Among 350 households of patients with Staphylococcus aureus skin infections, extra-nasal S. aureus colonization was common. USA300 MRSA appeared more transmissible among household members than other S. aureus strain types. Multiple S. aureus genetic backgrounds were present in many households.
Abstract
Background. The USA300 methicillin resistant Staphylococcus aureus (MRSA) genetic background has rapidly emerged as the predominant cause of community-associated S. aureus infections in the U.S. However, epidemiologic characteristics of S. aureus household transmission are poorly understood.
Methods. We performed a cross-sectional study of adults and children with S. aureus skin infections and their household contacts in Los Angeles and Chicago. Subjects were surveyed for S. aureus colonization of the nares, oropharynx, and inguinal region and risk factors for S. aureus disease. All isolates underwent genetic typing.
Results. We enrolled 1162 persons (350 index patients and 812 household members). The most common infection isolate characteristic was ST8/SCCmec IV, PVL+ MRSA (USA300) (53%). S. aureus colonized 40% (137/350) of index patients and 50% (405/812) of household contacts. A nares-only survey would have missed 48% of S. aureus and 51% of MRSA colonized persons. Sixty-five percent of households had >1 S. aureus genetic background identified and 26% of MRSA isolates in household contacts were discordant with the index patients' infecting MRSA strain type. Factors independently associated (P < .05) with the index strain type colonizing household contacts were recent skin infection, recent cephalexin use, and USA300 genetic background.
Conclusions. In our study population, USA300 MRSA appeared more transmissible among household members compared with other S. aureus genetic backgrounds. Strain distribution was complex; >1 S. aureus genetic background was present in many households. S. aureus decolonization strategies may need to address extra-nasal colonization and the consequences of eradicating S. aureus genetic backgrounds infrequently associated with infection.
The emergence of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections in the late 1990s [1] has resulted in a dramatic shift in the epidemiology of S. aureus infections. In the United States, the predominant CA-MRSA clone, USA300 MRSA, has become the most common cause of community-associated skin infection [2] and an endemic pathogen in many hospitals [3–5]. Data suggest that CA-MRSA infections have high attack rates in household contacts after an initial CA-MRSA infection occurs [6, 7].
In contrast to healthcare-associated (HA)–MRSA strain types, which have circulated in the healthcare setting for more than 40 years and rarely spread outside the hospital [8–10], there is evidence that CA-MRSA strain types frequently spread from person to person in households [11–14]. Although transmission of HA-MRSA may occur in part via asymptomatic carriers [8], less is known about MRSA and methicillin-susceptible S. aureus (MSSA) dissemination in community settings. Previous investigations of MRSA spread in households have been limited by relatively small sample size [15–21], lack of geographic diversity [16, 19–22], focus on HA-MRSA [23, 24], nares-only surveillance [17, 19, 20, 22, 25, 26], or lack of distinction among S. aureus genetic backgrounds at the molecular level [23].
Recent investigations suggest nares-only screening may underestimate S. aureus colonization prevalence because S. aureus has been found to colonize oropharyngeal [27–29] and inguinal areas [30–32] in persons irrespective of nasal colonization. Additionally, although USA300 MRSA is the most common genetic background causing CA–S. aureus skin infection [2], MRSA nasal colonization remains uncommon in the general population (<5%) [33].
To better understand the spread of USA300 MRSA and other S. aureus strain types in households, we studied S. aureus colonization in patients with skin infections and among their household members in 2 large US cities.
METHODS
We performed a cross-sectional investigation of children and adults with S. aureus skin infection and their household members. Patients were enrolled from Harbor-UCLA Medical Center in Torrance, California, and the University of Chicago Medical Center in Chicago, Illinois, from August 2008 to June 2010. Each center’s clinical microbiology laboratory was screened daily for new skin cultures growing S. aureus. Both inpatients and outpatients were eligible. S. aureus was identified by standard techniques (Vitek 2, bioMérieux). Patients were eligible for participation if they (1) had the culture taken from a skin infection, (2) were willing to provide informed consent, (3) had >1 household member who would participate, and (4) resided within 25 miles of the site’s medical center. Patients who lived in a group living facility or were homeless were ineligible. Infected patients were designated as “index patients.” This study was approved by each site’s institutional review board.
Home Visit
Consenting patients agreed to have a home visit within 21 days of enrollment during which all participating household members or their parent or guardian provided informed consent. Research personnel administered a standardized questionnaire on MRSA risk factors based on previously developed surveys of known or hypothesized CA-MRSA and MRSA risk factors [8, 13, 30, 34–43]. Survey questions for this study were refined using cognitive interviewing [44].
