Abstract
To assess the level of intra-patient diversity and evolution of HIV-1C non-structural genes in primary infection, viral quasispecies obtained by single genome amplification (SGA) at multiple sampling timepoints up to 500 days post-seroconversion (p/s) were analyzed. The mean intra-patient diversity was 0.11% (95% CI; 0.02 to 0.20) for vif, 0.23% (95% CI; 0.08 to 0.38) for vpr, 0.35% (95% CI; −0.05 to 0.75) for vpu, 0.18% (95% CI; 0.01 to 0.35) for tat exon 1 and 0.30% (95% CI; 0.02 to 0.58) for rev exon 1 during the time period 0 to 90 days p/s. The intra-patient diversity increased gradually in all non-structural genes over the first year of HIV-1 infection, which was evident from the vif mean intra-patient diversity of 0.46% (95% CI; 0.28 to 0.64), vpr 0.44% (95% CI; 0.24 to 0.64), vpu 0.84% (95% CI; 0.55 to 1.13), tat exon 1 0.35% (95% CI; 0.14 to 0.56 ) and rev exon 1 0.42% (95% CI; 0.18 to 0.66) during the time period of 181 to 500 days p/s. There was a statistically significant increase in viral diversity for vif (p = 0.013) and vpu (p = 0.002). No associations between levels of viral diversity within the non-structural genes and HIV-1 RNA load during primary infection were found. The study details the dynamics of the non-structural viral genes during the early stages of HIV-1C infection.
Introduction
The majority of HIV infections in the world, specifically those in sub-Saharan Africa, are attributable to subtype C cementing its status as a major public health concern. Insights into the primary phase of HIV-1C infection are important to better understand disease pathogenesis and early mechanisms of virus-host interactions affecting disease progression. A recent study showed that mutational patterns during the first 5 weeks of infection in a subtype B cohort indicated rapid viral population growth and included a slight decrease in viral genetic diversity over the first 20 to 40 days [1]. Numerous studies have linked viral diversity in HIV-1 structural genes with disease progression [2]–[8]. However, little is known about the diversity and evolution of non-structural genes, which are crucial for virus replication particularly during the early stages of the virus life cycle.
To our best knowledge no published reports to date have pertained to diversity and/or divergence of viral non-structural genes during primary HIV-1 subtype C infection. Salazar-Gonzales et al. showed that the transmitted/founder full length sequences from 12 subjects (9 infected with HIV subtype B and 3 subtype C) contained intact non-structural genes [9]. Michael et al. reported defective accessory genes (HXB2 sequence positions 4,961 to 6,346) in longitudinal HIV-1 subtype B infection [10]. A study on evolution of HIV-1 subtype B Tat and Rev demonstrated that viral mutations were identified within the predicted CTL epitopes suggesting that CTL-mediated selection plays an important role in viral escape from immune pressure and early viral evolution [11]. A longitudinal study on twins infected with HIV-1 subtype B perinatally showed discordant disease progression rates with a dramatic increase in tat gene sequence diversity in the sicker child over time [12].
Associations between long term non-progression and viral mutations (including deletions) in HIV-1 non-structural genes have been reported previously [13]–[19]. Yedavalli et al. found that HIV-1 Vif and Vpr were less conserved in their functional domains in a cohort of HIV-1 non-transmitting mothers [20].
A systematic approach to assess the evolution of HIV-1 non-structural genes during primary infection is critical to better understand the role these genes play in HIV-1 pathogenesis and their potential association with early disease progression. This seems particularly important for HIV-1C where a significant fraction of early infections have shown extended periods of high viral RNA load and rapid progression to loss of CD4 lymphocyte numbers and disease progression [21], [22]. This knowledge could inform HIV vaccine design as non-structural viral genes are potentially attractive vaccine candidates. The increased levels of viral replication in HIV-infected individuals rapidly progressing to AIDS can result in greater virus diversity. Although HIV-1 non-structural genes are thought to be relatively conserved, the extent of conservation has been understudied. In this study we assess viral diversity and evolution of five major non-structural HIV-1 genes vif, vpr, vpu, tat exon 1 and rev exon 1 during the primary phase of HIV-1 subtype C infection up to 500 days post-seroconversion (p/s). We addressed the following questions: (1) What are the phylogenetic relationships of HIV-1 nonstructural genes during primary HIV-1 infection? (2) What is the level of intra-patient diversity during primary infection? (3) How does viral diversity in non-structural HIV-1 genes change over time? and (4) Is HIV-1 RNA load in plasma associated with diversity of HIV-1 non-structural genes during primary HIV-1 infection?
Methods
Ethics Statement
This study was conducted according to the principles expressed in the Declaration of Helsinki, and was approved by the Human Research Development Committee of the Botswana Ministry of Health and by the Office of Human Research Administration at the Harvard School of Public Health. All study participants provided written informed consent for the collection of samples and subsequent analysis.
Study Subjects
Study subjects were enrolled in the primary HIV-1C infection cohort in Botswana and followed-up during 2004–2010 [23]. A subset of 20 adults (eight acutely infected individuals and 12 randomly selected seroconverters) included 5 males and 15 females (Table S1). Age of subjects at enrollment ranged from 20 to 53 years. Acutely infected subjects (patient code A through H) were identified before seroconversion within Fiebig stage II [24] by a negative HIV-1 serology combined with a positive HIV-1 RT-PCR test. Twelve seroconverters (two-digit patient code OC to QR) were identified within the early stage of HIV-1 infection and included 3 subjects within Fiebig stage IV, 4 subjects within Fiebig stage V, 2 subjects on the edge of Fiebig stage V and VI, and 3 subjects in Fiebig stage VI. The time of seroconversion (time 0) was estimated as the midpoint between the last seronegative test and the first seropositive test (approximately one week in most cases) for the acutely infected subjects and by mid-point of the corresponding Fiebig stage for the recently infected subjects [23]. Written informed consent was obtained from all study participants; ethical approval for this research was obtained from the Human Research Development Committee of the Botswana Ministry of Health and OHRA at Harvard School of Public Health.
