Abstract
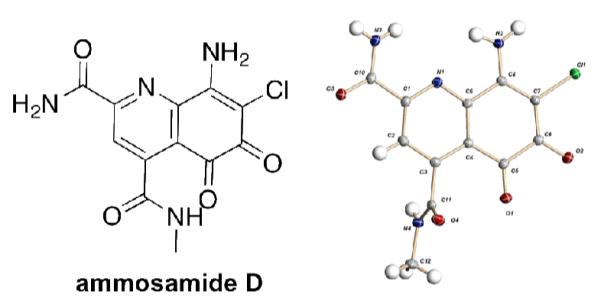
Ammosamide D (1), an oxidized analog of the ammosamide family, was isolated from a marine-derived Streptomyces variabilis. Pyrroloquinoline containing alkaloids are a growing class of natural products, with 1 being the first example of an oxidized analog resulting in a 5,6-dioxo-5,6-dihydroquinoline ring system. Attempts at chemical conversion of ammosamide B to ammosamide D revealed that a strong chemical oxidant is required. Ammosamide D has modest cytotoxicity to the MIA PaCa-2 pancreatic cancer cell line.
Cytotoxicity based screening for natural products has been the most succssful strategy for the discovery of bioactive natural products with new modes of action.1 In 2008 the Fenical laboratory reported the isolation, characterization and molecular target of the cytotoxic agents ammosamides A and B (2 – 3).2 These heteroaromatic alkaloids contain an unusual pyrroloquinoline moiety, which most likely derives from modification of tryptophan. Biological studies demonstrated that these compounds exert their cytotoxicity via covalent modification of myosin.3 The interesting biology and structural features of the ammosamides have led to considerable interest from the synthetic community.4 Recently, biosynthetic studies on the pyrrolquinoline containing natural product lymphostin (4) indicate that the pyrroloquinoline carbon skeleton is derived from tryptophan and further modified in an assembly line fashion.5 This study also showed that 4 and related analogs are potent inhibitors of mTOR. There are now a growing number of natural products with the pyrrolo[4,3,2-de]quinoline core from multiple organisms, including mycenarubin A (5) from a mushroom.6
In our continuing efforts to search for natural products from marine bacteria with selective cytotoxicity against cancer cell lines, we screened a library of 1500 natural products fractions against a panel of cancer cell lines from multiple tissue types.7 From this screen we obtained a series of fractions from a marine-derived Streptomyces variabilis (strain SNA-020)8 that exhibited modest selectivity and potency for the MIA PaCa-2 pancreatic cancer cell line. Analysis of the active fractions by LC–UV-MS showed the presence of chlorinated compounds with a UV-Vis profile with absorptions at λmax = 550, 420, 340, 280 and 235 nm. Based on this UV profile and MS, we could discern the presence of 2 and 3 in the active fractions. 2 and 3 exhibit activity against the colon tumor cell line HCT-1162, but when tested against MIA PaCa-2 cells, there was no cytotoxicity < 20 μM, suggesting the presence of additional active metabolites in the fraction. Further analysis revealed additional chlorine bearing molecules with complex UV profiles similar to 2 and 3, leading to the isolation of ammosamide D (1), which has modest cytotoxicity against MIA PaCa-2 (IC50 = 3.2 μM).
1 Was isolated as an orange solid. The positive ion HRESIMS at m/z 309.0224 [M+H]+, corresponds to a molecular formula of C12H9ClN4O3. 1H NMR in DMSO-d6 exhibited five singlets: δH 9.35, 9.17, 8.55, 8.02, 8.00, one quartet proton at δH 8.18 (J = 4.7 Hz) and one methyl doublet at δH 2.77 (J = 4.7 Hz) (Table 1). Addition of a 30 μL of D2O to the NMR sample lead to the disappearance of all signals in the 1H NMR with the exception of the singlet at δH 8.00 ppm and the methyl doublet, suggesting there are five exchangeable protons. The 13C NMR revealed the presence of 11 sp2 carbons and a sp3 carbon. The above data for 1, along with the UV spectrum, suggested an ammosamide like structure.
Table 1.
