Abstract
To determine how the lipid environment affects membrane protein structure and function, strains of Escherichia coli were developed in which normal phospholipid composition can be altered or foreign lipids can be introduced. The properties of LacY (lactose permease) were investigated as a function of lipid environment. Assembly of LacY in membranes lacking PE (phosphatidylethanolamine) results in misorientation of the N-terminal six-TM (transmembrane domain) helical bundle with loss of energy-dependent uphill transport and retention of energy-independent downhill transport. Post-assembly introduction of PE results in nearly native orientation of TMs and restoration of uphill transport. Foreign lipids with no net charge can substitute for PE in supporting native LacY topology, but restoration of uphill transport is dependent on native topology and the proper folding of a solvent-exposed domain. Increasing the positive charge density of the cytoplasmically exposed surface of LacY counters TM misorientation in the absence of neutral lipids, demonstrating that charge interactions between these domains and the surface of the membrane bilayer are determinants of TM orientation. Therefore membrane protein organization or reorganization is determined either during initial assembly or post-insertionally through direct interactions between the protein and the lipid environment, which affects the topogenic potency of opposing charged residues as topological signals independent of the translocon.
Keywords: lipid–protein interaction, lipid-sensitive domain, membrane organization, membrane protein, phospholipid, positive-inside rule
Introduction
The process by which polytopic membrane proteins are initially inserted into the membrane bilayer has been reviewed extensively [1,2]. Final orientation of TMs (transmembrane domains) and folding of membrane proteins is determined by the membrane-protein-insertion machinery (translocon) [3,4], topogenic sequences within the protein [5–7] and the properties of the lipid bilayer [5,8–10]. The primary driving force for membrane insertion of TMs is their hydrophobicity [11–13]. The orientation of TMs with respect to the plane of the lipid bilayer is largely determined by the positive-inside rule [14], which is based on the observed bias for exposure of positively charged extramembrane domains to the cytoplasm with neutral or negatively charged extramembrane domains oriented to the opposite side of the membrane. However, molecular genetic manipulation of membrane lipid composition has revealed a role for the properties of the lipid bilayer in determining TM orientation of polytopic membrane proteins [15]. Although gross lipid composition of individual membranes may not change dramatically, local changes of lipid composition within a given membrane do occur. In eukaryotic cells, lipid composition varies greatly between different membranes within the same cell [16], which exposes proteins to a varying lipid environment during intracellular trafficking. Understanding the effects of membrane lipid composition even in membranes where composition may not vary is of importance, since topogenic signals have evolved in the context of the host lipid composition [15], which varies between different organisms.
In the present paper, the molecular genetic manipulation of Escherichia coli membrane lipid composition is reviewed. The effects of changes in protein sequence and changes in membrane lipid composition on polytopic membrane protein TM orientation are integrated. The results strongly indicate that protein sequence and membrane lipid composition have co-evolved as synergistic determinants of membrane protein organization.
Genetic manipulation of membrane lipid composition
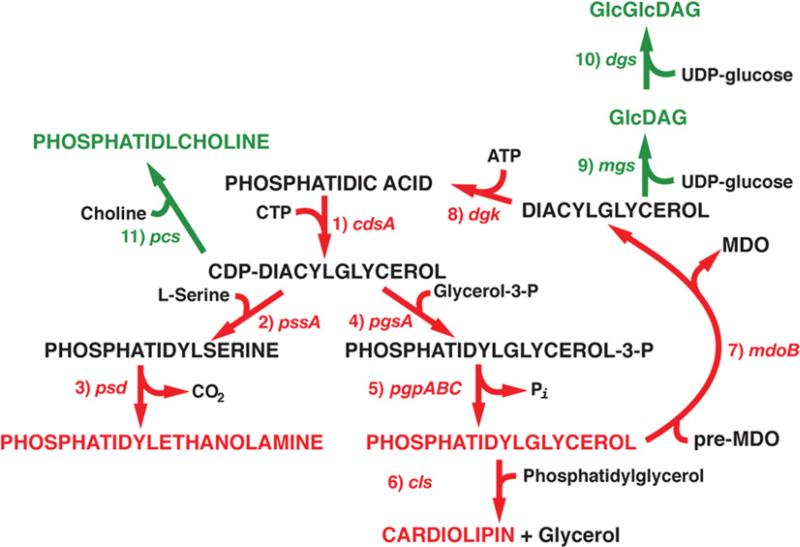
Surprisingly, large changes in E. coli membrane lipid composition are not lethal, even though there are significant changes in cell phenotype (see [17] for a detailed review and references therein). The major glycerophosphate-based lipids (phospholipids) of E. coli are zwitterionic PE (phosphatidylethanolamine) (~70%), anionic PG (phosphatidylglycerol) (~20%) and CL (cardiolipin; diphosphatidylglycerol) (~5%) (Figure 1). The remaining phospholipids are less than 5% of the total pool. The ratio of zwitterionic to anionic phospholipids can be varied by mutations in the pssA and pgsA genes. Null mutations in the pgsA gene are lethal, but when combined with a set of suppressor mutations, viable cells can be grown with ~95% PE and ~5% primarily phosphatidic acid and CDP-diacylglycerol. Null mutations in the pssA gene result in complete elimination of all amino-containing and zwitterionic phospholipids, with 100% of phospholipids being anionic. Both genes have been placed under the control of inducible promoters so that the ratio of zwitterionic to anionic phospholipids can be systematically varied. Cells with mutations in pgsA have been used to establish a role for anionic lipids as sites for organization of integral membrane proteins with amphitropic proteins to form membrane associated molecular machines responsible for cell division, DNA replication, protein secretion and membrane protein insertion.