Viral RNA Extraction and cDNA Synthesis
Viral RNA was isolated from plasma by QIAamp viral RNA Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. For viral loads >35,000 copies/ml viral RNA was isolated from 140 µl of plasma, for samples with viral loads <35,000 copies per ml, a volume of plasma containing 5,000 viral RNA copies was spun down at 24,000×g at 4 degrees for 1 hour, the supernatant was removed, the pellet was re-suspended in 140 µl of supernatant for RNA extraction. For samples with viral loads <5,000 copies per ml, the entire aliquot (1.5 mL) was spun down and the pellet was re-suspended in 140 µl of supernatant followed by RNA extraction. Viral RNA was recovered from spin columns in an elution volume of 60 µl. Reverse transcription to single-stranded cDNA was performed using SuperScript III (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions and primer OFM19 (5′ –GCA CTC AAG GCA AGC TTT GAG GCT TA - 3′; HXB2 coordinates 9632 to 9604).
Single Genome Amplification (SGA) and Sequencing
The single genome amplification was based on the method of limiting dilutions [25], and was used with minor modifications. A median (IQR) of 2.5 (2 to 4) timepoints were sequenced for each patient, and a median of 11.5 (10.4 to 13.5) sequences were generated per patient. The median (IQR) time range of sampling post-seroconversion was 174 (55–349) days. Briefly, the cDNA produced was diluted in 96 well plates with the aim to yield <30% positive reactions of PCR-amplified product. The targeted region spanned from HXB2 nt position 5,041 to 6,310, and included HIV-1 sequence encoding overlapping non-structural viral genes vif (nt positions 5,041 to 5,619; HXB2 numbering), vpr (nt positions 5,559 to 5,850), tat exon1 (nt positions 5,831 to 6,045), rev exon 1 (nt positions 5,970 to 6,045) and vpu (nt positions 6,062 to 6,310). The diluted cDNA corresponding to about 30% of positive PCR was used as a template for two rounds of nested PCR. The first round PCR was conducted using primers Vif1bw- 5′ – GGG TTT ATT ACA GAG ACA GCA GAG- 3′ (HXB2 coordinates 4900 to 4923) and OFM19, and second-round PCR primers Fvif- 5′ - AGA CCC TAT TTG GAA AGG ACC AGC - 3′ (HXB2 coordinates 4922 to 4945) and Rvpu- 5′-CTT CTT TCC ACA CAG GTA CCC CAT- 3′ (HXB2 coordinates 6366 to 6343). The amplicons were sequenced on both strands. Sequencing primers included Rvpu, Fvif, 5198L- 5′ - TCC AGG GCT CTA GGT TAG - 3′, 4679L- 5′ - GCC CAG GGT CTA CTT GTG - 3′; an additional primer 1466L- 5′ - TCA TTG CCA CTG TCT TCT GCT CTT - 3′ was used to increase coverage in cases primer 5198L failed. Amplicons were Exo-SAP purified [26], and sequenced directly using BigDye technology on the ABI 3730 DNA Analyzer. The sequence fragments were assembled and edited using SeqScape v2.6 (Applied Biosystems). The sequences generated were tested using HYPERMUT v 2.0 [27], and all hypermutated sequences were excluded from the analysis. A total of 6 out of 667 (0.88%) sequences were found to be hypermutated.
During the preliminary analysis of viral quasispecies a link was identified between sequences of subjects OI and OK. We ruled out a potential contamination by (1) similar clustering patterns observed in structural genes, env and gag, of the same subjects (env/gag unpublished data from same cohort); (2) similar branching topology of viral sequences obtained at multiple timepoints of sampling; (3) nucleic acid isolation, PCR amplification and sequencing were separated by place and time; and (4) proper quality controls were used in each experiment.
HIV-1 Subtyping
Nucleotide sequences were codon aligned using the MUSCLE algorithm implemented in Mega 5 [28] followed by manual adjustment in Bioedit [29]. For HIV-1 subtyping, three sequences per patient were randomly selected from the pool of generated viral quasispecies. The selection criteria included earliest timepoint available and sequence length with more than 90% coverage of the targeted region (5,041 to 6,310 nt position in HXB2). To determine phylogenetic relationships and clustering patterns of generated viral sequences, the phylogenetic tree reconstruction was performed using Neighbor joining, Maximum likelihood, and Bayesian methods. A standard set of HIV-1 subtype references from LANL was included in the analysis. The sequence CPZ.CM.98.CAM3.AF115393 was used as an outgroup. The online Rega-2 subtyping tool was used in parallel for confirmation [30].