1H and 13C NMR of 1 in DMSO-d6
| no. | δH, m (J Hz) | δ C | COSY | HMBC |
|---|---|---|---|---|
| 1 | 8.18 q (4.7) | H1a | C1a,C2 | |
| 1a | 2.77 d (4.7) | 25.9 | H1 | C2 |
| 2 | - | 166.3 | ||
| 2a | - | 146.8 | ||
| 3 | 8.00 s | 122.7 | C2,C2a,C4a,C5b | |
| 4 | - | 152.0 | ||
| 4a | - | 163.7 | ||
| 5a | - | 146.7 | ||
| 5b | - | 125.4 | ||
| 6 | - | 151.7 | ||
| 7 | - | 106.9 | ||
| 8 | - | 168.9 | ||
| 8a | - | 178.3 | ||
| CONH2-a | 9.17 s; | - | CONH2-b | C4a |
| CONH2-b | 8.02 s | CONH2-a | C4 | |
| NH2(6)-a | 9.35 s; | NH2(6)-b | C5a,C7,C8 | |
| NH2(6)-b | 8.55 s | NH2(6)-a | C5a,C5b,C6,C7 |
Analysis of the 2D data provided only a few correlations for assignment of the structure. A COSY correlation between the exchangeable 1H at δ 8.18 and the methyl doublet at δH 2.77 was suggestive of an N-methyl amide, a deviation from the previously reported ammosamides. Additionally, we observed four carbons shifted downfield of 160 ppm, whereas 2 and 3 only have two carbons downfield of 160 ppm (Table S1). Although the data suggested that 1 had the basic carbon framework as 3, there were clearly some differences. Further examination of the HMBC revealed correlations from the aromatic proton H3 at δH 8.00 to C2 at δC 166.3 and C4a at δC δc 163.7, which established the locations of two amide carbons, C4a and C2.
Due to the lack of NMR correlations, we turn our attention towards obtaining an X-ray crystal structure. Following a similar procedure as that of ammosamide A9 we were able to obtain small crystals. The X-ray assignment of 1 revealed that the pyrrole ring had been cleaved at C8a to give an N-methylamide and a ketone at C8a (Figure 2). The presence of carbonyls at C8 and C8a are consistent with the two additional downfield signals in the 13C NMR. Additionally, the crystal structure explained the presence of only five exchangeable protons. Thus, 1 contains a 5,6-dioxo-5,6-dihydroquinoline ring system.
Figure 2.
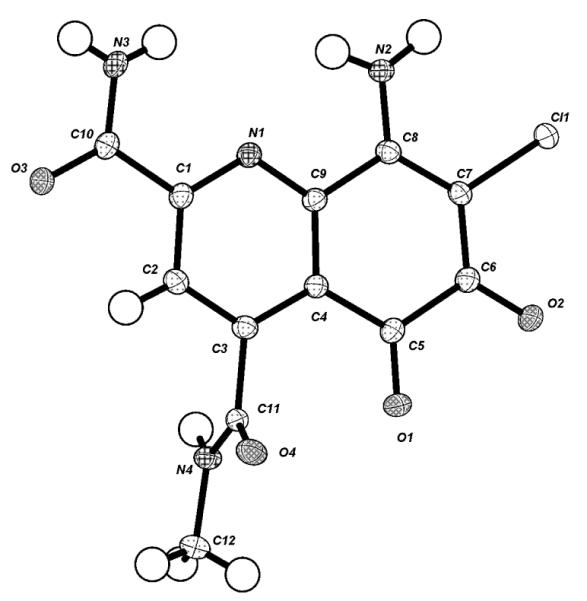
X-ray crystal structure of 1.
There is a significant change in the bond lengths of the ortho- quinone portion of 1 upon lose of aromaticity. The bond length from C-5b to C-8a has increased by 0.12 Å to 1.48 Å while C-5b to C-8a has increased by 0.14 Å to 1.53 Å. The 5,6-dioxo-5,6-dihydroquinoline ring system explains the dramatic hypsochromic UV shift from 520 nm in 3 to 475 nm in 1. Although we would anticipate an even much shorter λ for the non-aromatic ring system of 1. DFT calculations of the UV spectra of 1 and 3 reasonably predicted the λmax for both molecules (3 λcalculated 580 nm; 1 λcalculated 500 nm).10 This suggests that the pyridine-2,4-dicarboxyamide moiety, which exists in both structures plays a key determinant in the long wavelength UV transition.
It is clear from the previous isolation studies on 2 and 3 that these compounds are susceptible to oxidation. In particular, it was shown that the thiolactam moiety of 2 could be converted to the lactam upon storage or rapidly via treatment with H2O2 in MeOH. We envision that 1 results from oxidation of 3 to give the peroxy intermediate. This is mechanistically similar to the oxidation of flavin to flavin hydroperoxide.11 There are a number of pathways that by which the peroxy species could lead to the oxidatively ring opened product. One possiblity is elimination to form the acyliminium ion, which could then react with H2O to form the aminal and open to iminoquinone 6 (Figure 3). With this proposed mechanism in mind, there is a probability that 1 is an artifact of the isolation, via reaction with O2. To test this possibility we subjected 3 to a series of oxidation conditions to look for conversion to 1 or other ring open compounds (Figure 4).