Figure 1. Synthesis of native and foreign lipids in E. coli.
Pathways, genes encoding the enzyme responsible and major phospholipids naturally occurring in E. coli are indicated in red. Pathways, genes encoding the enzyme responsible and the major lipid synthesized by foreign enzymes in E. coli are indicated in green. 1. cdsA, CDP-diacylglycerol synthase; 2. pssA, phosphatidylserine synthase; 3. psd, phosphatidylserine decarboxylase; 4. pgsA, phosphatidylglycerophosphate synthase; 5. pgpABC, phosphatidylglycerophosphate phosphatase (any of three gene products catalyse this step [18]); 6. cls, CL synthase; 7. mdoB, PG:pre-MDO (membrane-derived oligosaccharide) sn-glycerol-1-phosphate transferase; 8. dgk, diacylglycerol kinase; 9. mgs, GlcDAG synthase (Acholeplasma laidlawii); 10. dgs, GlcGlcDAG synthase (Acholeplasma laidlawii); 11. pcs, PC synthase (Legionella pneumophila). Modified from [9] with permission. © 2009 The American Society for Biochemistry and Molecular Biology.
Cells completely lacking PE (pssA-null strains) are dependent on millimolar levels of bivalent cations (Ca2+ > Sr2+ > Mg2+, with Ba2+ being ineffective) to prevent cell lysis and possibly to support non-bilayer properties of CL; PE is the predominant phospholipid with non-bilayer properties in wild-type cells. Lack of PE also compromises late stages of cell division and final constriction both in E. coli and eukaryotic cells. In addition, PE is required to support the energy-dependent uphill transport of solutes catalysed by several amino acid and sugar permeases. Detailed analysis of the molecular basis for the lack of uphill transport function discussed below uncovered the involvement of membrane lipid composition as a determinant of topological organization of some membrane proteins and further extended the rules governing membrane protein folding and assembly.
E. coli membrane lipid composition can be further manipulated by introduction of foreign genes [9] that encode lipids not found in E. coli (Figure 1). Expression of such genes in cells lacking PE allows the study of the effects of lipids with a different, but overlapping, mixture of physical and chemical properties when compared with PE. PE shows non-bilayer-forming properties dependent on fatty acid composition, is charged but carries no net charge, and is capable of hydrogen-bonding through its amine-containing headgroup. PG and CL carry a net negative charge and have headgroup hydrogen-bonding ability, but PG is bilayer-prone whereas CL is non-bilayer prone in the presence of bivalent cations, which are prevalent in cells. Introduction of the pcs gene results in replacement of PE by 70% of total phospholipid as PC (phosphatidylcholine). PC is similar to PE in being net-charge-neutral, but is a bilayer-prone lipid. However, the quaternary amine of PC cannot hydrogenbond. Introduction of the mgs and dgs genes into PE-lacking cells results in 40% of the total glycerol-based lipids as either GlcDAG (monoglucosyl diacylglycerol) or GlcGlcDAG (diglucosyl diacylglycerol) respectively. Both carry no charge, have hydrogen-bonding properties, and, like PE and PC, can dilute the high negative charge density of the membrane surface resulting from PG and CL. However, they differ in that GlcDAG is non-bilayer-prone, whereas GlcGlcDAG is bilayer-prone.
