was described in this section and RK11 was described in the next section.
Clinical disease parameters in EIAV-infected horses with high functional avidity CTL
To determine whether high functional avidity CTL responses to Gag epitopes were related with virus control in the nine EIAV-infected horses, the number of days since the last clinical episode and RNA copies/ml plasma were evaluated (Table 5). The five horses with CTL recognizing Gag MA or CA optimal epitopes with high functional avidities (≤11 nM) had significantly more days since the last clinical episode (P < 0.05) than the four horses with low avidity CTL (>1000). There was no significant difference between the days post-infection in the horses with high functional avidity CTL and those with low avidity CTL (P > 0.05), indicating that having CTL with high functional avidity was not simply a matter of being infected with EIAV for a longer time (Table 5). The standard deviations for the means of the two groups did not overlap (Table 5). Furthermore, there was no significant difference in the total number of epitopes recognized between horses with high and low functional avidity CTL (P > 0.05). When all nine horses in Table 5 were considered, there was a significant inverse correlation (P < 0.05) between the number of days since the last clinical episode and CTL avidity to Gag peptides with a Spearman’s rank correlation coefficient of −0.81. The rank correlation coefficient of −0.45 between days post-infection and CTL avidity was not significant (P > 0.05). In plasma samples obtained 1 month after the CTL avidity was determined, the only horse with a relatively high number of viral RNA copies/ml was horse H614 (Table 5). This horse had low functional avidity CTL to a single Gag epitope which varied and escaped CTL recognition.
Table 5.
Number of CTL epitopes recognized and clinical disease episodes
| Horse no. | No. of CTL epitopes | No. of epitopes recognized by high avidity CTL (≤11 nM) | Days post-infection | Days since last clinical episode | Viral RNA copies/ml plasma |
|---|---|---|---|---|---|
| H593 | 2 | 1 | 1651 | 952 | <10 |
| H596 | 4 | 3 | 845 | 781 | 40 |
| H601 | 1 | 1 | 845 | 605 | <10 |
| H631 | 3 | 1 | 692 | 692 | 17 |
| A2140 | 1 | 1 | 1748 | 1402 Mean 885 (315 SD) |
<10 |
| H610 | 5 | 0 | 692 | 447 | <10 |
| H614 | 1 | 0 | 832 | 482 | 27,090 |
| H629 | 3 | 0 | 526 | 511 | <10 |
| A2147 | 7 | 0 | 1367 | 167 Mean 402 (158 SD) |
<10 |
Discussion
Most HIV-1-infected humans ultimately have immunodeficiency leading to death from opportunistic infections and malignancy (Sleasman and Goodenow, 2003). In contrast, most EIAV-infected horses control the viremia and clinical disease episodes within the first year of infection by specific lymphocyte immune responses including CTL (Mealey et al., 2001; Perryman et al., 1988). Therefore, the mechanisms EIAV-infected horses use for controlling viral load are of interest. The presence of CTL correlates with control of the initial viremia in infected horses (McGuire et al., 1994), whereas neutralizing antibody appears later (Carpenter et al., 1987; Montelaro et al., 1984; O’Rourke et al., 1988). Furthermore, CTL to Gag protein epitopes occur in most infected horses and are present in sufficient numbers in carriers to be involved in virus control (Chung et al., 2004; McGuire et al., 2000). Both the quantity and quality of Gag CTL responses could be important for controlling clinical disease episodes in EIAV carriers. CTL responses to several Gag epitopes could effectively control viremia because escape mutations would have to occur in all the recognized epitopes for a loss of viral control. Alternatively, high functional avidity CTL to a small number of relatively conserved epitopes could effectively control virus.
In this study, CTL from nine horses that had diverse MHC class I alleles and that had controlled clinical disease episodes for variable periods were used to identify optimal CTL epitopes in previously defined Gag EC. These optimal epitopes were then used to evaluate the functional avidity of the responding CTL. Twenty-four optimal epitopes were identified, but only six were recognized by high avidity CTL (≤11 nM); four were in conserved EC3 with three recognized by CTL from one horse (H596). Though there is support for either specificity or avidity being most important in antiviral CTL responses (Alexander-Miller et al., 1996; Derby et al., 2001; Yang et al., 2003), it may be that high avidity CTL responses to epitopes in relatively invariant sequences like EC3 may provide the most effective lentivirus control. CTL recognition of epitopes in a functionally important region would restrict the number of mutations resulting in viable virus and thereby the number of CTL escape variants. Therefore, immunization of horses with more conserved EC3 and other EC with similar properties may induce high avidity CTL to epitopes that vary less. Vaccine vectors expressing relatively small EC from minigenes could result in a marked increase in peptide–MHC class I formation and stimulate CTL more effectively (Anton et al., 1997). Using small EC might avoid problems caused by using full-length proteins which include stimulation of less effective CTL and of immune responses that cause adverse effects (Raabe et al., 1998; Wang et al., 1994; Woodberry et al., 1999).
