Abstract
Neurogenic transcription factors and evolutionarily conserved signalling pathways have been found to be instrumental in the formation of neurons1,2. However, the instructive role of microRNAs (miRNAs) in neurogenesis remains unexplored. We recently discovered that miR-9* and miR-124 instruct compositional changes of SWI/SNF-like BAF chromatin-remodelling complexes, a process important for neuronal differentiation and function3–6. Nearing mitotic exit of neural progenitors, miR-9* and miR-124 repress the BAF53a subunit of the neural-progenitor (np)BAF chromatin-remodelling complex. After mitotic exit, BAF53a is replaced by BAF53b, and BAF45a by BAF45b and BAF45c, which are then incorporated into neuron-specific (n)BAF complexes essential for post-mitotic functions4. Because miR-9/9* and miR-124 also control multiple genes regulating neuronal differentiation and function5,7–13, we proposed that these miRNAs might contribute to neuronal fates. Here we show that expression of miR-9/9* and miR-124 (miR-9/9*-124) in human fibroblasts induces their conversion into neurons, a process facilitated by NEUROD2. Further addition of neurogenic transcription factors ASCL1 and MYT1L enhances the rate of conversion and the maturation of the converted neurons, whereas expression of these transcription factors alone without miR-9/9*-124 was ineffective. These studies indicate that the genetic circuitry involving miR-9/9*-124 can have an instructive role in neural fate determination.
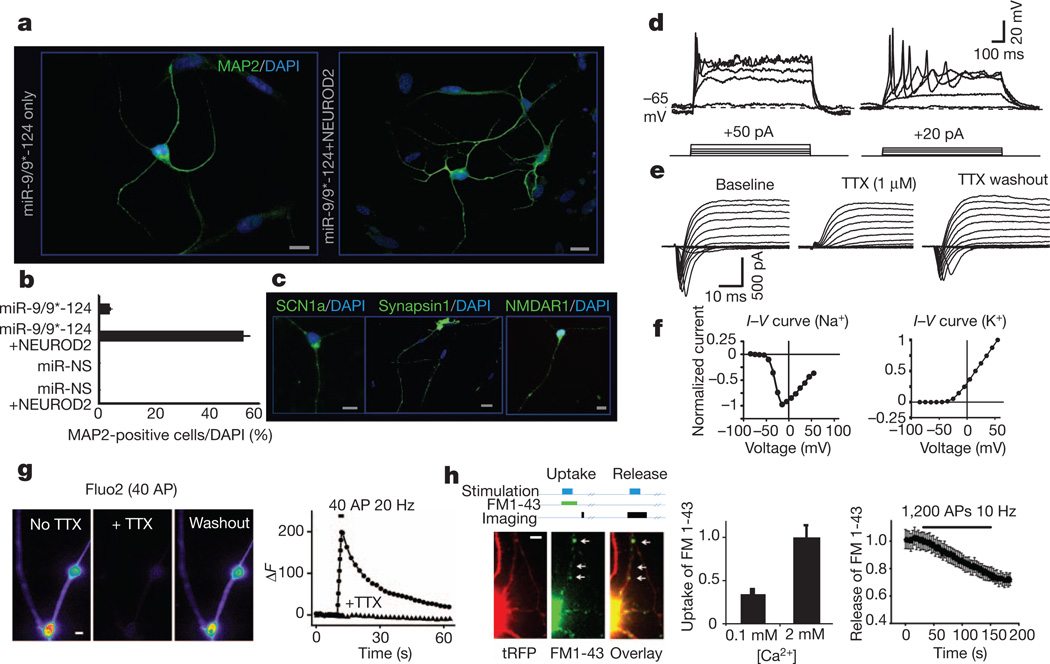
During the course of exploring the roles of miR-9/9* and miR-124 (ref. 5), we noted that these miRNAs induced neuronal morphologies in cultured cells. To explore this effect in greater detail, we prepared a single lentiviral vector that expresses both precursors of miR-9/9* and miR-124 along with a turbo red fluorescent protein (tRFP) marker, and infected human neonatal foreskin fibroblasts (Supplementary Fig. 1). The fibroblast culture was free of neural progenitors, keratinocytes or melanocytes (Supplementary Figs 2–4). Remarkably, fibroblasts expressing miR-9/9*-124 showed a rapid reduction in proliferation, displayed neuron-like morphologies (Supplementary Fig. 5) and expressed MAP2, a marker of post-mitotic neurons, within 4 weeks after infection (Fig. 1a, left). This was owing to synergism between miR-9/9* and miR-124, as expressing these miRNAs separately did not lead to the appearance of MAP2-positive cells (Supplementary Fig. 6). In light of the low percentage of MAP2-positive cells obtained with miRNAs only (less than 5%, Fig. 1b), we began adding neurogenic transcription factors and found that NEUROD2 (refs 14–17) was most effective at increasing the conversion frequency (Supplementary Fig. 7). We estimate that ~50% of these cells have acquired neuronal fates as indicated by MAP2 expression 30 days after infection (Fig. 1a, right, b). However, because cells detached, remained uninfected or died during the conversion process, a conservative estimate is that ~5% of the starting cells became neurons. Importantly, neither NEUROD2 alone nor non-specific miRNA (miR-NS) could convert fibroblasts into neurons (Fig. 1b), demonstrating the essential role of miR-9/9*-124 in this process. Synergism between miR-9/9* and miR-124 seemed to be crucial: expressing miR-9/9* and miR-124 individually with NEUROD2 failed to produce MAP2-positive cells (Supplementary Fig. 6). Using EdU-incorporation, we found that miR-9/9*-124-infected fibroblasts had exited the cell cycle 1 week after infection (Supplementary Fig. 8), consistent with the anti-proliferative role of these miRNAs5. Lastly, immunostains indicated that the induced neurons expressed SCN1a, a key contributor to neuronal excitability, as well as synapsin 1 and NMDA receptor 1 (Fig. 1c).