Intra-patient Diversity Analysis
The intra-patient mean pairwise distances were measured in Mega 5 using the Maximum Composite Likelihood (MCL) model, following nucleotide sequences alignment using the MUSCLE algorithm implemented in Mega 5 [28] and manual adjustment in Bioedit [29]. Mean values were calculated at each sampling timepoint and the averages of these were used in each time “bin” (0–90, 91–180, 181–500 days p/s). The viral diversity for each non-structural gene was computed as a mean value with 95% Confidence Interval (CI). HIV-1 RNA load in plasma was measured routinely at each study visit over the first year of follow-up. The study schedule included weekly visits for the first two months, bi-weekly for the next two months, and monthly for the next eight months for acutely infected individuals, or monthly for individuals identified within Fiebig stage IV-V. After the first year, quarterly study visits were scheduled for all subjects. Details of HIV-1 RNA load measurement have been presented elsewhere [21], [22], [31]. Mean values for the analyzed time intervals, 0–90 days p/s, 91–180 days p/s, and 181–500 days p/s, and for 100–300 days p/s as described previously, were also included [21]. Potential associations between intra-patient viral diversity and HIV-1 RNA load at the three time intervals, 0–90 days p/s, 91–180 days p/s, and 181–500 days p/s, were assessed.
Statistical Methods
Sigma plot version 11 was used for descriptive statistics to summarize medians, means, and standard deviations. Comparisons between groups were made using t-test and Mann Whitney Rank sum test for continuous and binary outcomes respectively. All reported p-values are 2-sided. Regression analysis was performed using linear regression and the Spearman Rank Test.
GenBank Accession Numbers
Sequences have been assigned GenBank accession numbers JQ895561–JQ896230.
Results
Inferred Phylogeny of Non-structural Viral Sequences
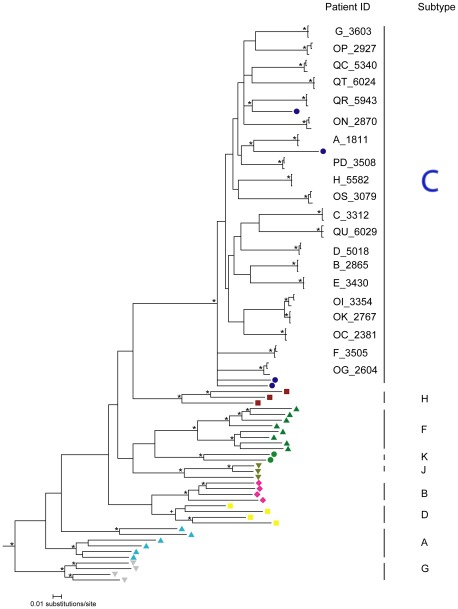
All generated sequences in this study clustered with HIV-1 subtype C reference sequences (Fig. 1). The subject-specific splits were supported by branching topology and clade credibility values of ≥0.99 in the Bayesian analysis. One epidemiologically linked pair, subjects OI and OK, was identified; contamination of samples from subjects OI/OK pair was ruled out.
Figure 1. HIV-1 subtyping by analysis of phylogenetic relationships of HIV-1 non-structural genes.
The analyzed region of HIV-1 genome corresponded to nucleotide positions 5,041 to 6,310 in HXB2. Three sequences were randomly selected for each study subject (see Methods). A phylogenetic tree was inferred by Mr. Bayes using GTR model. The convergence was reached after 10 M MCMC run. The consensus tree was visualized in Figtree v.1.3.1 [54]. Clade credibility values of >0.95 shown by asterisk, Subtype D cluster showed the support of 0.93 indicated by + symbol. HIV-1 subtype C reference sequences are shown as blue circles. All non-subtype C group M reference sequences are shown at the bottom of the phylogenetic tree. SIV sequence (CPZ.CM98.CAM3.AF115393) was used as an outgroup.
Good congruence and similar branching topology was observed between the Bayesian (Fig. 1), maximum likelihood (ML; Fig. S1), and neighbor joining (NJ; Fig. S2) tree reconstruction methods. The reliability of the branching topology and clustering patterns was supported by both the approximate likelihood ratio test (aLRT) for ML tree of ≥0.99 and the bootstrap (1,000 replicates) estimates of ≥99% in the NJ analysis. The observed splits support and phylogenetic relationships suggest that the HIV-1 non-structural gene region comprised of vif, vpr, vpu, tat exon 1 and rev exon 1 can be used reliably for HIV-1 subtyping. The HIV-1 subtyping results were congruent with results obtained using the Rega-2 HIV-1 subtyping tool (data not shown).
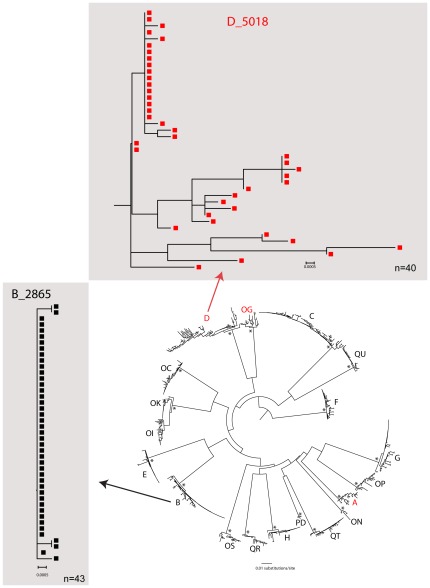
To address whether phylogeny of non-structural genes is consistent with multiplicity of HIV-1 transmission, the branching topology of generated viral quasispecies at early timepoints (0–90 days p/s) were analyzed and compared with branching patterns with HIV-1C structural genes, env and gag [32]. The inferred phylogeny in 3 (15%) subjects included in this study supported transmission of multiple viral variants, and was consistent with our previous findings in env/gag analysis (Fig. 2). For example, more diversified branching topology was observed in subject D (env/gag: transmission of multiple viral variants) compared to subject B (env/gag: transmission of single viral variant).