Lipid effects on folding of an extramembrane domain that is crucial for uphill transport function
On the basis of in vitro reconstitution of LacY (lactose permease) of E. coli into proteoliposomes, it has been recognized for over 35 years that PE is required for energy-dependent uphill accumulation of substrate, but not for energy-independent downhill equilibration of substrate [19]. Unexpectedly, PC does not substitute for PE in supporting uphill transport by LacY in proteoliposomes [19,20]. However, phosphatidylserine did substitute, suggesting a requirement for an ionizable amine to support uphill transport. With the construction of strains of E. coli with different lipid compositions, the physiological significance of lipid requirements for transport function could be tested in vivo. Using strains of E. coli either with or without PE, the requirement for PE for uphill transport and not for downhill transport was verified in vivo [21]. Surprisingly, cells with PC in place of PE also supported uphill transport [8]. Strains containing GlcDAG [22] or phosphatidylserine [23] in place of PE also supported uphill transport. However, GlcGlcDAG did not support uphill transport [24]. Part of the answer for the above lipid requirements was obtained using a monoclonal antibody (mAb 4B1) specific for the sequence and conformation of an epitope in the extramembrane domain P7 of LacY [25] that is exposed to the periplasm (Figure 2A). This antibody recognizes LacY in membranes and after SDS/PAGE and Western blotting, provided LacY initially exhibits uphill transport function [26,27]. Perturbation of P7 domain organization results in a lowering of the abnormally high pKa of a glutamate residue residing within TM X [25]. The monoclonal antibody recognizes LacY from PE-containing cells or from cells in which LacY was originally assembled in the absence of PE and then exposed to PE either in vitro during refolding on a solid support [26,27], in situ during or after insertion into isolated E. coli inside-out membrane vesicles [28] or in vivo after assembly [5,27] by interaction with non-native late-folding intermediates of LacY, like most conventional protein molecular chaperones [29]. Recognition was also observed for LacY from cells containing PC or GlcDAG, but not GlcGlcDAG [8], in place of PE. LacY was not recognized from cells containing only PG and CL [26,27]. The PC species used in most reconstitutions of LacY were those with high unsaturated fatty acid content [19,20], whereas E. coli-derived PC [8,19] is a mixture of species containing significant amounts of saturated fatty acids. Proper folding of the P7 domain requires PC species with at least one saturated fatty acid in combination with anionic lipid possessing hydrogen-bonding capability [8]. Therefore support of uphill transport function and recognition by mAb 4B1 appears to require a complex mixture of physical and chemical properties dictated by the hydrophilic headgroup and the hydrophobic backbone of the supporting lipid.
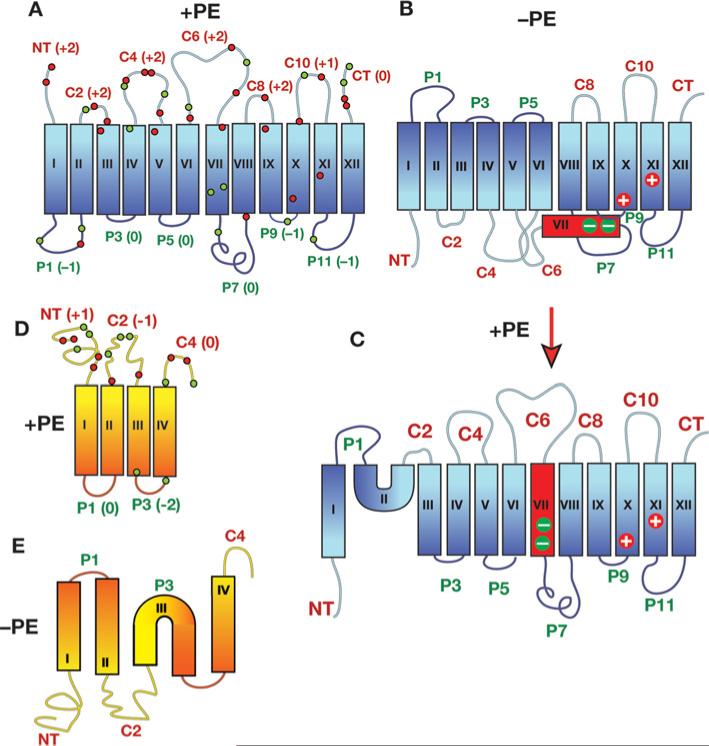
Figure 2. TM organization of secondary transporters as a function of membrane lipid composition.
The top of each structure faces the cytoplasm. Blue, red and yellow/orange rectangles denote TMs numbered sequentially by Roman numerals. N- and C-terminal domains are labelled as NT and CT respectively. Extramembrane domains are labelled according to their exposure to the cytoplasm (C) or periplasm (P) in PE-containing cells. (A) Topology organization of LacY as determined in PE-containing (+PE) cells is depicted. The net charge of each extramembrane domain is noted next to the domain name, and the approximate positions of charged residues throughout the protein are indicated by green (negative charge) and red (positive charge) dots. (B) Topology organization of LacY as determined in PE-lacking (–PE) cells is shown. TM VII (red) exposure to the periplasm results in the loss of salt bridges between the two aspartate residues in TM VII with positively charged amino acids in TM X and TM XI. (C) Topology organization of LacY as determined in cells where assembly of LacY initially occurred in the absence of PE followed by the induction of PE synthesis in the absence of new LacY synthesis. (D) Topology organization of PheP (topology of lipid-insensitive region TM V-CT not shown) as determined in PE-containing cells is shown with labelling of domains following that described in (A). (E) Topology organization of PheP as determined in cells lacking PE. Note that the topological organization of GabP is the same as that of PheP with a slightly different distribution of charged residues. Modified from [9] with permission. © 2009 The American Society for Biochemistry and Molecular Biology.