EIAV-infected horses have recurrent episodes soon after infection that may be associated with CTL escape variants as variation in a SU epitope resulted in escape from CTL (Mealey et al., 2003). Variation has been detected in EIAV Gag CTL epitopes, but there is no direct evidence that this resulted in escape from CTL (Mealey et al., 2003; Zhang et al., 1998). In the current study, CTL escape variants were identified from plasma viral RNA obtained from three of the nine horses. The Gag protein epitopes that varied were recognized by high (7.9 nM), moderate (71.3 nM), or low (>1000 nM) functional avidity CTL. The high and moderate avidity CTL recognizing epitopes that varied (KV9 and RK11) were from horse H631. RK11 was the only epitope among six recognized by high functional avidity CTL that varied in this study. No viral sequences could be detected in plasma from the other four horses with high avidity CTL preventing further evaluation of possible escape from CTL. In SIV-infected macaques, CTL escape occurs in epitopes recognized by high avidity CTL during acute infection (O’Connor et al., 2002). The major difference in this horse study was that the high avidity CTL were identified in carrier horses with relatively long-term infection and no detectable viremia.
The horses in this study segregated into two groups depending on whether they had high functional avidity CTL to Gag protein epitopes or not. There were five horses in the group that had high avidity CTL (≤11 nM); furthermore, one of these horses (H631) also had the only moderate avidity CTL (71.3 nM) to another epitope. The other group of four horses all had low avidity CTL (>1000 nM). Thus, the hypothesis that all the carrier horses would have high avidity CTL to optimal epitopes in the EC was not supported by these data. Since the horse groups were clearly segregated, it was of interest to see if clinical disease patterns varied between the two groups. The number of acute episodes that happened early after infection of these horses was not considered because it happened at a time when virus was obviously not controlled. Thus, another parameter, the number of days since the last clinical disease episode, was used in the analysis. The group with high functional avidity CTL had significantly more days since the last clinical episode than the group with low functional avidity CTL. However, high avidity CTL were not significantly associated with time of EIAV infection, suggesting that their induction was not time-dependent in these long-term carrier horses. Furthermore, when all nine horses were considered, there was a significant (P < 0.05) inverse correlation between the days since the last clinical episode and CTL avidity to Gag peptides. The significant association between high avidity CTL and less recent clinical disease episodes was remarkable since the horses had been infected for different times and were infected with different virus strains. It was expected that such heterogeneity would obscure any significant differences; even so, it will be necessary to repeat this potentially important observation.
The effectiveness of high functional avidity CTL in viral immune control is documented in mice models using LCMV (Ehl et al., 1997; Gallimore et al., 1998; Speiser et al., 1992), HIV-1 (Derby et al., 2001), and paramyxovirus simian virus 5 (Gray et al., 2001). Adoptive transfer of high or low functional avidity CTL into SCID mice demonstrated that high avidity CTL were 100- to 1000-fold more efficacious at clearing of vaccinia virus expressing HIV-1 gp160 than the low functional avidity CTL (Alexander-Miller et al., 1996). The association of high functional avidity CTL with control of EIAV episodes is different from a recent study demonstrating that in vitro inhibition of viral replication was more dependent on the epitope specificity than functional avidity (Yang et al., 2003). It is likely that CTL responses to particular epitopes are critical for lentiviral control (Mealey et al., 2003; Yang et al., 2003), however, CTL with high functional avidity to these epitopes should be more effective than CTL with low functional avidity. Another parameter that may be important is the number of viral epitopes recognized by CTL. CD4+ cell counts from HIV-1-seropositive patients were positively correlated with the number of HIV-1 viral peptides recognized by CTL, but not with the intensity of anti-HIV CD8+ CTL responses (Dalod et al., 1999). However, there was no significant difference in the number of Gag epitopes recognized by CTL from EIAV-infected horses in the high and low functional avidity groups in the current study.