Figure 2. HIV-1 subtype C phylogenetic relationship and diversity of HIV non-structural genes is consistent with the multiplicity of HIV-1 infection determined by analysis of the env/gag genes.
A maximum likelihood phylogenetic tree was reconstructed using Fastree2 (Price et al., 2010) using the GTR+G model for nucleotide substitution and visualized in Figtree v.1.1.3 [54]. Alternative likelihood ratio tests [55] >0.95 are shown by an asterisk. Subjects infected with multiple viral variants are colored red. Patient B and D subtrees (individual trees on grey background) show branching topology of earliest sampling (0–90 days p/s) and represent examples of single (subject B) and multiple (subject D) HIV-1 transmission.
Intra-patient Diversity and Diversification of HIV-1C Non-structural Genes During Primary Infection
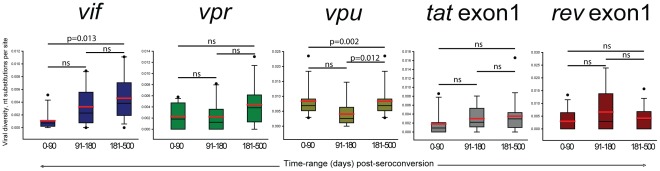
To assess the level of intra-patient diversity of HIV-1C non-structural genes in primary infection, we used viral quasispecies obtained by SGA at multiple sampling timepoints from seroconversion up to 500 days p/s. The mean intra-patient diversity was 0.11% (95% CI; 0.02 to 0.20) for vif, 0.23% (95% CI; 0.08 to 0.38) for vpr, 0.35% (95% CI; −0.05 to 0.75) for vpu, 0.18% (95% CI; 0.01 to 0.35) for tat exon 1 and 0.30% (95% CI; 0.02 to 0.58) for rev exon 1 during the time period 0 to 90 days p/s. The intra-patient diversity increased gradually in all non-structural genes (Fig. 3), which was evident from the vif mean intra-patient diversity of 0.46% (95% CI; 0.28 to 0.64), vpr 0.44% (95% CI; 0.24 to 0.64), vpu 0.84% (95% CI; 0.55 to 1.13), tat exon 1 0.35% (95% CI; 0.14 to 0.56) and 0.42% (95% CI; 0.18 to 0.66) for rev exon 1 during the time period of 181 to 500 days p/s. There was a significant increase in viral diversity calculated using the Mann-Whitney Rank Sum Test for vif (p = 0.013) and vpu (p = 0.002).
Figure 3. Individual distribution of pairwise distances for each of the non-stuctural genes,vif (HXB2 start 5041 to 5619), vpr (HXB2 start 5559 to 5850), vpu (HXB2 start 6062 to 6310), tat exon 1 (HXB2 start 5831 to 6045), and rev exon 1(HXB2 start 5970 to 6045).
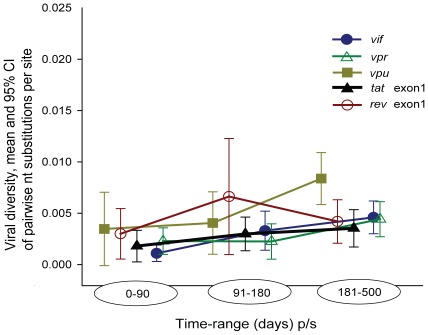
The intra-patient viral diversity of HIV-1C non-structural genes suggested a high level of conservation during primary infection (Fig. 4). A tight range of viral diversity was observed for each non-structural gene analyzed, at the earliest timepoint (0–90 days p/s): vif (0%–0.51%), vpr (0%–0.58%), vpu (0%–1.98%), tat exon 1 (0%–0.86%) and rev exon 1 (0%–1.33%). At later timepoints, over the first year of HIV1-C infection, an increase was also observed, the range of viral diversity was tight: vif (0%–1.11%), vpr (0%–1.30%), vpu (0.28%–2.35%), tat exon 1 (0%–1.66%) and rev exon 1 (0%–1.57%). To assess the change in diversity over time, slopes of changes were analyzed. The gradual increase observed by positive slopes of viral diversity (vif, 0.40%; vpr; 0.25%; vpu, 0.61%; tat exon1, 0.19%; and rev exon1, 0.05%) is consistent with ongoing viral evolution.
Figure 4. HIV-1C diversity, mean and 95% confidence intervals for non-structural genes vif, vpr, vpu, tat exon 1 and rev exon 1 over the first 500 days p/s.
Viral diversity for each subject was calculated using maximum composite likelihood model [56].
HIV-1 RNA Load and Diversity within HIV-1C Non-structural Genes
The potential association between intra-patient non-structural gene diversity and levels of viral RNA load during primary HIV-1C infection was examined. It was assumed that higher levels of viral replication during the early stages of infection could drive increased viral diversity at later timepoints. The levels of intra-patient diversity within vpr and HIV-1C RNA load at 0–90 days p/s showed a weak but statistically significant correlation (r2 = 0.587, p = 0.042). The intra-patient diversity within tat exon 1 and rev exon 1 at later timepoints (181–500 days p/s) was associated with HIV-1 RNA load at the baseline timepoint, r2 = 0.505, p = 0.044; and r2 = 0.577, p = 0.019, respectively. There was association between tat exon 1 diversity and HIV-1 RNA load at 181–500 days p/s, r2 = 0.516, p = 0.039. Thus we found only sporadic associations between levels of HIV-1 RNA load and intra-patient viral diversity within non-structural genes. This can be attributed to the relatively short region of analyzed viral genes, fewer timepoints analyzed, uneven number of generated quasispecies, or missing the region encoding the second exons for Tat and Rev.