Lipids as topological determinants
The molecular basis for lack of uphill transport by LacY in cells containing only PG and CL is due to a large topological inversion that also disrupts the structure of the P7 domain [5,27]. The SCAM™ (substituted cysteine accessibility method as applied to TM orientation) was used to map the organization of LacY as a function of membrane lipid composition. In this method [30], single cysteine replacements were made in a LacY derivative lacking normal cysteine residues. After synthesis in cells of various lipid compositions, the accessibility of the cysteine residues to a membrane-impermeable thiol-group-specific reagent linked to biotin was determined in whole cells or lysed cells. Cysteine residues in periplasmic domains should be accessible in whole cells and those in cytoplasmic domains should be accessible only after cell lysis, whereas those in TMs should not be accessible under either condition since the thiol-specific reagent reacts only with ionized cysteine residues in a hydrophilic environment. The method was extended further to differentiate locally restricted domains from TMs or domains that partially insert into the membrane without traversing the membrane (mini-loops) [5,30]. Use of mild alkaline treatment during derivatization can disrupt local structurally hindered cysteine residues within solvent-exposed domains, whereas strong alkaline treatment can release mini-loops from the membrane without exposing cysteine residues within TMs.
Assembly of LacY in PE-containing cells results in the expected orientation of all TMs [31] on the basis of all previous biochemical and structural information on LacY (Figure 2A). However, assembly in cells lacking PE (Figure 2B) results in the complete inversion of the N-terminal six-TM helical bundle with respect to the plane of the membrane bilayer and the last five TMs [5,27]. In addition TM VII no longer resides in the membrane, but exists as an extramembrane domain exposed to the periplasm [5]. TM VII is of low hydrophobicity due to two aspartate residues that are normally salt-bridged to positive residues in TMs X and XI [32,33]. Remarkably, downhill transport is not affected by this topological inversion, probably because the N-terminal bundle, with most of the substrate-binding sites [32], remains intact. However, this inversion results in a major disruption of secondary structure in the vicinity of domain P7, which contains the recognition site for mAb 4B1. Reconstitution of LacY isolated either from PE-containing or PE-lacking cells into proteoliposomes results in a topological organization dictated by the lipid composition of the proteoliposomes and not the lipid composition of the source of LacY [20]. Although PC species with high unsaturated fatty acid content used in proteoliposomes does not support uphill transport or P7 conformation [19,20], they do support wild-type topology as does total E. coli total phospholipid composition [20]. However, presence of PG and CL alone in liposomes results an inverted topology [20]. In vivo substitution of PE with similarly net-neutral PC [8], GlcDAG [34] or GlcGlcDAG [24] also supports wild-type topology. Therefore the relevant property of lipids that support wild-type topological organization is their net-charge-neutral headgroup character that dilutes out the high negative charge density of the membrane surface contributed by PG and CL [15].
Most remarkable is that topological organization and TM orientation, once attained, is dynamic and can change in response to changes in the lipid environment [5,27]. Using an inducer-regulated promoter for pssA gene expression, LacY was synthesized and fully assembled in the absence of PE. After termination of new LacY synthesis, PE synthesis was induced with PE returning to wild-type levels. Post-assembly synthesis of PE resulted in a regain of recognition by mAb 4B1 and uphill transport function, insertion of TM VII into the membrane, and a near complete return to native topological organization [5]. Only NT, TM I, P2 and TM II did not return to native organization (Figure 2C).
Properties of lipid-sensitive domains
The topological organization of amino acid permeases for phenylalanine (PheP) [35] and γ-aminobutyrate (GabP) [36] are also sensitive to the lipid composition (Figures 2D and 2E). In PE-lacking cells, the N-terminal TM hairpin of these two transporters is inverted with respect to the remainder of the protein and TM III exists as a mini-loop. This inversion also results in loss of uphill transport function. In the case of PheP, wild-type topological organization and function were restored by post-assembly synthesis of PE. For the amino acid permeases, the N-terminal extramembrane domains do not follow the positive-inside rule and are, in some cases, net negative on the cytoplasmic side (Figure 2D). Although the extramembrane domains of the N-terminal bundle of LacY follow the positive-inside rule, there are more negatively charged amino acids on the cytoplasmic surface of the N-terminal than the C-terminal half of LacY (Figure 2A).