To assess S. aureus colonization, research personnel obtained separate cultures from the nares and oropharynx from subjects using a dry rayon-tip applicator (CultureSwab, BD Diagnostic Systems). Inguinal cultures were obtained by the subject or their parent/guardian in private after being provided detailed instructions.
Cultures for Colonization
After collection, swabs were transported promptly to the site’s research laboratory and enriched in trypticase soy broth with 7% sodium chloride overnight at 37°C. The culture broth was plated onto BBL CHROMagar S. aureus media (BD Diagnostic Systems) and incubated for 24 hours at 37°C. Isolates were confirmed as S. aureus by positive catalase and StaphAureux tests (Remel).
Molecular Characterization of Isolates and Definition of Isolate Relatedness
Speciation of all S. aureus infection and colonization isolates was confirmed at the University of Chicago MRSA Research Laboratory. Genomic DNA was extracted from each isolate using the Qiagen DNeasy Blood and Tissue Kit following manufacturer’s instructions and modified by incubation with lysostaphin in resuspension buffer (at 37°C for 30 minutes) to facilitate S. aureus lysis [45]. Staphylococcus aureus speciation was confirmed using a polymerase chain reaction (PCR) assay specific for spa (encoding Protein A). Staphylococcus aureus isolates were characterized by multilocus sequence typing (MLST) [46] to determine the genetic background and by typing of the SCCmec element, the mobile genetic element that carries mecA [47]. SCCmec typing was performed by PCR as described [48], with type assignments using published guidelines [47]. Detection of genes encoding the Panton-Valentine leukocidin (PVL) was performed as described [49].
Two S. aureus isolates were considered indistinguishable if they shared the same MLST and SCCmec type and were concordant with respect to the presence or absence of the PVL genetic determinants. Based on a previous investigation demonstrating that ST8/PVL+/SCCmec IV is highly concordant with USA300 MRSA genetic background assessed by pulsed-field gel electrophoresis (M. David et al, unpublished data), isolates with these characteristics were categorized as USA300 MRSA.
Chart Abstraction and Criteria for CA S. aureus
We reviewed medical records of index patients using a standardized chart abstraction instrument that quantified recent hospitalizations, prior S. aureus infections, and comorbidities using a standard index [50]. We used the Centers for Disease Control and Prevention’s Active Bacterial Core surveillance case definition to classify each infection as CA or HA [51].
Statistical Analyses
Data were analyzed using SAS software (version 9.1.3; SAS Institute). Colonizing isolates that were indistinguishable from the index infection were considered the outcome of interest for the data analysis. Bivariate analysis was performed using χ2 or Fisher exact test, as appropriate. Multivariate modeling procedures [52] were performed to predict colonization of the index patient with their infecting strain type. Similar procedures accounting for clustering of household members were used to predict colonization of household members with the index patients’ infecting strain type. All variables with a P value ≤.10 in the bivariate analysis were included in a multivariate logistic regression analysis. Backward elimination was performed using the likelihood ratio test to identify the optimal model for the risk factors associated with colonization of the index patient. Backward elimination was performed using the Score test to find the best model of risk factors associated with colonization of household members. Models were examined for goodness of fit using the Hosmer-Lemeshow test. All variables were considered significant at the α = .05 level.
RESULTS
We screened 2097 patients with S. aureus skin infections and successfully contacted 877 by telephone or inpatient visits. Of these, 710/877 patients (80%) were eligible; among eligible patients, 502/710 (71%) verbally agreed to participate. Household visits were completed among 356 (71%) of those who agreed to participate. The remaining 146 households either never scheduled a household visit or were not present when research personnel arrived at the home. We enrolled 179 households in Los Angeles and 177 households in Chicago. Six households in Los Angeles were excluded from analysis because the patient was discharged to a long-term care facility (n = 1), unable to schedule a study visit (n = 1), or the index isolate was not confirmed as S. aureus during molecular characterization (n = 4).
Characteristics of Index Patients and Epidemiologic Case Definitions
Among the 350 households, the mean household size comprised 5.3 members (5.3 in Los Angeles and 5.4 in Chicago) and the mean number of household members enrolled was 3.4 (3.3 in Los Angeles and 3.5 in Chicago). Demographic, clinical, and behavioral characteristics of the 350 index patients are presented in Table 1. By epidemiologic categorization, 111 (32%) patients had CA-MRSA, 122 (35%) had HA-MRSA, 45 (13%) had CA-MSSA, and 72 (20%) had HA-MSSA infections (Table 1). Location of infection was head and neck in 16% (56/350), trunk in 16% (57/350), arm in 18% (63/350), buttocks/genitals in 19% (66/350), and leg in 38% (132/350). Of note, 18 patients (5%) had skin infections in >2 anatomic locations.
Table 1.