The potential difference in intra-patient diversity within HIV-1C non-structural genes between extended high viremics [21] and other patients during primary HIV-1C infection were also examined. Levels of intra-patient diversity between these two groups of subjects were compared at three time intervals, 0–90, 91–180, and 181–500 days p/s. We hypothesized a higher level of viral diversity within the HIV-1C non-structural genes in extended high viremics; however, no significant differences between groups were found (Fig. S3).
Discussion
This study performed a comprehensive analysis of the molecular evolution of HIV-1C non-structural genes during primary infection from seroconversion up to 500 days p/s. To our best knowledge, this is the first report on the extent and dynamics of viral diversity within HIV-1C non-structural genes during primary HIV-1C infection. Also recent studies suggest that HIV-1C infections may show unusually high HIV RNA levels for prolonged periods following initial infection.
A limited number of studies have addressed viral quasispecies in HIV-1C infection [33]–[35], although none has focused on the evolution of HIV-1C non-structural genes. While the diversity of HIV-1C non-structural genes has been addressed in previous cross-sectional studies [36]–[38], these studies were not powered to evaluate the extent of viral diversification over time. The use of the SGA method in this study allowed the assessment of longitudinal diversity and diversification of non-structural genes during primary HIV-1C infection.
Intra-patient nucleotide diversity increased over time in all non-structural genes studied, which is consistent with previously published data on env gp120 in primary HIV-1B [8] and HIV-1C [23] infection. The tight range observed in vif (0%–0.51%), vpr (0%–0.58%), vpu (0–1.98%), tat1 (0%–0.86%) and rev1 (0%–1.33%) at the earliest timepoint which increased slightly up to 500 days p/s vif (0%–1.11%),vpr (0%–1.30%),vpu (0.28%–2.35%), tat exon1 (0%–1.66%) and rev exon1 (0%–1.57%) highlights the high level of viral conservation in HIV-1 non-structural genes. Viral diversification in non-structural genes is lower than in structural HIV-1 genes, which is evident from comparison of slopes with the slope in HIV-1 Benv reported at 1% per year [8]. Previous studies have assessed whether viral mutations occur within CTL epitopes and/or flanking regions; however, most previous studies analyzed primarily structural viral genes, and evolution of CTL epitopes within accessory genes remains understudied [1], [39]–[44]. Therefore, it remains unclear to what extent selection pressure drives evolution of the HIV-1 non-structural genes. Although data from cross-sectional HIV-1C studies have shown a high level of conservation in major non-structural genes [36]–[38].The lower level of non-structural genes diversity during the primary infection is consistent with known viral homogeneity during this period [23], [45]–[48].
We found that the HIV-1 genome region encoding non-structural viral genes can be used reliably for HIV-1 subtyping, which is consistent with previous studies [36], [49]–[51]. The sporadic associations between non-structural HIV-1C gene diversity and viral RNA load in plasma is consistent with previous studies [4], [52], [53]. The viral diversity in HIV-1C non-structural genes was consistent with multiplicity of viral transmission and results in the env/gag analysis [32]. The analyzed timepoints allowed us to detect transmission of more than a single viral variant. However, we were not able to deduce the number of transmitted viral variants due to potential recombinations between transmitted variants, if samples were collected later than Fiebig stage IV.
The relatively small sample size and different number of generated viral quasispecies per timepoint and/or per subject are the major limitations in this study. Ideally, it would have been preferable to obtain at least 20 sequences for each patient at each timepoint. However, this task was challenging and difficult to achieve due to inter-subject heterogeneity and viral RNA as a template for amplification. An additional limitation of the study is the overlapping nature of the proteins in the region amplified, making it difficult to assign specific mutational events to specific proteins. Also, the second exon of tat and rev were not studied.
The study provides insights into the dynamics of the non-structural HIV-1C genes during the early stages of infection. The results of our study highlight differential diversity across HIV-1 genes and slower diversification of viral accessory genes over time. Apparently, the most likely reason is different selection pressure imposed by host immune response to the encoded viral gene products which may result in different evolutionary rates. Our results suggest (1) a high level of conservation of viral non-structural genes during primary HIV-1C infection; (2) a gradual increase in viral diversity of these genes over the first 500 days p/s; and (3) no associations between levels of viral diversity within the non-structural genes and HIV-1 RNA load during primary infection, which might be due to a relatively small sample size. To conclusively evaluate potential associations, larger and more in-depth studies might be warranted. This could yield valuable information to aid vaccine development, due to the increasing interest of non-structural genes as targets in vaccine design.
Supporting Information
HIV-1 subtyping. Phylogenetic relationship between HIV-1 non-structural genes. A phylogenetic tree was constructed using PhyML [49] using the GTR+I+G model for nucleotide substitution and visualized in Mega 5 [27]. Three sequences were used for each patient Subjects’ branches are labeled on the right with patient codes. aLRT >0.99 shown by asterisk. HIV-1 subtype C reference sequences are shown in blue, and all other HIV-1 group M (non-C) reference sequences are labeled at the bottom of the figure. SIV sequence (CPZ CM98.CAM3.AF115393) was used to root the tree.