To test whether the presence of negative residues in these domains makes the protein topology dependent on lipid composition, the mixture of charged residues on the cytoplasmic surface of the N-terminal six-TM bundle of LacY was altered, and topological organization as a function of lipid composition was determined. Increasing the net positive charge of the bundle surface by 1 in a position-independent manner (i.e. within C-3, C-4 or C-6), either by adding a positive charge or removing a negative charge, prevented inversion in PE-lacking cells [5]. Adding a positive and negative charge (no net change) did not prevent inversion. If increasing the positive charge of the N-terminal bundle prevents inversion in PE-lacking cells, then increasing the negative charge should induce inversion in PE-containing cells. This proved to be the case, but inversion only occurred after changing the net charge of the cytoplasmic surface of the bundle from + 6 to – 6. Therefore the main effect of net-neutral lipids is to dampen the translocation potential of negative residues in opposition to the positive-inside rule to allow the presence of negative residues in the cytoplasmic domain for functional purposes without affecting protein topology. This conclusion was supported further by results with PheP and CscB (sucrose permease) [37]. Topological inversion for PheP is prevented in PE-lacking cells by increasing the net positive charge in domains NT and C2. The topology of wild-type CscB is not sensitive to lipid composition. However, decreasing the net positive charges within the cytoplasmic domains of the N-terminal six-TM bundle results in inversion of the bundle first in PE-lacking cells and, upon further decreases in positive charges, inversion occurs in PE-containing cells. These results are consistent with previous results that indicate dominance for positive charge over negative charge as a topological determinant, but now demonstrate that the translocation potential of negative residues is greatly enhanced in the absence of PE [5].
PE and the positive-inside rule
Although the positive-inside rule is generally accepted [14,38], the cellular factors involved in its execution and how positively charged residues exert their effect on topology are not well established. The contribution of the translocon to making a topological decision is limited by the size of newly synthesized proteins, the time of protein residency within the translocon and the effective size of the translocation pore, which is still a matter of debate [4,39]. Since orientation of TMs within a membrane protein can be changed by the addition or removal of a single positively charged residue or by the introduction of one or many negatively charged residues depending on the absence or presence of PE respectively, the question of the relative topological potency of the charged residues in the context of different phospholipid compositions is raised. The translocation block provided by protonated positively charged amino acid residues within cytoplasmic segments serves as a primary factor determining TM topology; however, the effective charge of these domains can be affected by mechanisms that selectively protonate or deprotonate negatively charged amino acid residues depending of the presence or absence of PE [9,40]. Whatever the precise mechanism of protonation/deprotonation of these residues, PE and other neutral phospholipids clearly attenuate the topological effects of acidic residues [15]. Negative residues in the absence of PE exert an increased translocation potential that results in translocation of domains that now exhibit a lower effective net positive charge. The retention potential of positively charged residues is enhanced in the presence of PE or other net-neutral lipids by reducing the translocation potential of negative residues, thus providing a molecular basis for the operation of the positive-inside rule for domains containing a mixture of positive and negative residues. LacY and PheP initially misoriented by assembly in membranes lacking PE can be properly oriented by post-assembly exposure to PE due to strengthening of the positive retention potential to drive reorientation of TMs.
The membrane potential (outside-positive in E. coli) may actively promote translocation of acidic residues and impede translocation of basic residues and therefore contribute to the establishment of topology governed by the positive-inside rule [41,42]. However, in the endoplasmic reticulum membrane, where no detectable or defined membrane potential besides a Ca2+ gradient is found, proteins still follow the positive-inside rule [11]. The positive-inside rule also applies to obligate acidophiles, such as Sulfolobus acidocaldarius, where membrane potential is permanently reversed (i.e. inside-positive instead of negative), suggesting that the positive-inside rule may not be dependent on the polarity of the membrane potential [43]. Therefore the net effect of positive charge of cytoplasmic domains as determined by the membrane lipid composition may be dominant in retention of the positively charged residues on the cytoplasmic side of different membrane systems.
Effect of flexible domains between differentially lipid-sensitive domains
In the three proteins (LacY, PheP and GabP) demonstrated to date to contain a mixture of domains whose topologies are sensitive and insensitive to the lipid environment, a TM (Figures 2B, 2C and 2E) exists that apparently acts as a flexible molecular hinge between the lipid-sensitive and -insensitive domains. For LacY, TM VII exits the membrane in PE-lacking cells and TM II forms a mini-loop during reorientation of LacY after post-assembly introduction of PE [5]. TM III of PheP and GabP forms a mini-loop in PE-lacking cells [35,36]. Are such connecting flexible domains required in order for the flanking domains to respond differently to the lipid environment? This was tested for LacY by increasing the hydrophobicity of TM VII of LacY through replacement of Asp240 with isoleucine. This substitution prevented inversion and solvent-exposure of TM VII of wild-type LacY in cells lacking PE as well as the LacY derivative with a net – 6 charge for its cytoplasmic surface in PE-containing cells [5]. Therefore the unfavourable energetics of solvent-exposure of a hydrophobic domain can override the favourable energetics of inversion of the N-terminal bundle. This may explain why the majority of proteins are not sensitive to membrane lipid composition. Therefore lipid-dependent protein folding is dependent on both short-range interactions of charged extramembrane domains with the lipid environment and long-range co-operative interactions of TM helix packing. Unfortunately, predictions are difficult to make of domains that might act as flexible molecular switches allowing independent responses to the lipid environment.
Where is the final TM organization established?