A higher importance of CTL quality over quantity was suggested by the presence of high avidity CTL responses to only a single Gag epitope or a few epitopes in EIAV carrier horses with long-time clinical quiescence. CTL with high functional avidity require extremely low epitope concentration for recognition and may cause early lysis of infected target cells. HIV-1 Gag epitopes have been described which appear to be conserved because of constraints on variation due to involvement of the amino acids comprising the epitope in viral function (Goulder and Watkins, 2004). Furthermore, epitope mutations in conserved regions with functional constraint will result in viruses with decreased replication capacity and consequently decreased clinical disease (Friedrich et al., 2004; Kelleher et al., 2001). Stimulating CTL to these relatively invariant epitopes could stabilize CTL responses controlling virus replication and slow down or even prevent disease progression (Nowak et al., 1995). EIAV EC3 may be an example of such a region because mutations in a similar area of HIV-1 CA protein result in nonviable virus (Dorfman et al., 1994; Gitti et al., 1996; Momany et al., 1996; Zhang et al., 1996).
Conclusively, the number of optimal epitopes in four Gag CTL EC was determined, and the functional avidity of CTL recognizing these epitopes was evaluated. Even though all the EIAV carrier horses did not have high functional avidity CTL to Gag EC, those that did had significantly more days since the last clinical episode. In addition, there was a significant inverse correlation between the functional avidity of CTL and the days since the last clinical episode. CTL escape was documented in Gag protein epitopes from three horses, but only one of these was recognized by CTL with high functional avidity. Conserved EC3 had the most epitopes recognized by high functional avidity CTL, and this and similar regions can be used to induce CTL in horses with diverse MHC class I alleles. If protective CTL can be induced, it will help define the requirements for CTL to be protective against lentiviruses.
Materials and methods
EIAV-infected horses
The nine infected horses used in this study (Table 1) were previously inoculated with either EIAVWSU5 or EIAVPV and were used to initially identify EC recognized by CTL (Chung et al., 2004). Therefore, their use to identify CTL epitopes in the EC was appropriate. The origin of EIAVPV has been described (Rwambo et al., 1990). EIAVWSU5 (GenBank accession no. NP_056901) and EIAVPV (Gen-Bank accession no. AAC03760) have identical Gag MA and CA protein sequences. Autologous EK cells for CTL targets were obtained from kidney biopsies prior to infection of each horse and maintained as frozen cell lines (McGuire et al., 1994). The MHC class I ELA-A haplotypes of the horses used in this study were evaluated by lymphocyte microcytotoxicity to determine the ELA-A types using antisera A1–A10 and W11 (Table 1) (Bailey et al., 2000; Bernoco et al., 1987; Lazary et al., 1988; Terasaki et al., 1978). Furthermore, the MHC class I alleles of six of the nine horses (H593, H596, H610, H614, H631, A2147) were previously determined by sequence-based typing (Table 1) (Chung et al., 2003).
Synthetic peptides of EIAV Gag MA and CA proteins
EC were identified using 52 previously described peptides of 15 amino acids overlapping by 11 amino acids and 21 peptides of 16 amino acids overlapping by 12 amino acids with C-terminal OH and N-terminal H groups covering the entire EIAV Gag MA and CA proteins (Chung et al., 2004). For fine mapping of epitopes within the 15–16 amino acid peptides significantly recognized by CTL, additional peptides of 8–15 amino acids were synthesized (Sigma-Genosys, The Woodlands, TX). The peptides were dissolved in 100% dimethyl sulfoxide and stored at −20 °C before dilution and use for CTL epitope mapping.
PBMC isolation and stimulation of memory CTL for CTL assay
PBMC were isolated from the EIAV-infected horses by centrifugation on Histopaque (specific gravity 1.077), washed four times, and viability determined (Wyatt et al., 1988). Monocytes in PBMC were infected with EIAVWSU5 at a multiplicity of infection of one or incubated with peptides (103 nM final concentration) in 5 ml of RPMI 1640 with 20% FCS at 37 °C for 2 h with gentle mixing every 15 min (McGuire et al., 2000; Zhang et al., 1998). Stimulations were done in a 175 cm2 tissue culture flask containing RPMI 1640 with 10% fetal calf serum, 5 × 10−5 M 2-mercaptoethanol, and 20 mM Hepes. After incubation in a humidified chamber with 5% CO2 at 37 °C for 7 days, viable lymphocytes were counted using exclusion of trypan blue dye in a hemocytometer for use in the 51Cr release assay (McGuire et al., 2000).
