Abstract
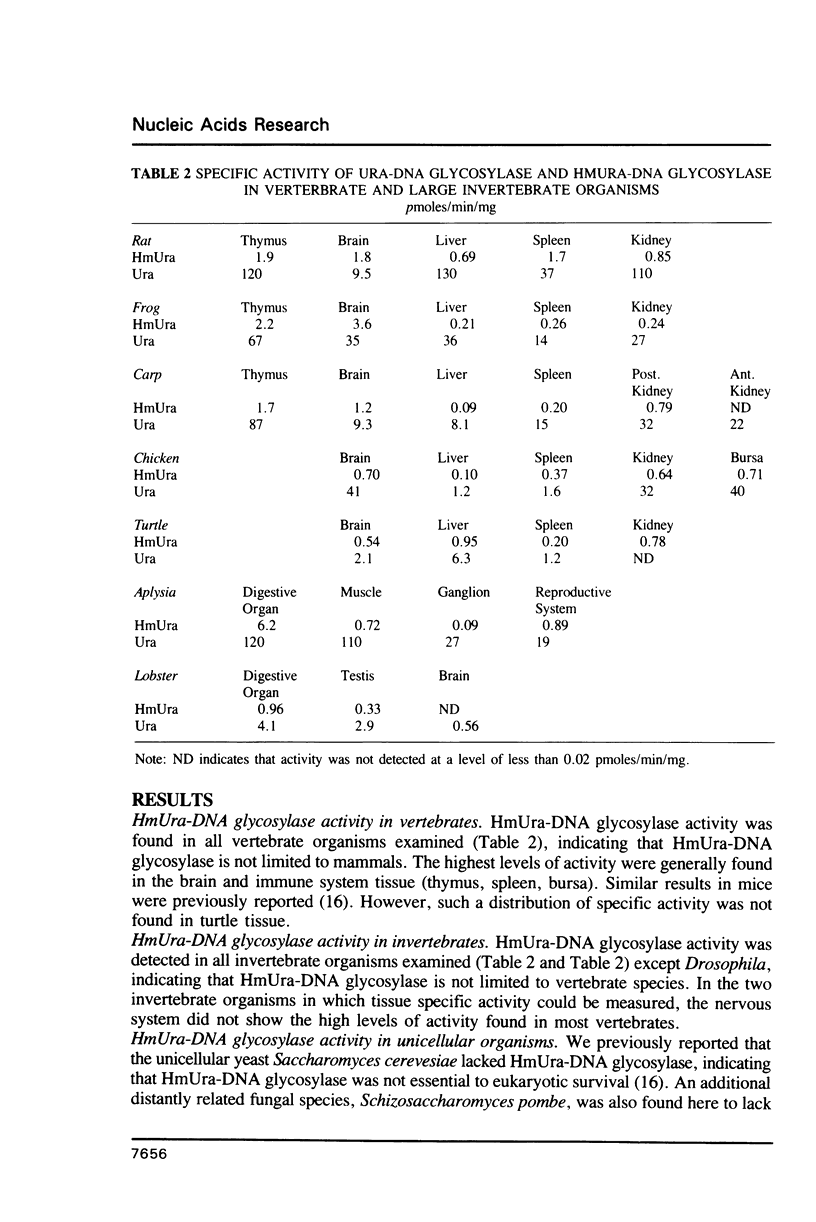
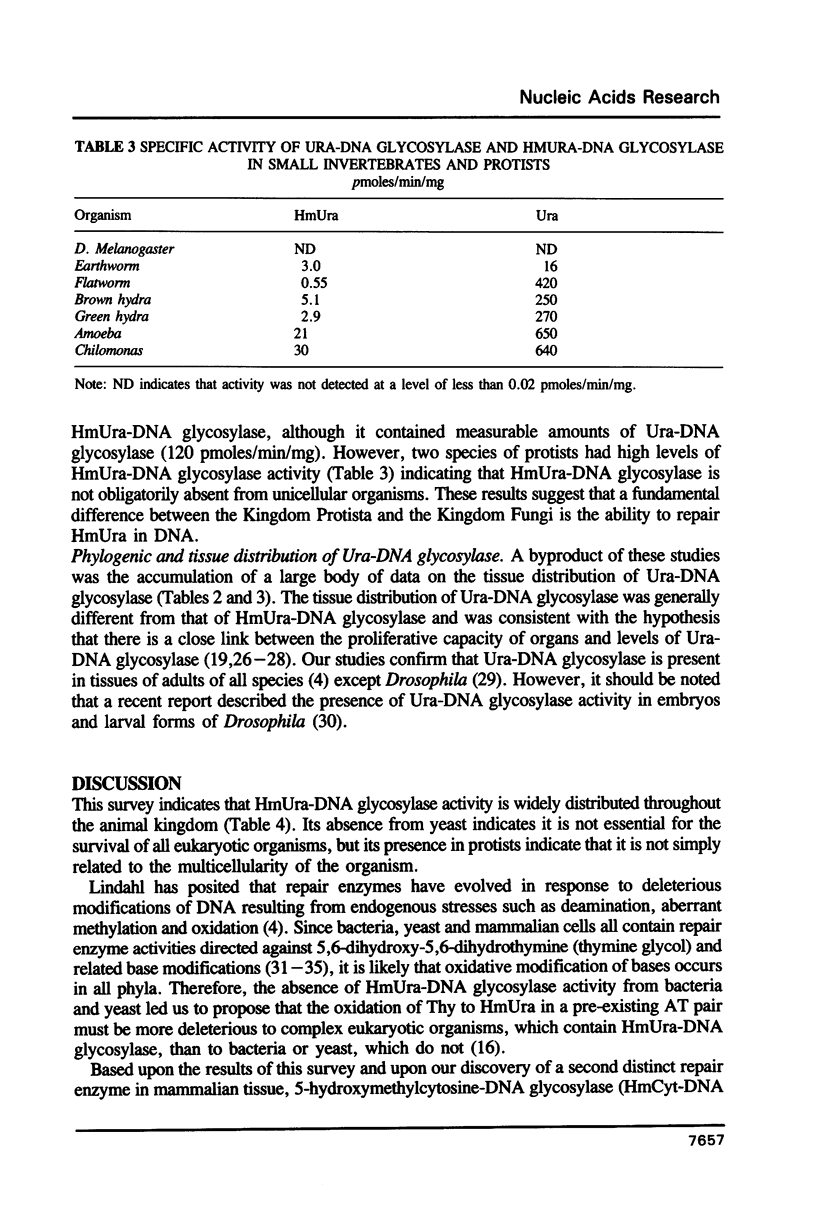
5-Hydroxymethyluracil (HmUra) is formed in DNA as a product of oxidative attack on the methyl group of thymine. It is also the product of the deamination of 5-hydroxymethylcytosine (HmCyt) which may be formed via oxidation of 5-methylcytosine (MeCyt). HmUra is removed from DNA by a DNA glycosylase which, together with HmCyt-DNA glycosylase, is unique among DNA repair enzymes in being present in mammalian cells but absent from bacteria and yeast. We found HmUra-DNA glycosylase activity in a wide variety of vertebrate and invertebrate animals (except Drosophila) and in protozoans. In most vertebrate organisms the highest specific activity was in nervous and immune system tissue. The phylogenetic distribution of HmUra-DNA glycosylase correlates with the presence of 5-methylcytosine (MeCyt) as a regulator of gene expression. This distribution of activity supports the contention that HmUra-DNA glycosylase aids in the maintenance of methylated sites in DNA.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammermann D., Steinbrück G., Baur R., Wohlert H. Methylated bases in the DNA of the ciliate Stylonychia mytilus. Eur J Cell Biol. 1981 Apr;24(1):154–156. [PubMed] [Google Scholar]
- Antequera F., Tamame M., Villanueva J. R., Santos T. DNA methylation in the fungi. J Biol Chem. 1984 Jul 10;259(13):8033–8036. [PubMed] [Google Scholar]
- Aprelikova O. N., Tomilin N. V. Activity of uracil-DNA glycosylase in different rat tissues and in regenerating rat liver. FEBS Lett. 1982 Jan 25;137(2):193–195. doi: 10.1016/0014-5793(82)80347-5. [DOI] [PubMed] [Google Scholar]
- Baltz R. H., Bingham P. M., Drake J. W. Heat mutagenesis in bacteriophage T4: the transition pathway. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1269–1273. doi: 10.1073/pnas.73.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein R. J., Levy D. D., Teebor G. W. 5-Hydroxymethyluracil-DNA glycosylase activity may be a differentiated mammalian function. Mutat Res. 1987 May;183(3):257–263. doi: 10.1016/0167-8817(87)90008-3. [DOI] [PubMed] [Google Scholar]
- Boorstein R. J., Teebor G. W. Mutagenicity of 5-hydroxymethyl-2'-deoxyuridine to Chinese hamster cells. Cancer Res. 1988 Oct 1;48(19):5466–5470. [PubMed] [Google Scholar]
- Breimer L. H., Lindahl T. DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J Biol Chem. 1984 May 10;259(9):5543–5548. [PubMed] [Google Scholar]
- Bron S., Luxen E., Venema G. Resistance of bacteriophage H1 to restriction and modification by Bacillus subtilis R. J Virol. 1983 Jun;46(3):703–708. doi: 10.1128/jvi.46.3.703-708.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T. C., Jiricny J. A specific mismatch repair event protects mammalian cells from loss of 5-methylcytosine. Cell. 1987 Sep 11;50(6):945–950. doi: 10.1016/0092-8674(87)90521-6. [DOI] [PubMed] [Google Scholar]