Demographics, Clinical Factors, and Bivariate Analysis of Risk Factors Associated With Colonization of the Index Patient by the Index Patient’s Infecting Strain Type
| Variable | All, n = 350 (%) | Colonized With Infecting Strain, n = 41 (%) | Not Colonized With Infecting Strain, n = 309 (%) | OR | 95% CI | P Value |
| Site | ||||||
| Chicago | 177 (51) | 22 (53) | 155 (50) | 1.15 | .60, 2.21 | .67 |
| Los Angeles | 173 (49) | 19 (46) | 154 (50) | … | … | … |
| Demographics | ||||||
| Gender | ||||||
| Female | 180 (51) | 26 (63) | 154 (50) | 1.75 | .89, 3.42 | .11 |
| Male | 170 (49) | 15 (37) | 155 (50) | … | … | … |
| Age | ||||||
| Older adult (>65 yr) | 15 (4) | 2 (5) | 13 (4) | 1.30 | .27, 6.22 | .74 |
| Adult (19–65 yr) | 180 (51) | 19 (46) | 161 (52) | Ref | … | … |
| Child (5–18 yr) | 55 (16) | 8 (20) | 47 (15) | 1.44 | .59, 3.50 | .42 |
| Younger child (<5 yr) | 100 (29) | 12 (29) | 88 (28) | 1.16 | .54, 2.49 | .71 |
| Ethnicity | ||||||
| African-American | 177 (51) | 24 (58) | 153 (50) | 1.20 | .34, 4.32 | .78 |
| Caucasian | 26 (7) | 3 (7) | 23 (7) | Ref | … | … |
| Hispanic | 121 (35) | 12 (29) | 109 (35) | 0.84 | .22, 3.23 | .81 |
| Other/mixed/unknown | 26 (7) | 2 (5) | 24 (8) | 0.64 | .10, 4.18 | .64 |
| Clinical factors | ||||||
| Charlson comorbidity score | ||||||
| Mean ± SD | 1 ± 2 | 1 ± 2 | 1 ± 2 | 0.92 | .78, 1.08 | .30 |
| Median (range) | 0 (0–14) | 0 (0–12) | 0 (0–14) | … | … | … |
| Comorbidities | ||||||
| Diabetes | 57 (16) | 5 (12) | 52 (17) | 0.69 | .26, 1.83 | .45 |
| HIV infection | 14 (5) | 0 (0) | 14 (14) | NA | NA | .39 |
| In the past 12 mo: | ||||||
| Had a previous skin infection | 217 (62) | 23 (56) | 194 (63) | 0.76 | .39, 1.46 | .41 |
| Undergone major surgery | 88 (25) | 12 (29) | 76 (24) | 1.27 | .62, 2.61 | .52 |
| Received dialysis | 8 (2) | 0 (0) | 8 (3) | NA | NA | .60 |
| Hospitalized | 172 (49) | 18 (44) | 154 (50) | 0.79 | .41, 1.52 | .48 |
| Days of hospitalization | ||||||
| Mean ± SD | 4 ± 18 | 3 ± 8 | 4 ± 19 | 0.99 | .97, 1.02 | .82 |
| Median (range) | 0 (0–320) | 0 (0–39) | 0 (0–320) | … | … | … |
| Any antibiotic exposure | 237 (67) | 24 (59) | 213 (69) | 0.64 | .33, 1.24 | .18 |
| Use of clindamycin | 35 (10) | 7 (18) | 28 (9) | 2.07 | .86, 5.09 | .16 |
| Use of TMP-SMX | 37 (11) | 3 (8) | 34 (11) | 0.65 | .19, 2.18 | .78 |
| Use of cephalexin | 17 (5) | 2 (5) | 15 (5) | 1.02 | .23, 4.65 | .99 |
| Use of immunosuppressant medications | 70 (20) | 9 (23) | 61 (20) | 1.15 | .52, 2.57 | .72 |
| Spent time living in a skilled nursing facility, rehabilitation center, or other type of group facility | 8 (2) | 2 (5) | 6 (2) | 2.68 | .52, 13.74 | .23 |
| Epidemiologic factors | ||||||
| Household density | ||||||
| Mean ± SD | 1.96 ± 1.08 | 1.94 ± 0.93 | 1.96 ± 1.10 | 0.99 | .73, 1.35 | .94 |
| Median (range) | 1.73 (0.40–9.0) | 1.66 (0.667–4.0) | 2.0 (0.40–9.0) | … | … | … |
| Homelessness in the past 12 mo | 14 (4) | 3 (7) | 11 (3) | 2.14 | .57, 8.01 | .22 |
| Cuts/scratches in the 30 d prior to index infection | 143 (41) | 19 (46) | 124 (40) | 1.29 | .67, 2.48 | .50 |
| Skin rash in the 90 d prior to index infection | 60 (18) | 14 (36) | 46 (16) | 2.97 | 1.44, 6.17 | .002a |