CTL assay
The 51Cr release assay was used to identify peptides recognized by CTL from EIAV-infected horses. EK target cells (3 × 104/well) were incubated in collagen-coated wells of 96-well plates at 37 °C with 5% CO2 for 24 h before labeling with 1.25 μCi of 51Cr and pulsing with 104 nM synthetic peptides in 50 μl per well of DMEM containing 5% fetal calf serum at 37 °C for 2 h. After washing three times with DMEM, effectors were incubated with target cells (effector-to-target cell ratio of 50:1) in 200 μl/well RPMI containing 10% fetal calf serum at 37 °C with 5% CO2 for 17 h (McGuire et al., 1994). Then, 100 μl of supernatant was removed from each well to determine 51Cr release. Percent specific lysis was calculated as [(E − S)/(M − S)] × 100, where E was the mean of three test wells, S was the mean spontaneous release from three wells without effector cells, and M was the mean maximal release from three wells with 2% Triton X-100. The standard error (SE) of percent specific lysis was calculated taking into consideration the variability of E, S, and M as described previously (Siliciano et al., 1985). Only assays with a spontaneous lysis of <30% were used. Specific lysis of peptide-pulsed target cells that was >10% and exceeded lysis of non-pulsed target cells by >2.5 SE was considered significant.
CTL functional avidity
The functional avidity of CTL was determined by stimulating PBMC with pools containing 103 nM of each optimal peptide epitope. CTL assays were then performed using target cells pulsed with 10-fold dilutions of peptides ranging from 100 to 104 nM, and functional avidity was calculated (Mealey et al., 2003). CTL functional avidity was defined as the peptide concentration (nM) that resulted in 50% maximal target cell specific lysis and was used to describe the effectiveness of CTL-mediated killing, encompassing TCR affinity and signaling, as well as the affinity of MHC–peptide binding (Alexander-Miller et al., 1996; Derby et al., 2001). Functional avidity was not determined if the maximal specific lysis of peptide-pulsed target cells was less than 10% above the specific lysis of non-pulsed target cells. CTL were considered to have high functional avidity if the peptide concentration resulting in 50% maximal lysis was ≤11 nM. This cut-off for high avidity was based on data being in three groups including 7 avidities ranging between 0.9 and 10.9 nM, one of 71.3 nM, and the remaining 17 were >1000 nM (Table 2). The high avidity cut-off of ≤11 nM is similar to the <5 nM cut-off used by others (Mealey et al., 2003; O’Connor et al., 2002). In the assays of CTL with high and moderate avidity, maximal lysis was reached expediting the calculations of avidity. In the cases of those CTL designated as having low avidity, it was not certain whether maximum lysis was obtained with 10,000 nM peptide or not. In those cases, the results at 10,000 nM were assumed to be maximal, and the EC50 was calculated. The EC50 was always >1000 nM, and this value was used as a minimum estimate of the EC50 for the low avidity CTL.
Cloning and sequencing of Gag MA and CA
Viral RNA amplification was attempted from all nine carrier horses but was successful in only three. Viral RNA was isolated using a QIAamp Viral RNA kit (Qiagen Inc., Chatsworth, CA) (Leutenegger et al., 2001) with 0.14 ml frozen EDTA plasma from horses H610 and H614 and 1.4 ml from H631. RNA was treated with DNase I on the spin column (DNase 1 set; Qiagen), eluted in 60 μl nuclease-free water, and frozen at −20 °C. First-strand cDNA was made using a primer (5′TGTCCTGGCTTCCCACAG) and a cDNA synthesis system (Invitrogen, Carlsbad, CA) with an annealing temperature of 42 °C. The first PCR was carried out in 25 μl reactions with 1.2 mM MgCl2, 0.5 mM of each dNTP, 10 μmol of each primer (forward: 5′ACAGAAGTCTTCTGGAGG3′, reverse: 5′CTTTAGTGGCCCTCCTTT3′), and Taq DNA polymerase (Invitrogen, Carlsbad, CA). Thirty-five cycles were done at 95 °C (30 s), 53 °C (30 s), and 72 °C (1 min), with a last extension reaction at 72 °C for 7 min. A second PCR was done with a nested primer set (forward: 5′TCTTCTGGAGGTGTTCCTGG3′, reverse: 5′CTTTAGTGGCCCTCCTTT3′) using the same conditions as in the first PCR.
Amplified DNA was separated on a 1% agarose gel (BioWhittaker Molecular Applications, Rockland, ME) for 2 h at 60 V in 0.04 M Tris–acetate buffer with 0.001 M EDTA. Bands with the appropriate number of base pairs were excised from the gel, and the DNA isolated using a gel extraction kit (Qiagen). Purified DNA was ligated into the Zero Blunt TOPO cloning vector (Invitrogen, Carlsbad, CA) for 5 min at room temperature, transformed into competent cells (Invitrogen), and plated on LB agar with 50 μg/ml kanamycin. Clones containing inserts of the predicted size were identified first by PCR, and then 10 clones from each amplification were selected for DNA sequencing using automated sequencer (ABI 377, Amersham Biosciences, Discataway, NJ). Thirteen variant EIAV Gag MA and CA sequences (GenBank accession no. AY742243–AY742255) obtained from the 3 horses, together with a published EIAVWSU5 Gag sequence (GenBank accession no. AF247394), were used for multiple alignments with CLUSTAL W (Chenna et al., 2003).