Figure 1. miRNA-induced transformation of human fibroblasts.
a, MAP2 expression in miR-9/9*-124 only (left) and miR-9/9*-124-NEUROD2 (right) converted cells. DAPI, 4′,6-diamidino-2-phenylindole. Scale bar, 20 µm. b, Quantification of MAP2-positive cells with processes at least three times the length of the cell body from ten random fields. The graph represents the percentage of MAP2-positive cells over DAPI-positive cells. miR-9/9*-124 only: n = 558; miR-9/9*-124-NEUROD2: n = 658 cells. The error bars are s.e.m. MAP2 signal was undetectable in fibroblasts infected with miR-NS, with or without NEUROD2. Scale bar, 20 µm. c, Expression of SCN1a, synapsin 1 and NMDAR1 in miR-9/9*-124-NEUROD2-converted cells. Scale bar, 20 µm. d, Representative traces of action potentials recorded in current clamp in miR-9/9*-124-NEUROD2-converted cells. Twelve out of 22 cells showed single action potentials and 2 cells showed repetitive firing. e, A representative example of a series of voltage steps applied to an miR-9/9*-124-NEUROD2-induced neuron held at−70 mV. An inward current was observed, blocked by 1 µM TTX, and reversed after TTX washout (n = 6). f, I–V curve for the peak inward (left) and outward (right) currents. g, An example of Ca2+ influx in induced neurons as measured by Fluo2-AM imaging. Images show the peak Fluo2-AM signal on stimulation before and during TTX application, and after TTX washout, respectively. The graph represents the changes in Fluo2 signal over time (circles, no TTX; triangles, with TTX). AP, action potential. Field stimulation is indicated by the black bar. Scale bar, 2 µm. h, An example of vesicle recycling measured by FM1-43 imaging in induced neurons. Top diagram illustrates the protocol of FM uptake or release experiments. The images represent the typical FM1-43 dye uptake signal (middle) in a converted cell marked by tRFP (left). The left graph shows the measurement of FM1-43 loading in 2 mM Ca2+, which was significantly reduced in low Ca2+ concentration (0.1 mM). The right graph quantifies FM1-43 release (destaining) during stimulations. FM1-43 signal was measured from n > 600 boutons (arrows indicate examples of boutons) from 4 cultures. All error bars are s.e.m. In some cases, the s.e.m. is too small to be resolved. Scale bar, 4 µm.
Using whole-cell patch recording, we found that injecting depolarizing current in induced neurons (cultured up to 8 weeks) could consistently trigger single action potentials and in some cases, repetitive firing (Fig. 1d). Moreover, their resting membrane potential (−34.1 ± 1.7 mV; Supplementary Fig. 9) was significantly more negative than that of control fibroblasts (−20.4 ± 0.6 mV, n = 4). Applying a series of voltage steps to the induced cells evoked large inward currents closely followed by outward currents, which were not observed in the fibroblasts (Supplementary Fig. 10). Importantly, adding 1 µM tetrodotoxin (TTX) completely and reversibly blocked the initial inward current, confirming that the current was due to voltage-gated sodium channels (Fig. 1e), as would be expected from the current–voltage (I–V) curve of inward currents (Fig. 1f, left). The I–V curve of outward currents showed the characteristics of voltage-gated potassium channels in neurons (Fig. 1f, right). Moreover, some of these cells exhibited postsynaptic currents, which could be reversibly blocked by 2,3-dihydroxy-6-nitro-7-sulfamoyl-benzo[f]quinoxaline-2,3-dione (NBQX) and 2-amino-5-phosphonopentanoic acid (APV) (Supplementary Fig. 11).
We examined the ability of cells converted by miR-9/9*-124- NEUROD2 to elicit a stimulation-dependent calcium influx using the calcium indicator Fluo2. Field stimulation triggered calciuminflux that could be abolished by adding TTX (Fig. 1g) or 200 µM Cd2+ (Supplementary Fig. 12), demonstrating the ability of converted cells to support activity-dependent Ca2+ influx through voltage-gated Ca2+ channels without any requirement for a pre-pulse. Activity-dependent uptake and release of the lipophilic dye FM1-43 was used to evaluate the ability to form functional presynaptic terminals18. We found that the induced cells were able to take up and release FM dyes in a stimulation-dependent and Ca2+-dependent manner (Fig. 1h).