(TIF)
HIV-1 subtyping. Phylogenetic relationship between HIV-1 non-structural genes. A phylogenetic tree was constructed from nucleotide alignments using Neighbor Joining (NJ) method. The evolutionary distances were computed using the Kimura 2-parameter method. The reliability of the branching topology was estimated from 1000 bootstrap replicates. Patient identifiers are shown to the right of the tree. Bootstrap values >99% are indicated by asterisks. SIV sequence (CPZ CM98 CAM3 AF115393) was used to root the tree.
(TIF)
Viremics Analysis; Kimura 2-parameter overall mean pairwise diversity of HIV-1C non-structural genes vif, vpr, vpu, tat exon1 and rev exon1 comparing two groups (high viremics – individuals with mean HIV-1 RNA load >100,000 copies/ml during the period 100–300 days p/s, and other subjects).
(TIF)
Subject demographics, time points of sampling, HIV-1 RNA load, CD4 count, and Fiebig stage at enrollment.
(DOCX)
Acknowledgments
We thank and are grateful to the participants of the Tshedimoso study in Botswana. Without their participation, this research would not have been possible. We thank the Botswana Ministry of Health, Gaborone City Council clinics, and the Gaborone VCT Tebelopele for collaboration. We are grateful to Gaseboloke Mothowaeng, Florence Modise, S’khatele Molefhabangwe, and Sarah Masole for their dedication and outstanding work in the clinic and outreach. We express thanks to Lauren Margolin, Jeannie Baca, Mary Fran McLane, Simani Gaseitsiwe, Carl Davis, Raylton Chikwati, Max Dougherty, Sandy Bolm, Lauren Buck and Iain J. MacLeod for excellent technical assistance. We thank Lauren Buck and Iain J. MacLeod for review of this manuscript. We acknowledge Lendsey Melton for excellent editorial and administrative assistance. We thank the Botswana Harvard AIDS Institute Partnership (Gaborone, Botswana) and Essex lab (Harvard School of Public Health, Boston, MA) where the laboratory and data analysis was performed.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The primary HIV-1 subtype C infection study in Botswana, the Tshedimoso study, was supported by NIH grant R01 AI057027. This work was supported in part by a Research Grant from the Office of Research and Development (ORD), University of Botswana (TKS). This work was also supported in part by the AAMC FIC (Association of American Medical Colleges Fogarty International Centre)/Ellison Overseas Fellowships in Global Health and Clinical Research (RR) and by the NIH grant D43 TW000004 (RR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Herbeck JT, Rolland M, Liu Y, McLaughlin S, McNevin J, et al. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J Virol. 2011;85:7523–7534. doi: 10.1128/JVI.02697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barroso H, Taveira N. Evidence for negative selective pressure in HIV-2 evolution in vivo. Infect Genet Evol. 2005;5:239–246. doi: 10.1016/j.meegid.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Iwami S, Nakaoka S, Takeuchi Y. Viral diversity limits immune diversity in asymptomatic phase of HIV infection. Theor Popul Biol. 2008;73:332–341. doi: 10.1016/j.tpb.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Mani I, Gilbert P, Sankale JL, Eisen G, Mboup S, et al. Intrapatient diversity and its correlation with viral setpoint in human immunodeficiency virus type 1 CRF02_A/G-IbNG infection. J Virol. 2002;76:10745–10755. doi: 10.1128/JVI.76.21.10745-10755.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller V. Commentary: Sifting through the maze of viral and host diversity and HIV/AIDS clinical progression. Int J Epidemiol. 2005;34:584–585. doi: 10.1093/ije/dyi090. [DOI] [PubMed] [Google Scholar]
- 6.Nowak MA, Anderson RM, McLean AR, Wolfs TF, Goudsmit J, et al. Antigenic diversity thresholds and the development of AIDS. Science. 1991;254:963–969. doi: 10.1126/science.1683006. [DOI] [PubMed] [Google Scholar]
- 7.Nowak MA, May RM. Mathematical biology of HIV infections: antigenic variation and diversity threshold. Math Biosci. 1991;106:1–21. doi: 10.1016/0025-5564(91)90037-j. [DOI] [PubMed] [Google Scholar]
- 8.Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michael NL, Chang G, d’Arcy LA, Ehrenberg PK, Mariani R, et al. Defective accessory genes in a human immunodeficiency virus type 1-infected long-term survivor lacking recoverable virus. J Virol. 1995;69:4228–4236. doi: 10.1128/jvi.69.7.4228-4236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guillon C, Stankovic K, Ataman-Onal Y, Biron F, Verrier B. Evidence for CTL-mediated selection of Tat and Rev mutants after the onset of the asymptomatic period during HIV type 1 infection. AIDS Res Hum Retroviruses. 2006;22:1283–1292. doi: 10.1089/aid.2006.22.1283. [DOI] [PubMed] [Google Scholar]
- 12.Hutto C, Zhou Y, He J, Geffin R, Hill M, et al. Longitudinal studies of viral sequence, viral phenotype, and immunologic parameters of human immunodeficiency virus type 1 infection in perinatally infected twins with discordant disease courses. J Virol. 1996;70:3589–3598. doi: 10.1128/jvi.70.6.3589-3598.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caly L, Saksena NK, Piller SC, Jans DA. Impaired nuclear import and viral incorporation of Vpr derived from a HIV long-term non-progressor. Retrovirology. 2008;5:67. doi: 10.1186/1742-4690-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang LJ, Zhang C. Infection and replication of Tat- human immunodeficiency viruses: genetic analyses of LTR and tat mutations in primary and long-term human lymphoid cells. Virology. 1995;211:157–169. doi: 10.1006/viro.1995.1388. [DOI] [PubMed] [Google Scholar]