| Pets in the home, currently | 156 (45) | 15 (38) | 141 (46) | 0.702 | .36, 1.38 | .32 |
| Incarceration in the past 12 mo | 9 (4) | 0 (0) | 9 (5) | - | - | .60 |
| Illicit drug use in the past 12 mo | 34 (10) | 1 (2) | 33 (11) | 0.21 | .03, 1.57 | .15 |
| >1 sexual partner in the past 12 mo | 25 (12) | 3 (12) | 22 (12) | 1.02 | .29, 3.70 | .99 |
| In the past 3 mo: | ||||||
| Showered at least once a day | 39 (12) | 4 (11) | 35 (12) | 0.91 | .30, 2.71 | .99 |
| Shared make-up with others | 20 (7) | 2 (5) | 18 (7) | 0.76 | .17, 3.42 | .99 |
| Shared bar soap with others | 193 (57) | 25 (63) | 168 (56) | 1.32 | .67, 2.60 | .42 |
| Shared clothes with others with washing | 21 (6) | 3 (7) | 18 (6) | 1.25 | .35, 4.44 | .73 |
| Shared towels with others | 157 (46) | 21 (51) | 136 (45) | 1.30 | .67, 2.49 | .43 |
| Wore clothes more than once without washing | 166 (49) | 23 (58) | 143 (47) | 1.48 | .76, 2.87 | .25 |
| Hand-washing frequency after using the bathroom | ||||||
| Mean ± SD | 2.6 ± 0.78 | 2.6 ± 0.80 | 2.6 ± 0.78 | 1.01 | .66, 1.54 | .97 |
| Median (range) | 3 (0–3) | 3 (0–3) | 3 (0–3) | |||
| Household cleaning scaleb | ||||||
| Mean ± SD | 16 ± 8 | 17 ± 8 | 16 ± 8 | 1.18 | .62, 2.20 | .61 |
| Median (range) | 18 (0–35) | 18 (0–33) | 18 (0–35) | |||
| Use of a gym | 27 (14) | 5 (14) | 22 (9) | 1.56 | .55, 4.42 | .38 |
| Participation in contact sports | 83 (24) | 11 (27) | 72 (23) | 1.21 | .58, 2.53 | .62 |
| Goes to day care | 25 (19) | 1 (7) | 24 (21) | 0.27 | .03, 2.19 | .30 |
| Use of public facilitiesc | 76 (22) | 12 (29) | 64 (21) | 1.58 | .77, 3.27 | .21 |
| Strain-specific factors | ||||||
| Infecting strain type categorization by the CDC case definition | ||||||
| CA-MRSA | 111 (32) | 15 (37) | 96 (31) | 1.28 | .65, 2.53 | .92 |
| HA-MRSA | 122 (35) | 17 (42) | 105 (34) | Ref | ||
| CA-MSSA | 45 (13) | 3 (7) | 42 (14) | 0.50 | .15, 1.70 | .29 |
| HA-MSSA | 72 (20) | 6 (15) | 66 (21) | 0.63 | .26, 1.56 | .28 |
| ST8-MRSA-Mec IV-PVL strain type | 186 (53) | 29 (71) | 157 (51) | 2.33 | 1.15, 4.75 | .02 |
| PVL presence | 266 (76) | 33 (81) | 233 (75) | 0.74 | .33, 1.67 | .56 |
| SCCmec type IV | 220 (63) | 30 (73) | 190 (62) | 1.71 | .82, 3.54 | .17 |
| Other household members colonized with the infection strain type | 81 (23) | 21 (51) | 60 (19) | 4.36 | 2.22, 8.55 | <.001d |
Statistically significant relationships are bolded; NA indicates cannot be calculated due to zero cell.
Abbreviations: CA-MRSA, community-associated methicillin-resistant Staphylococcus aureus; CA-MSSA, community-associated methicillin-susceptible S. aureus; CDC, Centers for Disease Control and Prevention; CI, confidence inverval; HA-MRSA, healthcare-associated methicillin-resistant S. aureus; HA-MSSA, healthcare-associated methicillin-susceptible S. aureus; HIV, human immunodeficiency virus; OR, odds ratio; PVL, Panton-Valentine leukocidin; Ref, reference group; SD, standard deviation; TMP-SMX, trimethoprim-sulfamethoxazole.
Variable significant in multivariable analysis (OR, 2.9 [1.4–6.2]; P = .006).
Household cleaning is a measure of the frequency of cleaning for common household items, with higher values representing more frequent cleaning.