Although the initial orientation of TM domains may be governed by interactions between the nascent polypeptide chain and the translocon, the final TM orientation depends on short-range interactions between the protein and the lipid environment and long-range interactions within the protein during final folding events [15]. The topogenic influence of lipids appears to be largely independent of other protein factors, including the translocon, since topological organization and transport function of LacY reconstituted into protein-free liposomes are determined solely by the phospholipid composition of the liposome independent of the lipid composition of the cells from which LacY was derived [20]. During reorientation of misoriented LacY by post-assembly introduction of PE, it is unlikely that the translocon is involved; however, the involvement of other proteins such as chaperones has not been ruled out. LacY forms a compact folded structure in PE-lacking cells on the basis of its function in downhill transport [21,31], and it must therefore exist as a structure free of association with the translocon. Utilization of the translocon would require reassociation and unfolding to effect reorientation. LacY is also in large molar excess, when overproduced, over the number of functional translocons. During initial synthesis and assembly of LacY, the hydrophobicity of TM VII and the charge nature of its preceding cytoplasmic domain determine final topology [5]. Since it is unlikely that the translocon can accommodate even five TMs in its aqueous environment [1], much of the N-terminal bundle of LacY must exist in a topologically undecided state outside the translocon until TM VII is synthesized [5]. Therefore all of these results are consistent with final topological decisions being made outside and independent of the translocon and dictated largely by lipid–protein interactions and the positive-inside rule as applied to a multiple TM unit.
Conclusions and perspectives
By artificially changing the steady-state membrane lipid composition, new principles governing membrane protein assembly have been uncovered. In addition, membrane protein structure during assembly and after assembly has been shown to be highly dynamic and not fixed or static. Changes in the charge nature of the extramembrane domains connecting TMs or changes in the net charge of the membrane surface synergistically can result in similar changes in the topological organization of a membrane protein, thus supporting charge interaction in a complementary manner between the cytoplasmic domains and the bilayer surface as a determinant of topological organization. Therefore proteins and lipids have co-evolved to establish a set of interdependent determinants of protein organization. Zwitterionic or net-neutral lipids are required to fulfil the positive-inside rule, which explains why positive residues are more potent topological determinants than negative residues under physiological conditions. During protein translation, LacY exists in a topologically undetermined state until at least TM VII has exited the ribosome. This is supported by the fact that the final topology of the N-terminal six-TM bundle is dependent on both the charge nature of C-6 and the hydrophobicity of TM VII. This fact disfavours a linear or sequential mode of TM insertion and demonstrates that the positive-inside rule can be executed in a retrograde manner and that long-range intramolecular interactions can influence final protein folding events. Subsequent to these findings, a similar conclusion was reached for EmrE of E. coli [44]. The results also definitively and systematically demonstrate that LacY and PheP can undergo TM flipping or changes in orientation after folding into a compact structure and that topology of the N-terminal TM bundle of LacY and the N-terminal TM hairpin of PheP or GabP are most probably determined outside the translocon. Therefore membrane protein organization is determined during initial assembly through interactions between the protein and the lipid environment, but is also dynamic and can change post-assembly, dependent on changes in the lipid environment. LacY, GabP and PheP are the best established examples of this dynamic aspect of native polytopic membrane proteins.
The fact that changes in lipid environment can have such a dramatic effect on membrane protein organization has important implications for membrane proteins in eukaryotic systems or in membranes where lipid composition is not uniform either spatially or temporally. Eukaryotic membrane proteins are exposed to various lipid environments as they move along the secretory pathway from the endoplasmic reticulum to the Golgi and finally to either the plasma membrane or other internal organelles. This affords the opportunity for initial latent activity to be activated at the site of final protein residency by the proper lipid environment. Local changes in lipid environment through interaction with specifically enriched lipid domains, lipid rafts and membrane fission or fusion sites could also activate or inactivate proteins. Lipid–protein interactions during polytopic protein biogenesis can contribute to folding anomalies induced by minor sequence perturbations in inherited topological disorders. Naturally occurring mutations of structurally important residues could lead to a different structural organization of the mutant protein within the changing lipid environment as the protein moves through the same or altered organelle trafficking route. Therefore changes in membrane lipid composition either locally or during intracellular movement of proteins along the organelle-based secretory pathway can lead to misinterpretation of existing topological signals or different interpretation of new ones. Incompatibility of lipids necessary for correct topogenesis and misinterpretation of topological signals by the ‘wrong’ lipid profile may be the basis for the inability to obtain active membrane proteins after expression in foreign host cells.
Clearly not all membrane proteins are sensitive to the lipid environment, since cells with large changes in membrane lipid composition are viable. Even in the case of solute permeases, partial transport function is retained in the absence of PE. Other functions, such as cell division, are also impaired in the absence of PE. The rescue of LacY function in vivo by PC [8] and GlcDAG [34] does not result in a completely wild-type structure as is evident from altered accessibility of some cysteine substitutions in the extramembrane domains. The availability of an extensive collection of mutants in which membrane lipid composition can be systematically controlled provides an important resource for the study of the physiologically relevant role of lipids in a wide range of biological processes. Since membrane protein sequence is ‘written’ for a given membrane environment and some topogenic signals may dominate over others within different lipid profiles, it is tempting to speculate that, during the course of evolution, both proteins and lipids co-evolved in the context of the lipid environment of membrane systems in which both are mutually dependent on each other.