- Cannon-Carlson S. V., Gokhale H., Teebor G. W. Purification and characterization of 5-hydroxymethyluracil-DNA glycosylase from calf thymus. Its possible role in the maintenance of methylated cytosine residues. J Biol Chem. 1989 Aug 5;264(22):13306–13312. [PubMed] [Google Scholar]
- Cannon S. V., Cummings A., Teebor G. W. 5-Hydroxymethylcytosine DNA glycosylase activity in mammalian tissue. Biochem Biophys Res Commun. 1988 Mar 30;151(3):1173–1179. doi: 10.1016/s0006-291x(88)80489-3. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Tait A., Goddard J. M. Methylated bases in DNA from Paramecium aurelia. Biochim Biophys Acta. 1974 Nov 20;374(1):1–11. doi: 10.1016/0005-2787(74)90194-4. [DOI] [PubMed] [Google Scholar]
- DUNN D. B., SMITH J. D. The occurrence of 6-methylaminopurine in deoxyribonucleic acids. Biochem J. 1958 Apr;68(4):627–636. doi: 10.1042/bj0680627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Linn S. 5,6-Saturated thymine lesions in DNA: production by ultraviolet light or hydrogen peroxide. Nucleic Acids Res. 1982 Jun 25;10(12):3781–3789. doi: 10.1093/nar/10.12.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Doetsch P. W., Helland D. E., Haseltine W. A. Mechanism of action of a mammalian DNA repair endonuclease. Biochemistry. 1986 Apr 22;25(8):2212–2220. doi: 10.1021/bi00356a054. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Wang R. Y. 5-Methylcytosine in eukaryotic DNA. Science. 1981 Jun 19;212(4501):1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- Evans H. H., Evans T. E., Littman S. Methylation of parental and progeny DNA strands in Physarum polycephalum. J Mol Biol. 1973 Mar 15;74(4):563–572. doi: 10.1016/0022-2836(73)90047-8. [DOI] [PubMed] [Google Scholar]
- Frenkel K., Cummings A., Solomon J., Cadet J., Steinberg J. J., Teebor G. W. Quantitative determination of the 5-(hydroxymethyl)uracil moiety in the DNA of gamma-irradiated cells. Biochemistry. 1985 Aug 13;24(17):4527–4533. doi: 10.1021/bi00338a007. [DOI] [PubMed] [Google Scholar]
- Friedberg E. C., Ganesan A. K., Minton K. N-Glycosidase activity in extracts of Bacillus subtilis and its inhibition after infection with bacteriophage PBS2. J Virol. 1975 Aug;16(2):315–321. doi: 10.1128/jvi.16.2.315-321.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroir L. E., Deutsch W. A. Drosophila deoxyuridine triphosphatase. Purification and characterization. J Biol Chem. 1987 Jan 5;262(1):130–134. [PubMed] [Google Scholar]