Figure 3.
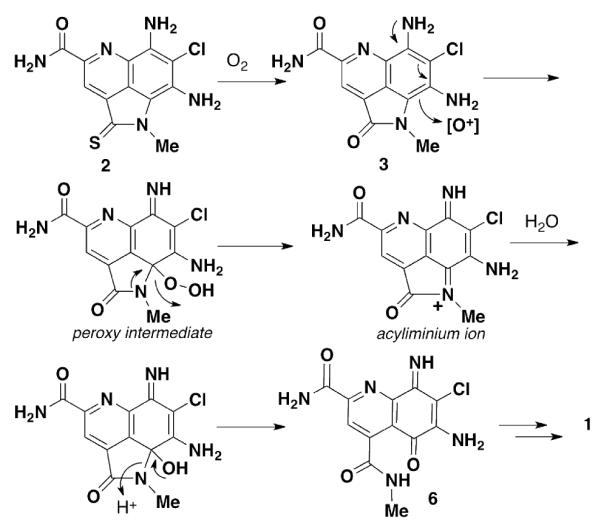
Proposed biosynthetic pathway from 3 to 1.
Figure 4.
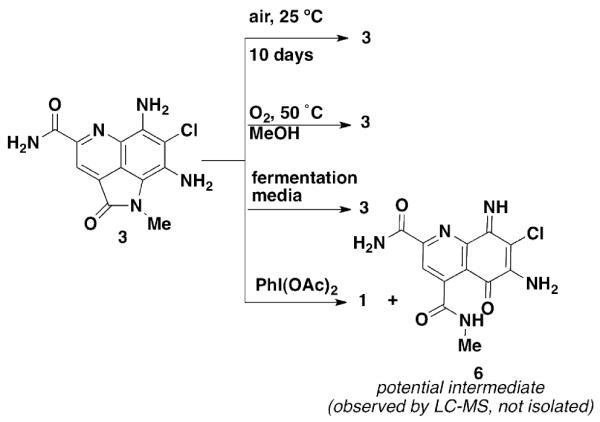
Attempts at conversion of 3 to 1 Table 1.
As a control, 3 was dissolved in 1:1 CH3OH/H2O and allowed to stand at 25 °C for 7 days exposed to air. These conditions were meant to mimic the solvent/air exposure that compounds receive during isolation conditions. Other than a small amount of decomposition, we observed only starting material. Under more forcing conditions, we placed 3 under an atmosphere of O2 in aq. methanol and heated to 50 °C. Under these conditions we still failed to see conversion of 3 to 1. In fact we found 3 to be incredibly stable under these conditions and only saw a small amount of decomposition. A third option we looked into was the possibility that 3 could be converted to 1 in the fermentation media. As the slightly basic (pH 8.0) fermentation media contains trace metals and μM concentrations of Fe, we thought these might play a role in mediating the oxidative ring opening. After addition of 50 μL of a 10 mM DMSO stock solution of 3 to the fermentation media and shaking for 7 days (the same time as for fermentation) we saw no conversion to 1.
We decided to undertake more forcing conditions and looked at using chemical oxidants such as PhI(OAc)2 and AgOAc that are frequently used to generate quinones.12,13 Treatment of 3 with PhI(OAc)2 in DMF at room temperature gave a 3:1 mixture of products, with the predominant product observed being 1 (LC-MS trace Figure S1) and a compound with MS data consistent with iminoquinone 6 or the C8 imine tautomer. Upon purification by reversed phase C18 HPLC, the only product observed is 1 in an overall 46% yield. It is not surprising that under the purification conditions potential intermediate 6 be converted to 1 by addition of H2O. As it required chemical intervention to convert 3 to 1, we propose that the opening of the pyrrole ring is not likely a simple artifact of the isolation, but is an enzyme catalyzed, biosynthetic product.
Based on previous reports of biological activity for ammosamides A and B (cytotoxicity) and lymphostin (mTOR inhibitor), we probed the activity of 1 in a variety of biological assays. Cytotoxicity measurements against the pancreatic cancer cell line MIA PaCa-2 revealed only modest cytotoxicity for 1, with an IC50 of 3.2 μM. 1 Was not found to be an mTOR inhibitor (tested up to 20 μM).
Supplementary Material
Figure 1.