Acknowledgments
Funding
This work was supported in whole or in part by the National Institutes of General Medical Sciences [grant number GM R37 20478] and the John Dunn Research Foundation (to W.D.)
Abbreviations used
- CL
cardiolipin
- CscB
sucrose permease
- GabP
γ -aminobutyrate
- GlcDAG
monoglucosyl diacylglycerol
- GlcGlcDAG
diglucosyl diacylglycerol
- LacY
lactose permease
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- PheP
phenylalanine permease
- TM
transmembrane domain
- SCAM™
substituted cysteine accessibility method as applied to TM orientation
References
- 1.Driessen AJ, Nouwen N. Protein Translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 2.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 3.Junne T, Kocik L, Spiess M. The hydrophobic core of the Sec61 translocon defines the hydrophobicity threshold for membrane integration. Mol. Biol. Cell. 2010;21:1662–1670. doi: 10.1091/mbc.E10-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skach WR. Cellular mechanisms of membrane protein folding. Nat. Struct. Mol. Biol. 2009;16:606–612. doi: 10.1038/nsmb.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogdanov M, Xie J, Heacock P, Dowhan W. To flip or not to flip: protein–lipid charge interactions are a determinant of final membrane protein topology. J. Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monne M, Hessa T, Thissen L, von Heijne G. Competition between neighboring topogenic signals during membrane protein insertion into the ER. FEBS J. 2005;272:28–36. doi: 10.1111/j.1742-4658.2004.04394.x. [DOI] [PubMed] [Google Scholar]
- 7.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 8.Bogdanov M, Heacock P, Guan Z, Dowhan W. Plasticity of lipid–protein interactions in the function and topogenesis of the membrane protein lactose permease from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15057–15062. doi: 10.1073/pnas.1006286107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogdanov M, Jun X, Dowhan W. Lipid–protein interactions drive membrane protein topogenesis in accordance with the positive-inside rule. J. Biol. Chem. 2009;284:9637–9641. doi: 10.1074/jbc.R800081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White SH, von Heijne G. Do protein–lipid interactions determine the recognition of transmembrane helices at the ER translocon? Biochem. Soc. Trans. 2005;33:1012–1015. doi: 10.1042/BST20051012. [DOI] [PubMed] [Google Scholar]
- 11.Harley CA, Holt JA, Turner R, Tipper DJ. Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity, and its distribution. J. Biol. Chem. 1998;273:24963–24971. doi: 10.1074/jbc.273.38.24963. [DOI] [PubMed] [Google Scholar]
- 12.Lee E, Manoil C. Mutations eliminating the protein export function of a membrane-spanning sequence. J. Biol. Chem. 1994;269:28822–28828. [PubMed] [Google Scholar]
- 13.White SH, Ladokhin AS, Jayasinghe S, Hristova K. How membranes shape protein structure. J. Biol. Chem. 2001;276:32395–32398. doi: 10.1074/jbc.R100008200. [DOI] [PubMed] [Google Scholar]
- 14.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dowhan W, Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu. Rev. Biochem. 2009;78:515–540. doi: 10.1146/annurev.biochem.77.060806.091251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 17.Dowhan W. Molecular genetic approaches to defining lipid function. J. Lipid Res. 2009;50:S305–S310. doi: 10.1194/jlr.R800041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y-H, Guan Z, Zhao J, Raetz CRH. Three phosphatidylglycerol-phosphate phosphatases in the inner membrane of Escherichia coli. J. Biol. Chem. 2011;286:5506–5518. doi: 10.1074/jbc.M110.199265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CC, Wilson TH. The phospholipid requirement for activity of the lactose carrier of Escherichia coli. J. Biol. Chem. 1984;259:10150–10158. [PubMed] [Google Scholar]
- 20.Wang X, Bogdanov M, Dowhan W. Topology of polytopic membrane protein subdomains is dictated by membrane phospholipid composition. EMBO J. 2002;21:5673–5681. doi: 10.1093/emboj/cdf571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogdanov M, Dowhan W. Phosphatidylethanolamine is required for in vivo function of the membrane-associated lactose permease of Escherichia coli. J. Biol. Chem. 1995;270:732–739. doi: 10.1074/jbc.270.2.732. [DOI] [PubMed] [Google Scholar]
- 22.Wikstrom M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander A, Dowhan W. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J. Biol. Chem. 2004;279:10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 23.Hawrot E, Kennedy EP. Phospholipid composition and membrane function in phosphatidylserine decarboxylase mutants of Escherichia coli. J. Biol. Chem. 1978;253:8213–8220. [PubMed] [Google Scholar]