- Gupta P. K., Sirover M. A. Cell cycle regulation of DNA repair in normal and repair deficient human cells. Chem Biol Interact. 1981 Jul;36(1):19–31. doi: 10.1016/0009-2797(81)90026-0. [DOI] [PubMed] [Google Scholar]
- Gupta P. K., Sirover M. A. Sequential stimulation of DNA repair and DNA replication in normal human cells. Mutat Res. 1980 Sep;72(2):273–284. doi: 10.1016/0027-5107(80)90042-1. [DOI] [PubMed] [Google Scholar]
- Gupta P. K., Sirover M. A. Stimulation of the nuclear uracil DNA glycosylase in proliferating human fibroblasts. Cancer Res. 1981 Aug;41(8):3133–3136. [PubMed] [Google Scholar]
- Hare J. T., Taylor J. H. One role for DNA methylation in vertebrate cells is strand discrimination in mismatch repair. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7350–7354. doi: 10.1073/pnas.82.21.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Kenny C., Berger L., Pratt K. Comparative study of DNA methylation in three unicellular eucaryotes. J Bacteriol. 1978 Sep;135(3):1156–1157. doi: 10.1128/jb.135.3.1156-1157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins S. A., Frenkel K., Cummings A., Teebor G. W. Definitive characterization of human thymine glycol N-glycosylase activity. Biochemistry. 1987 Mar 24;26(6):1683–1688. doi: 10.1021/bi00380a029. [DOI] [PubMed] [Google Scholar]
- Hollstein M. C., Brooks P., Linn S., Ames B. N. Hydroxymethyluracil DNA glycosylase in mammalian cells. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4003–4007. doi: 10.1073/pnas.81.13.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller T., Hood L. The growing immunoglobulin gene superfamily. Nature. 1986 Sep 4;323(6083):15–16. doi: 10.1038/323015a0. [DOI] [PubMed] [Google Scholar]
- Jiricny J., Hughes M., Corman N., Rudkin B. B. A human 200-kDa protein binds selectively to DNA fragments containing G.T mismatches. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8860–8864. doi: 10.1073/pnas.85.23.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KALLEN R. G., SIMON M., MARMUR J. The new occurrence of a new pyrimidine base replacing thymine in a bacteriophage DNA:5-hydroxymethyl uracil. J Mol Biol. 1962 Aug;5:248–250. doi: 10.1016/s0022-2836(62)80087-4. [DOI] [PubMed] [Google Scholar]
- Katcher H. L., Wallace S. S. Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry. 1983 Aug 16;22(17):4071–4081. doi: 10.1021/bi00286a013. [DOI] [PubMed] [Google Scholar]
- Lewis H. L., Muhleman D. R., Ward J. F. Serologic assay of DNA base damage. I. 5-Hydroxymethyldeoxyuridine, a radiation product of thymidine. Radiat Res. 1978 Aug;75(2):305–316. [PubMed] [Google Scholar]
- Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–192. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- Mamelak L., Boyer H. W. Genetic control of the secondary modification of deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1970 Oct;104(1):57–62. doi: 10.1128/jb.104.1.57-62.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Spear B. B. Macronuclear DNA of the hypotrichous ciliate Oxytricha fallax. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4992–4996. doi: 10.1073/pnas.75.10.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Steele R. E. Modified bases in the DNAs of unicellular eukaryotes: an examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems. 1978 Apr;10(1-2):37–53. doi: 10.1016/0303-2647(78)90027-8. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Salvini M., Durante M., Citti L., Nobili R. 5'-Methyl-cytosine in the macronuclear DNA of Blepharisma japonicum. Experientia. 1984 Dec 15;40(12):1401–1403. doi: 10.1007/BF01951912. [DOI] [PubMed] [Google Scholar]
- Sancar A., Sancar G. B. DNA repair enzymes. Annu Rev Biochem. 1988;57:29–67. doi: 10.1146/annurev.bi.57.070188.000333. [DOI] [PubMed] [Google Scholar]
- Teebor G. W., Boorstein R. J., Cadet J. The repairability of oxidative free radical mediated damage to DNA: a review. Int J Radiat Biol. 1988 Aug;54(2):131–150. doi: 10.1080/09553008814551591. [DOI] [PubMed] [Google Scholar]
- Teebor G. W., Frenkel K., Goldstein M. S. Ionizing radiation and tritium transmutation both cause formation of 5-hydroxymethyl-2'-deoxyuridine in cellular DNA. Proc Natl Acad Sci U S A. 1984 Jan;81(2):318–321. doi: 10.1073/pnas.81.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teebor G. W., Frenkel K. The initiation of DNA excision-repair. Adv Cancer Res. 1983;38:23–59. doi: 10.1016/s0065-230x(08)60186-4. [DOI] [PubMed] [Google Scholar]
- Teoule R., Cadet J. Radiation-induced degradation of the base component in DNA and related substances--final products. Mol Biol Biochem Biophys. 1978;27:171–203. doi: 10.1007/978-3-642-81196-8_9. [DOI] [PubMed] [Google Scholar]
- Urieli-Shoval S., Gruenbaum Y., Sedat J., Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982 Sep 6;146(1):148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Kirnos M. D. Structure of animal mitochondrial DNA (base composition, pyrimidine clusters, character of methylation). Biochim Biophys Acta. 1977 Mar 18;475(2):323–336. doi: 10.1016/0005-2787(77)90023-5. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Wallace S. S. AP endonucleases and DNA glycosylases that recognize oxidative DNA damage. Environ Mol Mutagen. 1988;12(4):431–477. doi: 10.1002/em.2860120411. [DOI] [PubMed] [Google Scholar]
- Wiebauer K., Jiricny J. In vitro correction of G.T mispairs to G.C pairs in nuclear extracts from human cells. Nature. 1989 May 18;339(6221):234–236. doi: 10.1038/339234a0. [DOI] [PubMed] [Google Scholar]