Use of public facilities is defined as use of a publicly available gym, locker room, shower, swimming pool, sauna, or Jacuzzi.
Variable significant in multivariable analysis (OR, 4.3 [2.1–8.6]; P < .001).
Strain Types of Isolates Infecting Index Patients
Among infecting isolates of index patients, the majority (233, 67%) were MRSA, 117 (33%) were MSSA, and 266 (76%) were PVL+. Among the 233 MRSA isolates, 220 (94.5%) contained SCCmec type IV, 6 (2.5%) contained SCCmec type II, 6 (2.5%) contained SCCmec type III, and 1 (0.5%) contained an untypeable SCCmec element. Among the infecting index subjects’ isolates, 230 (66%) were ST8 and 186 (53%) were USA300 MRSA. The genetic background of index patients’ isolates is summarized in Table 2.
Table 2.
Comparison of Index Infection Strains, Index Colonizing Strains, and Household Member Colonizing Strains Circulating in Households
| MRSA or MSSA | ST Type | SCCmec Type | PVL | Index Infecting Strain Type, n = 350 (%) | Index Colonizing Strain Type, n = 215 (%) | Household Member Colonizing Strain Type, n = 594 (%) | P Valuea |
| MRSA | 8 | IV | + | 186 (53) | 57 (27) | 178 (30) | <.001 |
| MRSA | 8 | IV | + | 8 (2) | 3 (1) | 6 (1) | .18 |
| MRSA | 8 | IV | – | 9 (3) | 0 (0) | 5 (1) | .02 |
| MRSA | 5 | IV | – | 3 (1) | 3 (1) | 3 (1) | .99 |
| MRSA | 5 | II | – | 3 (1) | 6 (3) | 8 (1) | .30 |
| MRSA | 239 | III | + | 6 (2) | 7 (3) | 7 (1) | .99 |
| MRSA | 1 | IV | + | 1 (1) | 0 (0) | 3 (1) | .99 |
| MRSA | 30 | IV | + | 1 (1) | 0 (0) | 3 (1) | .99 |
| MRSA | Miscb | Varied | Varied | 16 (3) | 15 (7) | 28 (4) | .59 |
| MSSA | 8 | None | + | 27 (8) | 9 (4) | 27 (5) | .03 |
| MSSA | 8 | None | – | 5 (1) | 3 (1) | 22 (4) | .11 |
| MSSA | 5 | None | – | 6 (2) | 23 (11) | 42 (7) | <.001 |
| MSSA | 15 | None | – | 3 (1) | 3 (1) | 33 (6) | .001 |
| MSSA | 30 | None | – | 5 (1) | 13 (6) | 47 (8) | <.001 |
| MSSA | 45 | None | – | 2 (1) | 12 (6) | 33 (6) | <.001 |
| MSSA | 72 | None | – | 1 (1) | 9 (4) | 20 (3) | <.001 |
| MSSA | 1 | None | + | 5 (1) | 0 (0) | 0 (0) | <.001 |
| MSSA | 188 | None | – | 4 (1) | 6 (3) | 12 (2) | .22 |
| MSSA | Miscb | None | Varied | 59 (17) | 46 (21) | 117 (20) | .22 |
Abbreviations: Misc, miscellaneous; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible S. aureus; PVL, Panton-Valentine leukocidin; SCCmec type, staphylococcal cassette chromosome mec type; ST type, sequence type by multilocus strain typing.
P value represents the proportion of the given strain type causing infection versus the proportion causing colonization in index and household members.
“Miscellaneous” strain types represent 106 other S. aureus strain types with different genotypes circulating in households.
Colonization Among Index Patients
Among index patients, 137/350 (40%) were colonized with S. aureus. Nasal colonization was present in 75 (22%) patients, oropharyngeal colonization in 64 (19%), and inguinal colonization in 76 (22%). Nonnasal colonization was found in 34% (117/350) of subjects, while 6% (24/350) were colonized only in the oropharynx and 8% (30/350) only in the inguinal region (Table 3). Including their infecting strain, 71% (247/350) of index patients had 1 S. aureus strain type, 24% (84/350) had 2 strain types, and 5% (19/350) had 3 strain types isolated from their body.
Table 3.