Determination of plasma viral load by real-time RT-PCR
A previously described quantitative real-time RT-PCR (Mealey et al., 2003) was used with modifications to determine plasma viral load by amplification of a 167 base pair segment (nucleotides 1637–1803) of the EIAVWSU5 (GenBank accession no. AF247394) gag gene. A standard RNA template was made and the copy numbers determined as described (Mealey et al., 2003). Primers 1637F (5′AGCCAGGACATTTATCTAGTCAATGTAGAGCACC3′) and 1803R (5′GTGCTGACTCTTCTGTTGTATCGGGAAAGTTTG3′), along with a previously reported gag-specific probe (Cook et al., 2002), were used in duplicate 25 μl reactions utilizing an iScript One-Step RT-PCR kit for probes (Bio-Rad Laboratories, Hercules, CA), 200 nM of each primer and probe, 40 U Rnasin, and 10 μl plasma RNA template. Reactions were done in an iCyler with the iQ Real-Time PCR Detection System and software (Bio-Rad) under the following conditions: 30 min at 50 °C, 3.5 min at 95 °C, 50 cycles at 95 °C for 15 s, and 60 °C for 1 min. Sensitivity was determined using a dilution series of the standard RNA transcripts. The dilutions containing 1 RNA copy was positive in 40% (4/10) of the replicates, while the dilutions containing 10 RNA copies or more were positive in 100% (10/10) of the replicates. The reliable detection limit, therefore, was somewhere between 43 and 428 RNA copies/ml when 140 μl plasma for the RNA extraction and 10 μl of the 60 μl eluate were used (Mealey et al., 2003).
Statistical analysis
The total number epitopes recognized by CTL, the number of days post-infection, and the number of days since the last clinical episode (non-progression period) were compared for horses with high functional avidity CTL and those with low functional avidity CTL using the Mann–Whitney U test. Significance was determined by P < 0.05. In addition, a Spearman’s rank correlation coefficient was determined for days since the last clinical episode and avidity for all horses as well as the days post-infection and avidity with significance determined by P < 0.05.
Acknowledgments
This research was supported in part by U.S. Public Health Service, National Institutes of Health grants AI24291, AI47660, and AI01575, and Morris Animal Foundation grant D01EQ-09. The authors acknowledge the assistance of Emma Karel.
References
- Alexander-Miller MA, Leggatt GR, Berzofsky JA. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc Natl Acad Sci USA. 1996;93:4102–4107. doi: 10.1073/pnas.93.9.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton LC, Yewdell JW, Bennink JR. MHC class I-associated peptides produced from endogenous gene products with vastly different efficiencies. J Immunol. 1997;158:2535–2542. [PubMed] [Google Scholar]
- Bailey E, Marti E, Fraser DG, Antczak DF, Lazary S. Immunogenetics of the Horse. In: Bowling AT, Ruvinsky A, editors. The Genetics of the Horse. CABI Publishing Inc; UK: 2000. pp. 123–155. [Google Scholar]
- Bernoco D, Byrns G, Bailey E, Lew AM. Evidence of a second polymorphic ELA class I (ELA-B) locus and gene order for three loci of the equine major histocompatibility complex. Anim Genet. 1987;18:103–118. doi: 10.1111/j.1365-2052.1987.tb00749.x. [DOI] [PubMed] [Google Scholar]
- Carpenter S, Evans LH, Sevoian M, Chesebro B. Role of the host immune response in selection of equine infectious anemia virus variants. J Virol. 1987;61:3783–3789. doi: 10.1128/jvi.61.12.3783-3789.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thomson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung C, Leib SR, Fraser DG, Ellis SA, McGuire TC. Novel classical MHC class I alleles identified in horses by sequencing clones of reverse transcription-PCR products. Eur J Immunogenet. 2003;30:387–396. doi: 10.1111/j.1365-2370.2003.00420.x. [DOI] [PubMed] [Google Scholar]
- Chung C, Mealey RH, McGuire TC. CTL from EIAV carrier horses with diverse MHC class I alleles recognize epitope clusters in Gag matrix and capsid proteins. Virology. 2004;327:144–154. doi: 10.1016/j.virol.2004.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggins L. Carriers of equine infectious anemia virus. J Am Vet Med Assoc. 1984;184:279–281. [PubMed] [Google Scholar]