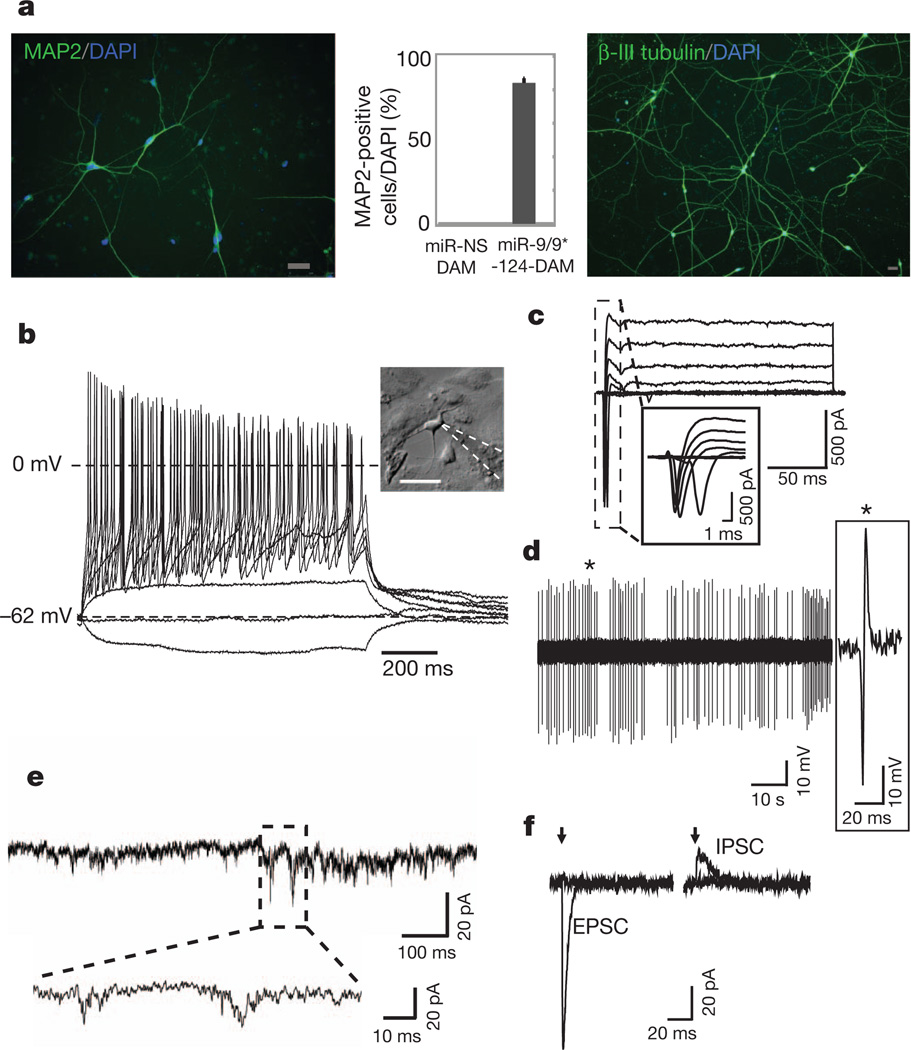
Because the miR-9/9*-124-NEUROD2-induced cells only occasionally showed repetitive action potentials, we sought to optimize the maturation of the cells by introducing additional neurogenic factors. Because ASCL1 and MYT1L were previously shown to be important for converting mouse embryonic fibroblasts into functionally mature neurons19, we expressed miR-9/9*-124 together with NEUROD2, ASCL1 and MYT1L (DAM). We found that the miR-9/9*-124-DAM-converted cells were positive for MAP2 expression in approximately 80% of the cells remaining on the coverslips (Fig. 2a, representing ~10% of the initially plated cells), and showed extensive neurite outgrowth as illustrated by β-III tubulin staining (Fig. 2a, right). In addition, miR-9/9*-124-DAM resulted in complete exit from cell cycle as assayed by EdU pulsing for 4 days (0/176 positive) whereas nearly all control cells were positive for EdU(97/107). Importantly, DAM factors with miRNA-NS failed to produce neurons as assayed by MAP2 staining (Supplementary Fig. 13). About 80% of the miR-9/9*-124-DAM-converted cells were able to fire repetitive action potentials in response to current injections, and showed typical sodium and potassium currents during voltage clamp depolarizations (Fig. 2b–c and Supplementary Figs 9 and 14). Among recorded cells, we also observed spontaneously active cells (2/21) (Fig. 2d). Spontaneous excitatory postsynaptic currents (EPSCs) were seen in 10 out of 14 induced cells (Fig. 2e) without co-cultured primary neurons (Supplementary Figs 9 and 15a). Furthermore, the induced cells exhibited evoked EPSCs and inhibitory postsynaptic currents (IPSCs) in response to local stimulation (Fig. 2f). Importantly, neuronal identity was stable after the removal of exogenous expression of miR-9/9*-124 and DAM after 3 weeks of induction, as they still stained positive for SV2 and synapsin 1 (Supplementary Fig. 16).
Figure 2. Additional neural factors enhance the conversion to neurons.
a, MAP2 (left) and β-III tubulin (right) immunostaining of miR-9/9*-124-DAM-converted cells. Scale bar, 40 µm. The graph represents the percentage of MAP2-positive cells over DAPI-positive cells. The error bars are s.e.m.; n = 150 cells. b, A representative current clamp recording from a cell with typical neuronal morphology (see inset; scale bar, 50 µm). Voltage deflections were elicited by somatic current injections of various amplitudes (Δ = 5 pA). c, A representative voltage clamp recording of the net current at various membrane potentials (−40 to +20 mV, ΔV = 10 mV, Vhold = −90 mV). d, A representative trace of spontaneously active cells recorded in cell-attached mode. e, A representative trace demonstrating spontaneous EPSCs. f, Representative traces of evoked postsynaptic currents (left, EPSC; right, IPSC) obtained in response to local field stimulation with single current pulses (1 ms) of various amplitudes (left, 0.25 and 0.3 mA; right, 0.3 and 0.4 mA) at different membrane holding potentials (left, −70 mV; right, 0 mV). The arrows indicate the time when stimulation was applied. Stimulation artefacts were eliminated for clarity.