Comparison of Staphylococcus aureus Body Colonization by Epidemiologic Infection Type
| Index Patients | Household Members | |||||||||||
| Colonization Site | All, n = 350 (%) | CA-MRSA, n = 111 (%) | HA-MRSA, n = 122 (%) | CA-MSSA, n = 45 (%) | HA-MSSA, n = 72 (%) | P Value | All, n = 812 (%) | CA-MRSA, n = 268 (%) | HA-MRSA, n = 258 (%) | CA-MSSA, n = 100 (%) | HA-MSSA, n = 186 (%) | P Value |
| Any body site | ||||||||||||
| Any S. aureus | 137 (40) | 45 (41) | 40 (33) | 18 (40) | 34 (47) | .007 | 405 (50) | 134 (50) | 125 (49) | 45 (45) | 101 (54) | <.001 |
| MSSA | 86 (25) | 27 (24) | 17 (14) | 15 (33) | 27 (40) | .13 | 267 (33) | 86 (32) | 73 (28) | 29 (29) | 79 (42) | <.001 |
| MRSA | 62 (18) | 21 (19) | 27 (22) | 3 (7) | 11 (15) | <.001 | 177 (22) | 61 (23) | 61 (24) | 22 (22) | 33 (17) | <.001 |
| Nasal | ||||||||||||
| Any S. aureus | 75 (22) | 23 (21) | 20 (16) | 11 (24) | 21 (30) | .21 | 205 (25) | 61 (23) | 68 (26) | 25 (25) | 51 (28) | <.001 |
| MSSA | 47 (14) | 15 (14) | 5 (4) | 10 (22) | 17 (24) | .06 | 116 (14) | 35 (13) | 34 (13) | 11 (11) | 15 (8) | .002 |
| MRSA | 28 (8) | 8 (7) | 15 (12) | 1 (2) | 4 (6) | .001 | 89 (11) | 26 (10) | 34 (13) | 14 (14) | 36 (20) | .007 |
| Oropharynx | ||||||||||||
| Any S. aureus | 64 (19) | 23 (21) | 19 (16) | 8 (18) | 14 (19) | .049 | 236 (30) | 81 (31) | 72 (28) | 22 (22) | 61 (34) | <.001 |
| MSSA | 36 (11) | 12 (11) | 6 (5) | 7 (16) | 11 (15) | .41 | 160 (20) | 55 (21) | 42 (16) | 13 (13) | 50 (28) | <.001 |
| MRSA | 28 (8) | 11 (10) | 13 (11) | 1 (2) | 3 (4) | .002 | 76 (10) | 26 (10) | 30 (12) | 11 (11) | 11 (6) | <.001 |
| Inguinal region | ||||||||||||
| Any S. aureus | 76 (22) | 18 (16) | 28 (23) | 7 (17) | 23 (32) | .005 | 154 (19) | 53 (20) | 50 (19) | 18 (18) | 33 (18) | <.001 |
| MSSA | 41 (12) | 10 (9) | 11 (9) | 5 (12) | 15 (21) | .18 | 77 (10) | 26 (10) | 21 (8) | 11 (11) | 19 (10) | .11 |
| MRSA | 35 (10) | 8 (7) | 17 (14) | 2 (5) | 8 (11) | .004 | 77 (10) | 27 (10) | 29 (11) | 7 (7) | 14 (8) | <.001 |
| Colonization at >1 body site | ||||||||||||
| Any S. aureus | 63 (18) | 16 (14) | 20 (16) | 7 (16) | 20 (28) | .67 | 150 (18) | 50 (18) | 48 (18) | 14 (14) | 38 (20) | <.001 |
| MSSA | 42 (12) | 12 (11) | 8 (7) | 6 (13) | 16 (22) | .13 | 107 (13) | 36 (13) | 27 (10) | 11 (11) | 33 (18) | .003 |
| MRSA | 31 (9) | 7 (6) | 16 (13) | 1 (2) | 7 (10) | .002 | 75 (9) | 22 (8) | 29 (11) | 9 (9) | 15 (8) | .007 |
| Nonnasal | ||||||||||||
| Any S. aureus | 117 (34) | 37 (33) | 37 (31) | 14 (31) | 29 (41) | .007 | 332 (41) | 115 (44) | 100 (39) | 34 (34) | 83 (44) | <.001 |
| MSSA | 69 (20) | 21 (19) | 15 (12) | 11 (24) | 22 (31) | .20 | 217 (27) | 75 (28) | 56 (22) | 23 (23) | 75 (40) | <.001 |
| MRSA | 52 (15) | 17 (15) | 23 (19) | 3 (7) | 9 (13) | <.001 | 133 (16) | 47 (18) | 48 (19) | 15 (15) | 23 (12) | <.001 |
| Nasal only | ||||||||||||
| Any S. aureus | 20 (6) | 8 (8) | 3 (2) | 2 (9) | 5 (7) | .42 | 73 (9) | 19 (7) | 25 (10) | 11 (11) | 18 (10) | .14 |
| MSSA | 13 (4) | 5 (5) | 0 (0) | 2 (9) | 4 (6) | .15 | 37 (5) | 11 (4) | 13 (5) | 5 (5) | 11 (6) | .26 |
| MRSA | 7 (2) | 3 (3) | 3 (3) | 0 (0) | 1 (1) | .57 | 36 (4) | 8 (3) | 12 (5) | 6 (6) | 7 (4) | .41 |