- 15.Iversen AK, Shpaer EG, Rodrigo AG, Hirsch MS, Walker BD, et al. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J Virol. 1995;69:5743–5753. doi: 10.1128/jvi.69.9.5743-5753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lum JJ, Cohen OJ, Nie Z, Weaver JG, Gomez TS, et al. Vpr R77Q is associated with long-term nonprogressive HIV infection and impaired induction of apoptosis. J Clin Invest. 2003;111:1547–1554. doi: 10.1172/JCI16233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mologni D, Citterio P, Menzaghi B, Zanone Poma B, Riva C, et al. Vpr and HIV-1 disease progression: R77Q mutation is associated with long-term control of HIV-1 infection in different groups of patients. AIDS. 2006;20:567–574. doi: 10.1097/01.aids.0000210611.60459.0e. [DOI] [PubMed] [Google Scholar]
- 18.Papathanasopoulos MA, Patience T, Meyers TM, McCutchan FE, Morris L. Full-length genome characterization of HIV type 1 subtype C isolates from two slow-progressing perinatally infected siblings in South Africa. AIDS Res Hum Retroviruses. 2003;19:1033–1037. doi: 10.1089/088922203322588396. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Mikhail M, Dyer WB, Zaunders JJ, Kelleher AD, et al. First demonstration of a lack of viral sequence evolution in a nonprogressor, defining replication-incompetent HIV-1 infection. Virology. 2003;312:135–150. doi: 10.1016/s0042-6822(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 20.Yedavalli VR, Ahmad N. Low conservation of functional domains of HIV type 1 vif and vpr genes in infected mothers correlates with lack of vertical transmission. AIDS Res Hum Retroviruses. 2001;17:911–923. doi: 10.1089/088922201750290032. [DOI] [PubMed] [Google Scholar]
- 21.Novitsky V, Ndung’u T, Wang R, Bussmann H, Chonco F, et al. Extended high viremics: a substantial fraction of individuals maintain high plasma viral RNA levels after acute HIV-1 subtype C infection. AIDS. 2011;25:1515–1522. doi: 10.1097/QAD.0b013e3283471eb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novitsky V, Wang R, Bussmann H, Lockman S, Baum M, et al. HIV-1 subtype C-infected individuals maintaining high viral load as potential targets for the “test-and-treat” approach to reduce HIV transmission. PLoS One. 2010;5:e10148. doi: 10.1371/journal.pone.0010148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novitsky V, Lagakos S, Herzig M, Bonney C, Kebaabetswe L, et al. Evolution of proviral gp120 over the first year of HIV-1 subtype C infection. Virology. 2009;383:47–59. doi: 10.1016/j.virol.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- 25.Liu SL, Rodrigo AG, Shankarappa R, Learn GH, Hsu L, et al. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- 26.Dugan KA, Lawrence HS, Hares DR, Fisher CL, Budowle B. An improved method for post-PCR purification for mtDNA sequence analysis. J Forensic Sci. 2002;47:811–818. [PubMed] [Google Scholar]
- 27.Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G –> A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 30.REGA HIV-1 & 2 Automated Subtyping Tool (Version 2.0). 1 Available: http://jose.med.kuleuven.be/genotypetool/html/subtypinghiv.html. Accessed 2011 Nov. [Google Scholar]
- 31.Novitsky V, Woldegabriel E, Kebaabetswe L, Rossenkhan R, Mlotshwa B, et al. Viral load and CD4+ T-cell dynamics in primary HIV-1 subtype C infection. J Acquir Immune Defic Syndr. 2009;50:65–76. doi: 10.1097/QAI.0b013e3181900141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novitsky V, Wang R, Margolin L, Baca J, Rossenkhan R, et al. Transmission of single and multiple viral variants in primary HIV-1 subtype C infection. PLoS One. 2011;6:e16714. doi: 10.1371/journal.pone.0016714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rong L, Gilchrist MA, Feng Z, Perelson AS. Modeling within-host HIV-1 dynamics and the evolution of drug resistance: trade-offs between viral enzyme function and drug susceptibility. J Theor Biol. 2007;247:804–818. doi: 10.1016/j.jtbi.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rousseau CM, Daniels MG, Carlson JM, Kadie C, Crawford H, et al. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J Virol. 2008;82:6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell CM, Connell BJ, Capovilla A, Venter WD, Stevens WS, et al. Molecular characterization of the HIV type 1 subtype C accessory genes vif, vpr, and vpu. AIDS Res Hum Retroviruses. 2007;23:322–330. doi: 10.1089/aid.2006.0181. [DOI] [PubMed] [Google Scholar]
- 37.Romani B, Glashoff R, Engelbrecht S. Molecular and phylogenetic analysis of HIV type 1 vpr sequences of South African strains. AIDS Res Hum Retroviruses. 2009;25:357–362. doi: 10.1089/aid.2008.0251. [DOI] [PubMed] [Google Scholar]
- 38.Scriba TJ, Treurnicht FK, Zeier M, Engelbrecht S, van Rensburg EJ. Characterization and phylogenetic analysis of South African HIV-1 subtype C accessory genes. AIDS Res Hum Retroviruses. 2001;17:775–781. doi: 10.1089/088922201750237059. [DOI] [PubMed] [Google Scholar]