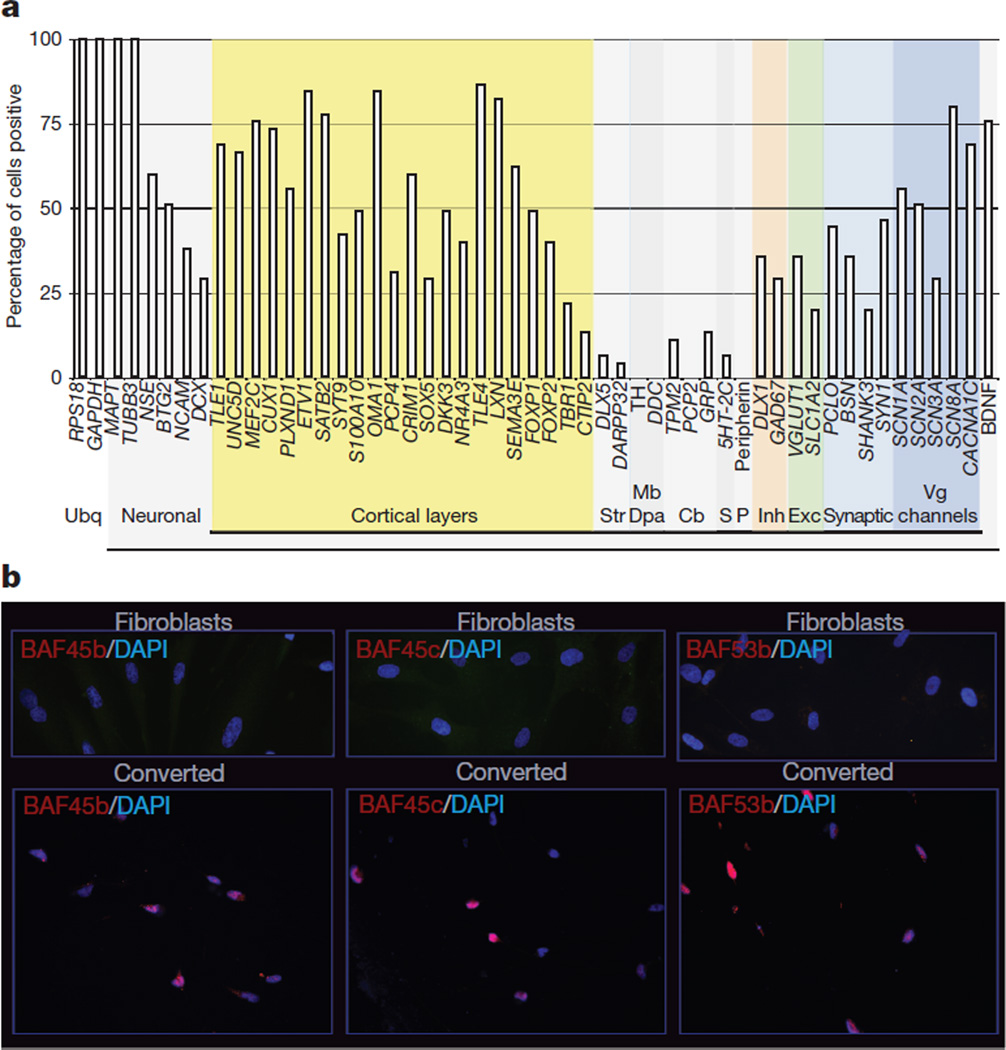
We next performed single-cell analysis to characterize the types of neurons in miR-9/9*-124-DAM-induced cells. From randomly collected single cells 4 weeks post-infection, we analysed a total of 45 induced neurons (based on MAPT and TUBB3 co-expression) for genes expressed in different types of neurons. We found that most induced cells were positive for genes expressed in cortical layers (Fig. 3a and Supplementary Fig. 17). Interestingly, we did not detect a peripheral nervous system marker (peripherin) or dopaminergic/noradrenergic markers (DDC, TH). Striatal markers (DLX5 and DARPP32 (also known as PPP1R1B)), the serotonergic marker 5HT-2C (also known as HTR2C), and cerebellar genes (PCP2, GRP, TPM2) were expressed in only a small number of cells. The miR-9/9*-124-DAM-induced cells seemed to be heterogeneously excitatory (VGLUT1 (also known as SLC17A7) and SLC1A2) and inhibitory (GAD67 (also known as GAD1) and DLX1) (Fig. 3a), which is further supported by immunostains of VGLUT1 and GABA (Supplementary Fig. 18). Moreover, by 4 weeks post-infection, the induced cells already expressed genes important for synaptic structure and function, including SYN1, BSN, PCLO and SHANK3 (Fig. 3a).
Figure 3. Characterization of induced neurons and nBAF subunit expression.
a, Multiplex quantitative polymerase chain reaction (qPCR) of 45 induced neurons for genes of specific brain regions and cell types. Cb, cerebellum; Dpa, dopaminergic; Exc, excitatory; Inh, inhibitory; Mb, midbrain; P, peripheral nervous system; S, serotonergic; Str, striatum; Ubq, ubiquitous; Vg, voltage gated. b, Fibroblasts stained negative for neuron-specific subunits of BAF complexes (top) whereas induced neurons expressed BAF45b, BAF45c and BAF53b (bottom). Scale bar, 20 µm.
miR-9* and miR-124 target separate sites in the 3′ untranslated region (UTR) of BAF53a, a subunit of BAF complexes resulting in repression of BAF53a and activation of BAF53b, which is involved in an evolutionarily conserved program of neural development20. Remarkably, we found that all of the nBAF subunits (BAF53b, 45b and 45c) were induced in the converted cells (Fig. 3b). In embryonic stem cells, BAF complexes function across the genome at several thousand sites to control placement of polycomb repressive complex 2 and the H3K27me3 repressive mark21,22. Hence, one role of the miRNAs might be to induce stable epigenetic changes involving polycomb function across the genome.