| Oropharynx only | ||||||||||||
| Any S. aureus | 24 (6) | 13 (12) | 5 (5) | 4 (9) | 2 (3) | .009 | 125 (15) | 47 (17) | 34 (13) | 11 (11) | 33 (18) | <.001 |
| MSSA | 13 (4) | 6 (6) | 2 (2) | 3 (7) | 2 (3) | .35 | 92 (11) | 33 (12) | 23 (9) | 7 (7) | 29 (16) | <.001 |
| MRSA | 11 (2) | 7 (6) | 3 (3) | 1 (2) | 0 (0) | .08 | 33 (4) | 14 (5) | 11 (4) | 4 (4) | 4 (2) | .03 |
| Inguinal region only | ||||||||||||
| Any S. aureus | 30 (8) | 8 (8) | 17 (14) | 3 (7) | 7 (10) | .14 | 57 (7) | 18 (6) | 18 (7) | 9 (9) | 12 (6) | .23 |
| MSSA | 18 (4) | 4 (4) | 0 (0) | 2 (5) | 5 (7) | .41 | 31 (4) | 9 (3) | 10 (4) | 6 (6) | 6 (3) | .65 |
| MRSA | 12 (4) | 4 (4) | 17 (14) | 1 (2) | 2 (3) | .34 | 26 (3) | 9 (3) | 8 (3) | 3 (3) | 6 (3) | .36 |
P values represent comparisons between groups within a given row. P values <.05 signify that 1 group has a proportion that was statistically significantly different from other groups.
Abbreviations: CA-MRSA, community-associated methicillin-resistant Staphylococcus aureus; CA-MSSA, community-associated methicillin-susceptible S. aureus; HA-MRSA, healthcare-associated methicillin-resistant S. aureus; HA-MSSA, healthcare-associated methicillin-susceptible S. aureus.
Among index patients, 12% (41/350) had >1 colonizing strain types concordant with their infecting isolate and 27% (96/350) were colonized with an S. aureus strain type discordant from their infecting isolate. Among index patients infected with MRSA, 14% (32/233) carried a concordant strain type and 25% (57/233) carried a discordant S. aureus strain type. Of these 57 discordant S. aureus strain types, 28% (16/57) were discordant MRSA strain types. Among those infected with an MSSA strain, 8% (9/117) carried a concordant strain type and 39% (46/117) carried a discordant S. aureus strain type. Of these 46 discordant S. aureus strain types, 72% (33/46) were colonized with other MSSA strain types. Among patients infected with USA300 MRSA, concordant carriage occurred in 16% (29/186) and 23% (43/186) carried a discordant S. aureus strain type. Of these 43 discordant strain types, 23% (10/43) were non-USA300 MRSA strain types.
Factors associated with colonization with the infecting strain type in bivariate analysis are described in Table 1. In the multivariable model, independent predictors of colonization with the infecting strain type were skin rash in the prior 90 days (odds ratio [OR], 2.9 [1.4–6.2]; P = .006) and having ≥1 household members colonized with the infection strain type (OR, 4.3 [2.1–8.6]; P < .001).
Household Contacts’ Colonization
Among the 826 household members, 14 declined the body colonization swabs. Demographics and comorbidities of the 812 household contacts are summarized in Table 4. Of note, 24% (197/812) of household members reported a skin infection in the prior 12 months and 50% (405/812) were colonized with S. aureus. Colonization site data are provided in Table 3. Overall, 33% (267/812) of household members were colonized with MSSA and 22% (177/812) with MRSA. Forty-one percent (333/812) were colonized with 1 strain type, 8% (63/812) with 2 strain types, and 1% (9/812) with 3 strain types.
Table 4.