- 39.Altfeld M, Addo MM, Eldridge RL, Yu XG, Thomas S, et al. Vpr is preferentially targeted by CTL during HIV-1 infection. J Immunol. 2001;167:2743–2752. doi: 10.4049/jimmunol.167.5.2743. [DOI] [PubMed] [Google Scholar]
- 40.Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, et al. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim GJ, Lee HS, Hong KJ, Kim SS. Dynamic correlation between CTL response and viral load in primary human immunodeficiency virus-1 infected Koreans. Virol J. 2010;7:239. doi: 10.1186/1743-422X-7-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura T, Brumme ZL, Brockman MA, Rosato P, Sela J, et al. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J Virol. 2010;84:7581–7591. doi: 10.1128/JVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Novitsky V, Rybak N, McLane MF, Gilbert P, Chigwedere P, et al. Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific elispot-based cytotoxic T-lymphocyte responses for AIDS vaccine design. J Virol. 2001;75:9210–9228. doi: 10.1128/JVI.75.19.9210-9228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wood N, Bhattacharya T, Keele BF, Giorgi E, Liu M, et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delwart E, Magierowska M, Royz M, Foley B, Peddada L, et al. Homogeneous quasispecies in 16 out of 17 individuals during very early HIV-1 primary infection. AIDS. 2002;16:189–195. doi: 10.1097/00002030-200201250-00007. [DOI] [PubMed] [Google Scholar]
- 46.Gottlieb GS, Heath L, Nickle DC, Wong KG, Leach SE, et al. HIV-1 variation before seroconversion in men who have sex with men: analysis of acute/early HIV infection in the multicenter AIDS cohort study. J Infect Dis. 2008;197:1011–1015. doi: 10.1086/529206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Verhofstede C, Demecheleer E, De Cabooter N, Gaillard P, Mwanyumba F, et al. Diversity of the human immunodeficiency virus type 1 (HIV-1) env sequence after vertical transmission in mother-child pairs infected with HIV-1 subtype A. J Virol. 2003;77:3050–3057. doi: 10.1128/JVI.77.5.3050-3057.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu T, Mo H, Wang N, Nam DS, Cao Y, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 49.Bibollet-Ruche F, Loussert-Ajaka I, Simon F, Mboup S, Ngole EM, et al. Genetic characterization of accessory genes from human immunodeficiency virus type 1 group O strains. AIDS Res Hum Retroviruses. 1998;14:951–961. doi: 10.1089/aid.1998.14.951. [DOI] [PubMed] [Google Scholar]
- 50.Jacobs GB, Nistal M, Laten A, van Rensburg EJ, Rethwilm A, et al. Molecular analysis of HIV type 1 vif sequences from Cape Town, South Africa. AIDS Res Hum Retroviruses. 2008;24:991–994. doi: 10.1089/aid.2008.0077. [DOI] [PubMed] [Google Scholar]
- 51.Wieland U, Seelhoff A, Hofmann A, Kuhn JE, Eggers HJ, et al. Diversity of the vif gene of human immunodeficiency virus type 1 in Uganda. J Gen Virol 78 (Pt. 1997;2):393–400. doi: 10.1099/0022-1317-78-2-393. [DOI] [PubMed] [Google Scholar]
- 52.Bello G, Casado C, Garcia S, Rodriguez C, del Romero J, et al. Plasma RNA viral load is not associated with intrapatient quasispecies heterogeneity in HIV-1 infection. Arch Virol. 2004;149:1761–1771. doi: 10.1007/s00705-004-0322-y. [DOI] [PubMed] [Google Scholar]
- 53.Chamberland A, Sylla M, Boulassel MR, Baril JG, Cote P, et al. Effect of antiretroviral therapy on HIV-1 genetic evolution during acute infection. Int J STD AIDS. 2011;22:146–150. doi: 10.1258/ijsa.2010.010292. [DOI] [PubMed] [Google Scholar]
- 54.Rambaut A. 2008. FigTree v1.1.2. http://tree.bio.ed.ac.uk/software/figtree.
- 55.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 56.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HIV-1 subtyping. Phylogenetic relationship between HIV-1 non-structural genes. A phylogenetic tree was constructed using PhyML [49] using the GTR+I+G model for nucleotide substitution and visualized in Mega 5 [27]. Three sequences were used for each patient Subjects’ branches are labeled on the right with patient codes. aLRT >0.99 shown by asterisk. HIV-1 subtype C reference sequences are shown in blue, and all other HIV-1 group M (non-C) reference sequences are labeled at the bottom of the figure. SIV sequence (CPZ CM98.CAM3.AF115393) was used to root the tree.
(TIF)
HIV-1 subtyping. Phylogenetic relationship between HIV-1 non-structural genes. A phylogenetic tree was constructed from nucleotide alignments using Neighbor Joining (NJ) method. The evolutionary distances were computed using the Kimura 2-parameter method. The reliability of the branching topology was estimated from 1000 bootstrap replicates. Patient identifiers are shown to the right of the tree. Bootstrap values >99% are indicated by asterisks. SIV sequence (CPZ CM98 CAM3 AF115393) was used to root the tree.
(TIF)
Viremics Analysis; Kimura 2-parameter overall mean pairwise diversity of HIV-1C non-structural genes vif, vpr, vpu, tat exon1 and rev exon1 comparing two groups (high viremics – individuals with mean HIV-1 RNA load >100,000 copies/ml during the period 100–300 days p/s, and other subjects).
(TIF)
Subject demographics, time points of sampling, HIV-1 RNA load, CD4 count, and Fiebig stage at enrollment.
(DOCX)