In addition to BAF53a, miR-9/9* and miR-124 also target other genes essential for neurogenesis and neuronal functions23 including components of the REST complex such as REST and CoREST8,9,24–26, and PTBP-1 (ref. 7). We found that human fibroblasts expressed BAF53a, which could be repressed by miR-9/9*-124 (Supplementary Fig. 19). However, prolonging the expression of BAF53a only incompletely blocked neuronal conversion of fibroblasts, as assayed by MAP2 staining (data not shown). Prolonging the expression of REST, CoREST or PTBP1 yielded similar results (data not shown). These findings indicate that in inducing cell fate transformations, the miRNAs miR-9/9* and miR-124 operate programmatically on multiple targets.
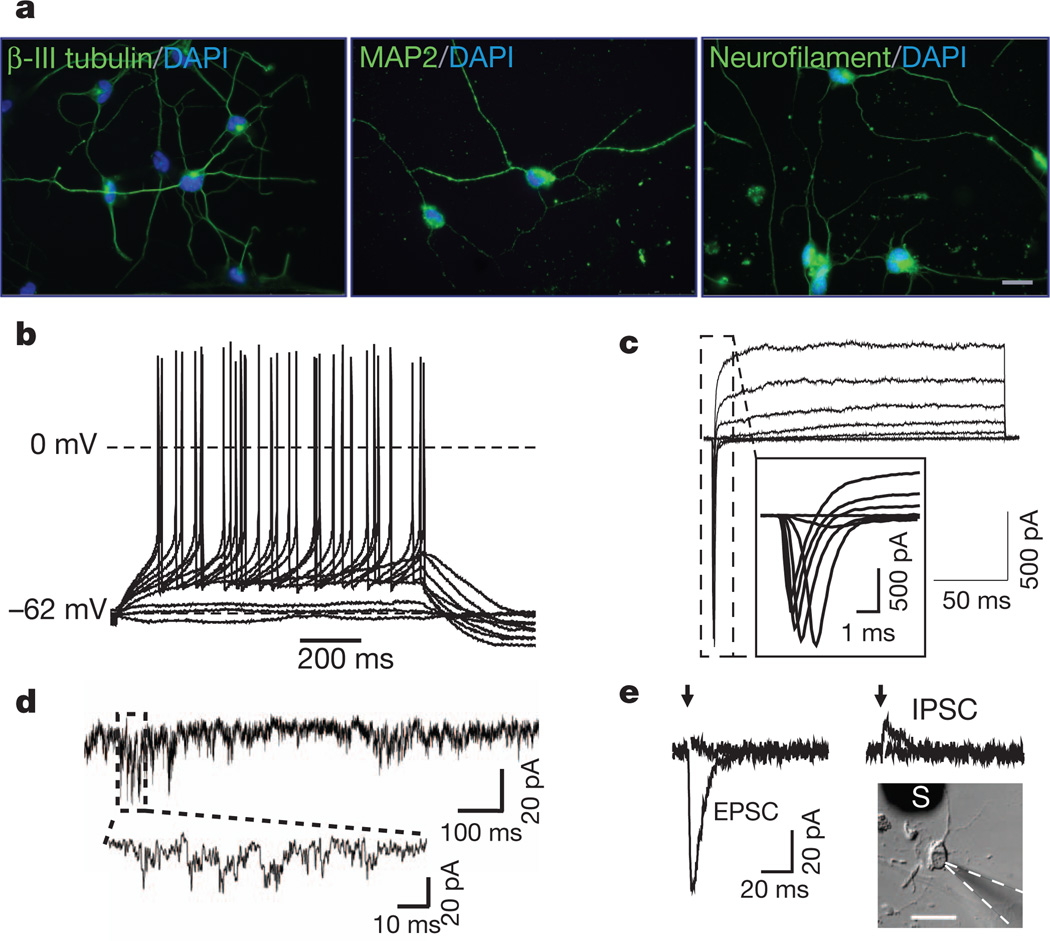
Lastly, we asked whether our approach could be effective in converting adult fibroblasts. We found that adult human dermal fibroblasts (from a 30-year-old female) could be converted into neurons (Fig. 4a), albeit more slowly. Recordings from adult cells converted by miR-9/9*-124-DAM 6 weeks after infection showed that they were able to generate action potentials (Fig. 4b). They also demonstrated typical voltage-gated sodium and potassium currents (Fig. 4c), spontaneous EPSCs (Fig. 4d and Supplementary Fig. 15b) and evoked EPSCs and IPSCs (Fig. 4e) without co-cultured primary neurons.