- Cook RF, Cook SJ, Li FL, Montelaro RC, Issel CJ. Development of a multiplex real-time reverse transcriptase-polymerase chain reaction for equine infectious anemia virus (EIAV) J Virol Methods. 2002;105:171–179. doi: 10.1016/s0166-0934(02)00101-5. [DOI] [PubMed] [Google Scholar]
- Dalod M, Dupuis M, Deschemin JC, Sicard D, Salmon D, Delfraissy JF, Venet A, Sinet M, Guillet JG. Broad, intense anti-human immunodeficiency virus (HIV) ex vivo CD8(+) responses in HIV type 1-infected patients: comparison with anti-Epstein–Barr virus responses and changes during antiretroviral therapy. J Virol. 1999;73:7108–7116. doi: 10.1128/jvi.73.9.7108-7116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derby M, Alexander-Miller M, Tse R, Berzofsky J. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J Immunol. 2001;166:1690–1697. doi: 10.4049/jimmunol.166.3.1690. [DOI] [PubMed] [Google Scholar]
- Dorfman T, Burkovsky A, Ohagen A, Hoglund S. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehl S, Klenerman P, Aichele P, Hengartner H, Zinkernagel RM. A functional and kinetic comparison of antiviral effector and memory cytotoxic T lymphocyte populations in vivo and in vitro. Eur J Immunol. 1997;27:3404–3413. doi: 10.1002/eji.1830271240. [DOI] [PubMed] [Google Scholar]
- Friedrich TC, Frye CA, Yant LJ, O’Connor DH, Kriewaldt NA, Benson M, Vojnov L, Dodds EJ, Cullen C, Rudersdorf R, Hughes AL, Wilson N, Watkins DI. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J Virol. 2004;78:2581–2585. doi: 10.1128/JVI.78.5.2581-2585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimore A, Dumrese T, Hengartner H, Zinkernagel RM, Rammensee HG. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J Exp Med. 1998;187:1647–1657. doi: 10.1084/jem.187.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Structure of the amino acid terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- Goulder PJ, Watkins DI. HIV and SIV CTL escape: implications for vaccine design. Nat Rev, Immunol. 2004;4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- Gray PM, Parks GD, Alexander-Miller MA. A novel CD8-independent high-avidity cytotoxic T-lymphocyte response directed against an epitope in the phosphoprotein of the paramyxovirus simian virus 5. J Virol. 2001;75:10065–10072. doi: 10.1128/JVI.75.21.10065-10072.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SA, Cook SJ, Lichtenstein DL, Issel CJ, Montelaro RC. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SA, Li F, McKeon BM, Sr, Cook SJ, Issel CJ, Montelaro RC. Immune responses and viral replication in long-term inapparent carrier ponies inoculated with equine infectious anemia virus. J Virol. 2000;74:5968–5981. doi: 10.1128/jvi.74.13.5968-5981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain KA, Issel CJ, Schnorr KL, Rwambo PM, Montelaro RC. Antigenic analysis of equine infectious anemia virus (EIAV) variants by using monoclonal antibodies: epitopes of glycoprotein gp90 of EIAV stimulate neutralizing antibodies. J Virol. 1987;61:2956–2961. doi: 10.1128/jvi.61.10.2956-2961.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher AD, Long C, Holmes EC, Allen RL, Wilson J, Conlon C, Workman C, Shaunak S, Olson K, Goulder P, Brander C, Ogg G, Sullivan JS, Dyer W, Jones I, McMichael AJ, Rowland-Jones S, Phillips RE. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med. 2001;193:375–386. doi: 10.1084/jem.193.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono Y, Kobayashi K, Fukunaga Y. Antigenic drift of equine infectious anemia virus in chronically infected horses. Arch Gesamte Virusforsch. 1973;41:1–10. doi: 10.1007/BF01249923. [DOI] [PubMed] [Google Scholar]
- Kono Y, Hirasawa K, Fukunaga Y, Taniguchi T. Recrudescence of equine infectious anemia by treatment with immunosuppressive drugs. Natl Inst Anim Health Q (Tokyo) 1976;16:8–15. [PubMed] [Google Scholar]