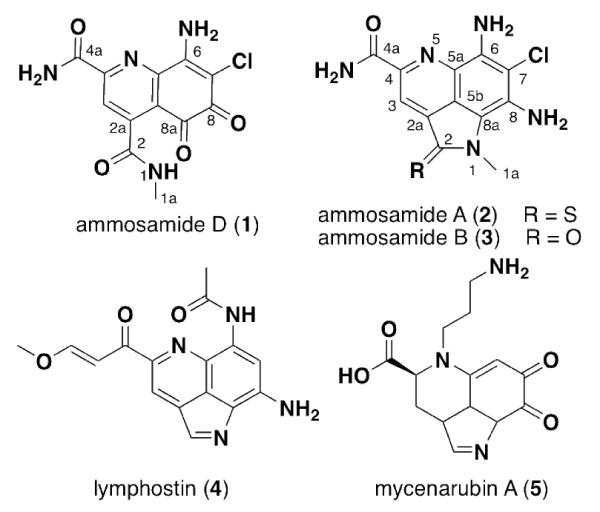
Structures of new compound 1 and other pyrrolo[4,3,2-de]quinoline natural products
Acknowledgment
The authors thank Bruce Posner and Shuguang Wei (University of Texas Southwestern Medical Center, Biochemistry) for cytotoxicity screening. We acknowledge the following grants for funding this project: NIH 5PO1CA095471 and the Welch Foundation I-1689. JBM is a Chilton/Bell Foundation Endowed Scholar.
Footnotes
Supporting Information Available General procedures, bioassay protocols, chemical derivatization, data tables, NMR spectra and X-ray crystal data.
References
- 1.Shoemaker RH. Nat. Rev. Cancer. 2006;6:813. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 2.Hughes CC, MacMillan JB, Gaudencio SP, Jensen PR, Fenical W. Angew. Chem. Int. Ed. 2009;48:725. doi: 10.1002/anie.200804890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes CC, MacMillan JB, Gaudencio SP, Fenical W, LaClair JJ. Angew. Chem. Int. Ed. 2009;48:728. doi: 10.1002/anie.200804107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Hughes CC, Fenical W. J. Am. Chem. Soc. 2010;132:2528. doi: 10.1021/ja9106572. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Reddy PVN, Banerjee B, Cushman M. Org. Lett. 2010;12:3112. doi: 10.1021/ol101215x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.For isolation see Nagata H, Ochiai K, Aotani Y, Ando K, Yoshida M, Takahashi I, Tamaoki TJ. Antibiot. 1997;50:537. doi: 10.7164/antibiotics.50.537. Aotani Y, Nagata H, Yoshida MJ. Antibiot. 1997;50:543. doi: 10.7164/antibiotics.50.543. For biosynthesis see: Miyanaga A, Janso JE, McDonald L, He M, Liu H, Barbieri L, Eustáquio AS, Fielding EN, Carter GT, Jensen PR, Feng X, Leighton M, Koehn FE, Moore BS. J. Am. Chem. Soc. 2011;133:13311. doi: 10.1021/ja205655w.
- 6.Peters S, Spiteller P. Eur. J. Org. Chem. 2007;1571 [Google Scholar]
- 7.Hu Y, Espindola AP, Stewart N. Biorg. Med. Chem. 2011;19:5183. doi: 10.1016/j.bmc.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bacterial strain SNA-020 was isolated from a sediment sample collected at Sweetings Cay, Bahamas (N 26° 33′27″, W 77° 51′15″) using a starch based isolation media. The phylogeny was established using 16S rRNA analysis with the universal bacterial primers F27 and R1492. The strain showed 99.9% identity to Streptomyces variabilis. The 16S gene sequence is deposited in the NCBI as JQ815387).
- 9.1 was dissolved in methanol and water in a 1 dram vial and allowed to stand for 30 days, after which small crystals were obtained.
- 10.The theoretical UV λmax was calculated using DFT calculations at the B3LYP/6-31+G level in the GAUSSIAN 09 software package.
- 11.a) Kemal C, Bruce TC. Proc. Nat. Acad. Sci. U.S.A. 1976;73:995. doi: 10.1073/pnas.73.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mager HIX, Berends W. Tetrahedron. 1976;32:2303. [Google Scholar]
- 12.Nicolaou KC, Sugita K, Baran PS, Zhong Y-L. Angew. Chem. Int. Ed. 2001;40:207. doi: 10.1002/1521-3773(20010105)40:1<207::AID-ANIE207>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 13.Adams R, Nagarkatti AS. J. Am. Chem. Soc. 1950;72:4601. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