Figure 4. Conversion of adult fibroblasts by miR-9/9*-124-DAM.
a, Immunostaining of β-III tubulin (left), MAP2 (middle) and neurofilament (right) in dermal fibroblasts of a 30-year-old individual converted by miR-9/9*-124-DAM. Scale bar, 20 µm. b, A representative current clamp recording. Voltage deflections were elicited by somatic current injections of various amplitudes (Δ = 2 pA). c, A representative voltage clamp recording of the net current at various membrane potentials (−40 to +20 mV, ΔV = 10 mV, Vhold = −90 mV). d, A representative trace of spontaneous EPSCs. e, Representative traces of evoked postsynaptic currents obtained from converted adult cells (see inset; scale bar, 50 µm) in response to local field stimulation with single current pulses (1 ms) of various amplitudes (left, 0.2 and 0.25 mA; right, 0.4 and 0.5 mA). Evoked EPSPs and IPSCs were recorded at −70 mV and +30 mV in the presence of picrotoxin and NBQX/APV, respectively. S, stimulator.
Our studies show that activating a neural developmental regulatory circuit involving miRNAs in human fibroblasts can surprisingly induce their conversion into neurons, indicating an instructive role for this circuitry in human neurogenesis. In our study, neurogenic transcription factors, delivered either singly (NEUROD2), or in combination (NEUROD2, ASCL1 and MTYL1), seem to function synergistically with the neurogenic activities of miR-9/9*-124. This raises the possibility of inducing various types of neurons using miR-9/9*-124 together with different sets of transcriptional factors.
METHODS SUMMARY
Transduction of human fibroblasts
A synthetic cluster of miR-9/9* and miR-124 validated previously to express miR-9* and miR-124 (ref. 5) was inserted downstream of tRFP in the pLemiR lentiviral construct carrying a puromycin selection cassette (Open Biosystems) driven by either a CMV promoter or a doxycycline-responsive promoter. A non-silencing sequence, which produces miRNA-NS, was used as a control (Open Biosystems). Each transcription factor was cloned downstream of the EF1 α promoter in a separate lentiviral construct. Typically, infected human fibroblasts were maintained in fibroblast media for 3–4 days before selection with appropriate antibiotics in Neuronal Media (ScienCell) supplemented with VPA (1 mM) and basic FGF (20 ng ml−1). dbcAMP (500 µM) was added 10 days later to enhance cell survival. Human BDNF and NT3 (10 ng ml−1; Peprotech) were added to the media after 3–4 weeks. Media were changed every 4 days.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
METHODS
Plasmid construction and viral preparation
We have previously constructed a synthetic cluster that expresses the precursors of both miR-9/9* (NCBI and miRBASE accession numbers MIMAT0000441 and MIMAT0000442) and miR-124 (accession number MIMAT0000422) and validated its ability to generate mature miRNAs of both. Here we cloned this cluster downstream of a tRFP marker into pLemiR (Open Biosystems), driven by either a CMV promoter or a doxycycline-responsive promoter. A non-silencing sequence in pLemiR (miR-NS, which produces non-specific miRNA) was used as a control (Open Biosystems). cDNA of each neural transcription factor used in this study was cloned downstream of the EF1α promoter in a separate lentiviral construct with blasticidin or neomycin selection. For doxycycline experiments, human fibroblasts were first infected with a lentiviral construct expressing rtTA under the EF1α promoter and stably selected with hygromycin. Infectious lentiviruses were collected 36–60 h after transfection of Lenti-X 293T cells (Clontech) with appropriate amounts of lentiviral vectors, psPAX2 and pMD2.G (Addgene) using Fugene HD (Roche).
Cell culture
All fibroblast cultures (human neonatal foreskin fibroblasts (ATCC, PCS-201-010) and adult dermal fibroblast (ScienCell)) were maintained in fibroblast media (Dulbecco’s Modified Eagle Medium; Invitrogen) containing 10% fetal bovine serum (FBS; Omega Scientific), β-mercaptoethanol (Sigma-Aldrich), non-essential amino acids, sodium pyruvate, GlutaMAX, and penicillin/streptomycin (all from Invitrogen). The day before lentiviral infection, human fibroblasts were seeded onto gelatin-coated 24-well tissue culture dishes (MidSci). Next day, cells were infected with filtered viral supernatants in the presence of polybrene (8 µg ml−1) overnight. Fresh media were then replaced for 2–3 days with appropriate antibiotics to select for infected cells. Four days after infection, the media was changed to Neuronal Media (ScienCell) supplemented with VPA (1 mM), basic FGF (20 ng ml−1) with media changes every 4 days. Our experiments to optimize the conversion protocol indicated that VPA and bFGF are beneficial for conversion for the first 2–3 weeks. We also added dbcAMP (0.5 mM, Sigma) after 2 weeks to the media as we found it enhanced cell survival. Human BDNF and NT3 (10 ng ml−1, Peprotech) were added to the media after 3–4 weeks to promote the survival of the induced cells. To facilitate immunostaining and electrophysiological studies in some experiments, cells were trypsinized (0.05% trypsin, Invitrogen) at about 10 days after infection and re-plated onto poly-ornithine (Sigma-Aldrich)/laminin (Roche)/fibronectin (Sigma-Aldrich)-coated glass coverslips.