- Kuroda MJ, Schmitz JE, Charini WA, Nickerson CE, Lifton MA, Lord CI, Forman MA, Letvin NL. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J Immunol. 1999;162:5127–5133. [PubMed] [Google Scholar]
- Lazary S, Antczak DF, Bailey E, Bell TK, Bernoco D, Byrns G, McClure JJ. Joint report of the Fifth International Workshop on Lymphocyte Alloantigens of the Horse, Baton Rouge, Louisiana, 31 October–1 November 1987. Anim Genet. 1988;19:447–456. doi: 10.1111/j.1365-2052.1988.tb00836.x. [DOI] [PubMed] [Google Scholar]
- Leutenegger CM, Higgins J, Matthews TB, Tarantal AF, Luciw PA, Pedersen NC, North TW. Real-time TaqMan PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res Hum Retroviruses. 2001;17:243–251. doi: 10.1089/088922201750063160. [DOI] [PubMed] [Google Scholar]
- Matano T, Kobayashi M, Igarashi H, Takeda A, Nakamura H, Kano M, Sugimoto C, Mori K, Iida A, Hirata T, Hasegawa M, Yuasa T, Miyazawa M, Takahashi Y, Yasunami M, Kimura A, O’Connor DH, Watkins DI, Nagai Y. Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J Exp Med. 2004;199:1709–1718. doi: 10.1084/jem.20040432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire TC, Tumas DB, Byrne KM, Hines MT, Leib SR, Brassfield AL, O’Rourke KI, Perryman LE. Major histocompatibility complex-restricted CD8+ cytotoxic T lymphocytes from horses with equine infectious anemia virus recognize Env and Gag/PR proteins. J Virol. 1994;68:1459–1467. doi: 10.1128/jvi.68.3.1459-1467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire TC, Leib SR, Lonning SM, Zhang W, Byrne KM, Mealey RH. Equine infectious anaemia virus proteins with epitopes most frequently recognized by cytotoxic T lymphocytes from infected horses. J Gen Virol. 2000;81:2735–2739. doi: 10.1099/0022-1317-81-11-2735. [DOI] [PubMed] [Google Scholar]
- Mealey RH, Fraser DG, Oaks JL, Cantor GH, McGuire TC. Immune reconstitution prevents continuous equine infectious anemia virus replication in an Arabian foal with severe combined immunodeficiency: lessons for control of lentiviruses. Clin Immunol. 2001;101:237–247. doi: 10.1006/clim.2001.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mealey RH, Zhang B, Leib SR, Littke MH, McGuire TC. Epitope specificity is critical for high and moderate avidity cytotoxic T lymphocytes associated with control of viral load and clinical disease in horses with equine infectious anemia virus. Virology. 2003;313:537–552. doi: 10.1016/s0042-6822(03)00344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, Lee BM, Gorbalenya AE, Tong L, McClure J, Ehrlich LS, Summers MF, Carter C, Rossmann MG. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- Montelaro RC, Ball JM, Rushlow KE. Equine retroviruses. In: Levy JA, editor. The Retroviridae. Plenum Press; New York, NY: 1993. pp. 257–360. [Google Scholar]
- Montelaro RC, Parekh B, Orrego A, Issel CJ. Antigenic variation during persistent infection by equine infectious anemia virus, a retrovirus. J Biol Chem. 1984;259:10539–10544. [PubMed] [Google Scholar]
- Novitsky V, Cao H, Rybak N, Gilbert P, McLane MF, Gaolekwe S, Peter T, Thior I, Ndung’u T, Marlink R, Lee TH, Essex M. Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J Virol. 2002;76:10155–10168. doi: 10.1128/JVI.76.20.10155-10168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak MA, May RM, Phillips RE, Rowland-Jones S, Lalloo DG, McAdam S, Klenerman P, Koppe B, Sigmund K, Bangham CRM, McMichael AJ. Antigenic oscillations and shifting immunodominance in HIV-1 infections. Nature. 1995;375:606–611. doi: 10.1038/375606a0. [DOI] [PubMed] [Google Scholar]
- O’Connor DH, Allen TM, Vogel TU, Jing P, DeSouza IP, Dodds E, Dunphy EJ, Melsaether C, Mothe B, Yamamoto H, Horton H, Wilson N, Hughes AL, Watkins DI. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat Med. 2002;8:493–499. doi: 10.1038/nm0502-493. [DOI] [PubMed] [Google Scholar]
- Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Cerundolo V, Hurley A, Markowitz M, Ho DD, Nixon DF, McMichael AJ. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- O’Rourke K, Perryman LE, McGuire TC. Antiviral, anti-glycoprotein and neutralizing antibodies in foals with equine infectious anaemia virus. J Gen Virol. 1988;69:667–674. doi: 10.1099/0022-1317-69-3-667. [DOI] [PubMed] [Google Scholar]
- Perryman LE, O’Rourke KI, McGuire TC. Immune responses are required to terminate viremia in equine infectious anemia lentivirus infection. J Virol. 1988;62:3073–3076. doi: 10.1128/jvi.62.8.3073-3076.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raabe ML, Issel CJ, Cook SJ, Cook RF, Woodson B, Montelaro RC. Immunization with a recombinant envelope protein (rgp90) of EIAV produces a spectrum of vaccine efficacy ranging from lack of clinical disease to severe enhancement. Virology. 1998;245:151–162. doi: 10.1006/viro.1998.9142. [DOI] [PubMed] [Google Scholar]
- Rwambo PM, Issel CJ, Hussain KA, Montelaro RC. In vitro isolation of a neutralization escape mutant of equine infectious anemia virus (EIAV) Arch Virol. 1990;111:275–280. doi: 10.1007/BF01311062. [DOI] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Sellon DC, Fuller FJ, McGuire TC. The immunopathogenesis of equine infectious anemia virus. Virus Res. 1994;32:111–138. doi: 10.1016/0168-1702(94)90038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano RF, Keegan AD, Dintzis RZ, Dintzis HM, Shin HS. The interaction of nominal antigen with T cell antigen receptors: I. Specific binding of multivalent nominal antigen to cytolytic T cell clones. J Immunol. 1985;135:906–914. [PubMed] [Google Scholar]
- Sleasman JW, Goodenow MM. 13. HIV-1 infection. J Allergy Clin Immunol. 2003;111:S582–S592. doi: 10.1067/mai.2003.91. [DOI] [PubMed] [Google Scholar]
- Speiser DE, Kyburz D, Stubi U, Hengartner H, Zinkernagel RM. Discrepancy between in vitro measurable and in vivo virus neutralizing cytotoxic T cell reactivities. Low T cell receptor specificity and avidity sufficient for in vitro proliferation or cytotoxicity to peptide-coated target cells but not for in vivo protection. J Immunol. 1992;149:972–980. [PubMed] [Google Scholar]
- Terasaki PI, Bernoco D, Park MS, Ozturk G, Iwaki Y. Microdroplet testing for HLA-A, -B, -C, and -D antigens. The Phillip Levine Award Lecture. Am J Clin Pathol. 1978;69:103–120. doi: 10.1093/ajcp/69.2.103. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Rushlow KE, Issel CJ, Cook RF, Cook SJ, Raabe ML, Chong YH, Costa L, Montelaro RC. Enhancement of EIAV replication and disease by immunization with a baculovirus-expressed recombinant envelope surface glycoprotein. Virology. 1994;199:247–251. doi: 10.1006/viro.1994.1120. [DOI] [PubMed] [Google Scholar]
- Woodberry T, Gardner J, Mateo L, Eisen D, Medveczky J, Ramshaw IA, Thomson SA, Ffrench RA, Elliott SL, Firat H, Lemonnier FA, Suhrbier A. Immunogenicity of a human immunodeficiency virus (HIV) polytope vaccine containing multiple HLA A2 HIV CD8(+) cytotoxic T-cell epitopes. J Virol. 1999;73:5320–5325. doi: 10.1128/jvi.73.7.5320-5325.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt CR, Davis WC, McGuire TC, Perryman LE. T lymphocyte development in horses: I. Characterization of monoclonal antibodies identifying three stages of T lymphocyte differentiation. Vet Immunol Immunopathol. 1988;18:3–18. doi: 10.1016/0165-2427(88)90032-3. [DOI] [PubMed] [Google Scholar]
- Yang OO, Sarkis PT, Trocha A, Kalams SA, Johnson RP, Walker BD. Impacts of avidity and specificity on the antiviral efficiency of HIV-1-specific CTL. J Immunol. 2003;171:3718–3724. doi: 10.4049/jimmunol.171.7.3718. [DOI] [PubMed] [Google Scholar]
- Zhang WH, Hockley D, Nermut MV, Morikawa Y, Jones IM. Gag–Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J Gen Virol. 1996;77:743–751. doi: 10.1099/0022-1317-77-4-743. [DOI] [PubMed] [Google Scholar]
- Zhang W, Lonning SM, McGuire TC. Gag protein epitopes recognized by ELA-A-restricted cytotoxic T lymphocytes from horses with long-term equine infectious anemia virus infection. J Virol. 1998;72:9612–9620. doi: 10.1128/jvi.72.12.9612-9620.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]