- 24.Wikstrom M, Kelly A, Georgiev A, Eriksson H, Rosen-Klement M, Bogdanov M, Dowhan W, Wieslander A. Lipid-engineered Escherichia coli membranes reveal critical lipid head-group size for protein function. J. Biol. Chem. 2009;284:954–965. doi: 10.1074/jbc.M804482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J, Wu J, Carrasco N, Kaback HR. Identification of the epitope for monoclonal antibody 4B1 which uncouples lactose and proton translocation in the lactose permease of Escherichia coli. Biochemistry. 1996;35:990–998. doi: 10.1021/bi952166w. [DOI] [PubMed] [Google Scholar]
- 26.Bogdanov M, Sun J, Kaback HR, Dowhan W. A phospholipid acts as a chaperone in assembly of a membrane transport protein. J. Biol. Chem. 1996;271:11615–11618. doi: 10.1074/jbc.271.20.11615. [DOI] [PubMed] [Google Scholar]
- 27.Bogdanov M, Umeda M, Dowhan W. Phospholipid-assisted refolding of an integral membrane protein: minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J. Biol. Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- 28.Bogdanov M, Dowhan W. Phospholipid-assisted protein folding: phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. EMBO J. 1998;17:5255–5264. doi: 10.1093/emboj/17.18.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogdanov M, Dowhan W. Lipid-assisted protein folding. J. Biol. Chem. 1999;274:36827–36830. doi: 10.1074/jbc.274.52.36827. [DOI] [PubMed] [Google Scholar]
- 30.Bogdanov M, Zhang W, Xie J, Dowhan W. Transmembrane protein topology mapping by the substituted cysteine accessibility method (SCAM™): application to lipid-specific membrane protein topogenesis. Methods. 2005;36:148–171. doi: 10.1016/j.ymeth.2004.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogdanov M, Heacock PN, Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abramson J, Iwata S, Kaback HR. Lactose permease as a paradigm for membrane transport proteins. Mol. Membr. Biol. 2004;21:227–236. doi: 10.1080/09687680410001716862. [DOI] [PubMed] [Google Scholar]
- 33.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 34.Xie J, Bogdanov M, Heacock P, Dowhan W. Phosphatidylethanolamine and monoglucosyldiacylglycerol are interchangeable in supporting topogenesis and function of the polytopic membrane protein lactose permease. J. Biol. Chem. 2006;281:19172–19178. doi: 10.1074/jbc.M602565200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Bogdanov M, Pi J, Pittard AJ, Dowhan W. Reversible topological organization within a polytopic membrane protein is governed by a change in membrane phospholipid composition. J. Biol. Chem. 2003;278:50128–50135. doi: 10.1074/jbc.M309840200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Campbell HA, King SC, Dowhan W. Phospholipids as determinants of membrane protein topology: phosphatidylethanolamine is required for the proper topological organization of the γ-aminobutyric acid permease (GabP) of Escherichia coli. J. Biol. Chem. 2005;280:26032–26038. doi: 10.1074/jbc.M504929200. [DOI] [PubMed] [Google Scholar]
- 37.Vitrac H, Bogdanov M, Heacock P, Dowhan W. Lipids and topological rules of membrane protein assembly: balance between long- and short-range lipid–protein interactions. J. Biol. Chem. 2011 doi: 10.1074/jbc.M110.214387. doi:10.1074/jbc.M110.214387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Heijne G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- 39.Higy M, Junne T, Spiess M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry. 2004;43:12716–12722. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- 40.Gbaguidi B, Hakizimana P, Vandenbussche G, Ruysschaert JM. Conformational changes in a bacterial multidrug transporter are phosphatidylethanolamine-dependent. Cell. Mol. Life Sci. 2007;64:1571–1582. doi: 10.1007/s00018-007-7031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersson H, von Heijne G. Membrane protein topology: effects of Δμ H+ on the translocation of charged residues explain the ‘positive inside’ rule. EMBO J. 1994;13:2267–2272. doi: 10.1002/j.1460-2075.1994.tb06508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao G, Kuhn A, Dalbey RE. The translocation of negatively charged residues across the membrane is driven by the electrochemical potential: evidence for an electrophoresis-like membrane transfer mechanism. EMBO J. 1995;14:866–875. doi: 10.1002/j.1460-2075.1995.tb07068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Vossenberg JL, Albers SV, van der Does C, Driessen AJ, van Klompenburg W. The positive inside rule is not determined by the polarity of the ΔΨ (transmembrane electrical potential). Mol. Microbiol. 1998;29:1125–1127. doi: 10.1046/j.1365-2958.1998.01001.x. [DOI] [PubMed] [Google Scholar]
- 44.Seppala S, Slusky JS, Lloris-Garcera P, Rapp M, von Heijne G. Control of membrane protein topology by a single C-terminal residue. Science. 2010;328:1698–1700. doi: 10.1126/science.1188950. [DOI] [PubMed] [Google Scholar]