Immunofluorescence
The following antibodies were used for the immunofluorescence studies: mouse anti-MAP2 (Sigma-Aldrich, 1:750), chicken anti-MAP2 (Abcam, 1:30,000), mouse anti-β-III tubulin (Covance, 1:30,000), rabbit anti-VGLUT1 (Synaptic Systems, 1:2,000), rabbit anti-SCN1a (Abcam, 1:1,000), rabbit anti-NMDAR1 (Abcam, 1:2,000), rabbit anti-neurofilament 200 (Sigma-Aldrich, 1:2,000), mouse anti-SV2 (Developmental Studies Hybridoma Bank, 1:100), rabbit anti-GABA (Sigma, 1:2,000) and rabbit anti-synapsin1 (Cell Signaling, 1:200). Antibodies against BAF subunits were generated in our laboratory and used as the following concentrations: BAF45b (1:250), BAF45c (1:1,000) and BAF53b (1:500). The secondary antibodies were goat anti-rabbit or mouse IgG conjugated with Alexa-488, -568 or -647 (Invitrogen). For SCN1a and BAF53b staining, biotinylated secondary antibodies were detected using TSA amplification kit (Invitrogen). EdU incorporation assay was performed according to the manufacturer’s protocols (Invitrogen). Images were captured using a Leica DM5000B microscope with Leica Application Suite (LAS) Advanced Fluorescence 1.8.0 and a Leica DMI4000B microscope with LAS v.2.8.1.
Electrophysiology
Recordings were performed on fibroblasts 5–8 weeks after infection for both miR-9/9*-124-NEUROD2 and miR-9/9*-124-DAM converted cells, which were co-cultured with mouse glia. Data were acquired in whole-cell mode at room temperature (25 °C) using an Axopatch 200B amplifier (Molecular Devices) or EPC 10 amplifier (HEKA) and sampled at 5 kHz with a 2 kHz low-pass filter. Recording pipette resistance was 2–6 MΩ. Intrinsic neuronal properties were studied using the following solutions (in mM): extracellular, 140 NaCl, 2.5 KCl, 2.5 CaCl2, 2 MgCl2, 1 NaH2PO4, 20 glucose, 10 HEPES, pH 7.4; intracellular, 120 KGluc, 20 KCl, 4 NaCl, 4 Mg2ATP, 0.3 NaGTP, 10 Na2PCr, 0.5 EGTA, 10 HEPES, pH 7.25. Synaptic activity was measured using the same extracellular solution, supplemented with 50 µM APV, and the following intracellular solution (in mM): 135 CsMeS, 5 CsCl, 10 HEPES, 0.5 EGTA, 1 MgCl2 4Mg2ATP, 0.4 NaGTP, 5 QX-314, pH 7.4 CsOH. Correspondingly, EPSCs were measured at −70 mV whereas IPCSs were measured at either 0 or +30 mV(with 10 µM NBQX and 50 µM APV, ECl− approximately −79 mV). Evoked postsynaptic currents were elicited by a stimulating electrode (CBAEC75, FHC) positioned 100–150 µm aside from the cell soma through which brief (1 ms) unipolar current pulses of various amplitudes (0.1–0.9 mA, Δ = 0.05–0.1 mA) were applied. Recordings were filtered at 2 kHz and digitally sampled at 10 kHz. Data were collected and initially analysed with Clampfit 10 or the Patchmaster software (HEKA). Further analysis was performed using IgorPRO and MS Excel. Series resistance was left uncompensated owing to the fragility of the cells, but was corrected in the current clamp calculations. The liquid junction potential was calculated to be 15 mV (Clampfit) and corrected in calculating resting membrane potentials according to published methods27.
Calcium imaging
Cells were loaded with Fluo2-AM (5 µM, TEFLABS) in Tyrode solution (150 mM NaCl, 4 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM glucose, 10 mM HEPES, 310–315 mOsm, with pH at 7.35) for 30 min in a 37 °C incubator. After two washes with Tyrode, cells were imaged using a filter cube (excitation 470 ± 20 nm and emission 535 ± 50 nm). In some cases, 1 µM TTX or 200 µM CdCl2 was superfused. All images were converted to TIFF files and analysed off-line with Metamorph or ImageJ. All error bars represent s.e.m. For analysis of FM-positive puncta, 1.3 µm in diameter regions of interest were used to cover functional boutons. Photobleaching was corrected by fitting the pre-stimulation baseline to a linear curve.
FM1-43 imaging
Cells were superfused with Tyrode solution. Switching of super-fusion solution was carried out with a precision of <2 s. Solutions contained 10 µM NBQX and 50 µM d-APV (Tocris Bioscience) to prevent possible recurrent activity and synaptic plasticity. All experiments were performed at room temperature and neurons were stimulated with platinum electrodes. Putative presynaptic boutons were stained with 8 µM FM1-43 (Molecular Probes) using field stimulation for 120 s at 10 Hz, followed by 60 s without stimulation to maximize the loading. In some experiments, 0.1 mM CaCl2 was used to test the calcium dependency. After 10 min of washing with dye-free Tyrode’s solution, individual boutons were destained by field stimulation. FM1-43 dyes were excited at 470 nm (D470-40x; Chroma) and their emission was collected at 535 nm (535/50m). tRFP was excited at 535 nm (535/50ex) and its emission was collected at 580 nm (580 lp). All images were taken at a frame rate of 1–3 Hz by a Cascade 512B camera.
Single-cell qPCR
Single cells were collected by clone FACS sorting using a BD influx sorter (BD Biosciences) in 10 µl of a pre-amplification mix containing 40 nM of all primers for genes of interest, and the following components of the CellsDirect One-Step qRT–PCR Kit (Invitrogen): 2× Reaction Mix, SuperScript III RT/Platinum Taq Mix. After sorting, samples were reverse transcribed and pre-amplified for 18 cycles. Pre-amplified samples were diluted (3×) with TE buffer and stored at −20 °C. Sample and assay (primer pairs) preparation for 96.96 Fluidigm Dynamic arrays was done according to the manufacturer’s recommendation. Briefly, sample was mixed with 20× DNA binding dye sample loading reagent (Fluidigm), 20× EvaGreen (Biotium) and TaqMan Gene Expression Master Mix (Applied Biosystems). Assays were mixed with 2× assay loading reagent (Fluidigm) and TE to a final concentration of 5 µM. The 96.96 Fluidigm Dynamic Arrays (Fluidigm) were primed and loaded on an IFC Controller HX (Fluidigm) and qPCR experiments were run on a Biomark System for Genetic Anaylsis (Fluidigm). Data were collected and analysed using the Fluidigm Real-Time PCR Analysis software (v.2.1.3 and v.3.0.2). Melting curves were used to determine specificity of each reaction. Further data analysis was performed using Microsoft Excel. In addition to collected single-cell material, every experiment contained samples for four standard dilutions of a mixed human cDNA library. The collected cells were confirmed based on RSG18 (18S small ribosomal subunit) and GAPDH co-expression. Of these, induced neurons were identified by co-expression of two general neuronal genes MAPT and TUBB3 for further analysis of genes specific to brain regions and cell types.
Supplementary Material
Acknowledgements
We thank I. Graef and A. Cho for helpful suggestions and reagents, A. Kuo and W. Ho for technical help, and X. Bao and P. Khavari for their generous gift of reagents. A.S.Y. is a fellow of the Helen Hay Whitney Foundation. A.X.S. is funded by the Agency of Science, Technology and Research of Singapore (A*STAR). L.L. is supported by the Stanford Medical Scientist Training Program, National Institutes of Mental Health (NIMH) F30MH093125, and the Frances B. Nelson predoctoral fellowship. A.S. is supported by the CIRM post-doctoral fellowship. T.P. is supported by a Swiss National Science Foundation SNSF fellowship for advanced researchers (PA00P3_134196). R.E.D. is supported by the NIH Director’s Award, and awards from the Simon’s Foundation and the CIRM. R.E.D. is also grateful for funding from B. and F. Horowitz, M. McCafferey, B. and J. Packard, P. Kwan and K. Wang. R.W.T. is supported by grants from the Simons, Mathers and Burnett Family Foundations. This work was supported by grants from the Howard Hughes Medical Institute (G.R.C.) and the NIH (HD55391, AI060037 and NS046789 to G.R.C., and NS24067, GM58234 and MH064070 to R.W.T.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions A.S.Y., A.X.S., and G.R.C. generated the hypotheses and designed experiments. A.S.Y. and A.X.S. performed experiments, generated data in all figures and Supplementary Data. A.S. and L.L. designed and performed experiments for Figs 1, 2 and 4 and Supplementary Data. T.P. designed and performed experiments in Fig. 3a. Y.L. generated data presented in Fig. 1. C.L.-M. performed experiments for Supplementary Data. A.S.Y., A.X.S., L.L., A.S., Y.L., T.P., R.W.T., R.E.D. and G.R.C. wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Hansen DV, Lui JH, Parker PR, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 2.Hansen DV, Rubenstein JL, Kriegstein AR. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron. 2011;70:645–660. doi: 10.1016/j.neuron.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu J, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56:94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 5.Yoo AS, Staahl BT, Chen L, Crabtree GR. MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature. 2009;460:642–646. doi: 10.1038/nature08139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136:200–206. doi: 10.1016/j.cell.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nature Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiorano NA, Mallamaci A. Promotion of embryonic cortico-cerebral neuronogenesis by miR-124. Neural Develop. 2009;4:40. doi: 10.1186/1749-8104-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang X, et al. A simple array platform for microRNA analysis and its application in mouse tissues. RNA. 2007;13:1803–1822. doi: 10.1261/rna.498607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin CH, et al. The dosage of the neuroD2 transcription factor regulates amygdala development and emotional learning. Proc. Natl Acad. Sci. USA. 2005;102:14877–14882. doi: 10.1073/pnas.0506785102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormick MB, et al. neuroD2 and neuroD3: distinct expression patterns and transcriptional activation potentials within the neuroD gene family. Mol. Cell. Biol. 1996;16:5792–5800. doi: 10.1128/mcb.16.10.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson JM, et al. NeuroD2 is necessary for development and survival of central nervous system neurons. Dev. Biol. 2001;234:174–187. doi: 10.1006/dbio.2001.0245. [DOI] [PubMed] [Google Scholar]
- 17.Ince-Dunn G, et al. Regulation of thalamocortical patterning and synaptic maturation by NeuroD2. Neuron. 2006;49:683–695. doi: 10.1016/j.neuron.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 18.Ryan TA, et al. The kinetics of synaptic vesicle recycling measured at single presynaptic boutons. Neuron. 1993;11:713–724. doi: 10.1016/0896-6273(93)90081-2. [DOI] [PubMed] [Google Scholar]
- 19.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parrish JZ, Kim MD, Jan LY, Jan YN. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006;20:820–835. doi: 10.1101/gad.1391006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl Acad. Sci. USA. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signaling and by regulating polycomb function. Nature Cell Biol. doi: 10.1038/ncb2285. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr. Opin. Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laneve P, et al. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 2010;38:6895–6905. doi: 10.1093/nar/gkq604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andres ME, et al. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl Acad. Sci. USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barry PH. JP Calc, a software package for calculating liquid junction potential corrections in patch-clamp, intracellular, epithelial and bilayer measurements and for correcting junction potential measurements. J. Neurosci. Methods. 1994;51:107–116. doi: 10.1016/0165-0270(94)90031-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.