Bivariate Analysis of Risk Factors Associated With an Index Patient’s Infecting Strain Colonizing a Household Member in at Least 1 Body Site
| Variable | All, n = 812 (%) | Colonized With Index Infection Strain, n = 111 (%) | Not Colonized With Index Infection Strain, n = 701 (%) | OR | 95% CI | P value |
| Site | ||||||
| Chicago | 422 (52) | 54 (49) | 368 (53) | 0.99 | .94, 1.04 | .63 |
| Los Angeles | 390 (48) | 57 (51) | 333 (48) | … | … | … |
| Demographics | ||||||
| Gender | ||||||
| Female | 510 (63) | 63 (57) | 447 (65) | 0.96 | .92, 1.01 | .13 |
| Male | 294 (37) | 48 (43) | 246 (36) | … | … | … |
| Age | ||||||
| Older adult (>65 yr) | 36 (4) | 5 (5) | 31 (5) | 1.01 | .90, 1.13 | .85 |
| Adult (19–65 yr) | 492 (61) | 67 (60) | 425 (61) | Ref | … | … |
| Child (5–18 yr) | 175 (22) | 27 (24) | 148 (21) | 1.02 | .95, 1.08 | .59 |
| Younger child (<5 yr) | 109 (13) | 12 (11) | 97 (14) | 0.97 | .91, 1.03 | .33 |
| Ethnicity | ||||||
| African-American | 412 (51) | 53 (48) | 359 (51) | 1.06 | .97, 1.16 | .18 |
| Caucasian | 48 (6) | 3 (3) | 45 (6) | Ref | … | … |
| Hispanic | 300 (37) | 50 (45) | 250 (36) | 1.11 | 1.001, 1.22 | .03 |
| Other/mixed/unknown | 52 (6) | 5 (5) | 47 (7) | 1.02 | .91, 1.14 | .69 |
| Clinical factors | ||||||
| Charlson comorbidity score | ||||||
| Mean ± SD | 0 ± 1 | 0 ± 1 | 0 ± 1 | 0.98 | .96, 1.00 | .16 |
| Median (range) | 0 (0–7) | 0 (0–6) | 0 (0–7) | … | … | … |
| Comorbidities | ||||||
| Diabetes | 71 (9) | 9 (8) | 62 (9) | 0.99 | .91, 1.07 | .73 |
| HIV infection | 8 (2) | 1 (1) | 7 (2) | 0.99 | .82, 1.19 | .91 |
| In the past 12 mo | ||||||
| Had a previous skin infection | 197 (25) | 45 (41) | 152 (23) | 1.12 | 1.04, 1.19 | <.001a |
| Undergone major surgery | 59 (7) | 4 (4) | 55 (8) | 0.94 | .87, 1.01 | .07 |
| Received dialysis | 1 (0.12) | 0 (0) | 1 (0.14) | NA | NA | .99 |
| Hospitalized | 101 (13) | 15 (14) | 86 (13) | 1.01 | .93, 1.08 | .88 |
| Days of hospitalization | ||||||
| Mean ± SD | 0.5 ± 2 | 0.5 ± 2 | 0.5 ± 2 | 1.00 | .98, 1.01 | .93 |
| Median (range) | 0 (0–30) | 0 (0–21) | 0 (0–30) | … | … | … |
| Any antibiotic exposure | 248 (31) | 43 (39) | 205 (30) | 1.05 | .99, 1.11 | .11 |
| Use of clindamycin | 12 (2) | 4 (4) | 8 (1) | 1.22 | .89, 1.67 | .22 |
| Use of TMP-SMX | 11 (1) | 4 (4) | 7 (1) | 1.23 | .96, 1.58 | .09 |
| Use of cephalexin | 15 (2) | 7 (6) | 8 (1) | 1.39 | 1.06, 1.78 | .02b |
| Use of immunosuppressant medications | 78 (10) | 14 (13) | 64 (9) | 1.04 | .95, 1.14 | .42 |
| Spent time living in a skilled nursing facility, rehabilitation center, or other type of group facility | 19 (2) | 2 (2) | 17 (3) | 0.95 | .84, 1.07 | .41 |
| Epidemiologic factors | ||||||
| Household density | ||||||
| Mean ± SD | 2.0 ± 0.98 | 2.2 ± 0.92 | 2.0 ± 0.98 | 1.01 | .99, 1.04 | .33 |
| Median (range) | 2.0 (0.33–6.0) | 2.2 (0.71–6.0) | 2.0 (0.33–6.0) | … | … | … |
| Homelessness in the past 12 mo | 40 (5) | 5 (5) | 35 (5) | 0.99 | .89, 1.11 | .99 |
| Cuts/scratches in the 30 d prior to index infection | 308 (38) | 51 (46) | 257 (37) | 1.05 | .99, 1.11 | .07 |
| Skin rash in the 90 d prior to index infection | 87 (11) | 18 (17) | 69 (10) | 1.09 | .99, 1.18 | .06 |
| Pets in the home currently | 339 (42) | 47 (43) | 292 (42) | 1.01 | .95, 1.06 | .83 |
| Incarceration in the past 12 mo | 21 (3) | 3 (4) | 18 (3) | 1.02 | .87, 1.19 | .86 |
| Illicit drug use in the past 12 mo | 66 (11) | 11 (14) | 55 (11) | 1.05 | .96, 1.15 | .26 |
| >1 sexual partner in the past 12 mo | 73 (12) | 9 (12) | 64 (12) | 0.99 | .92, 1.07 | .80 |
| In the past 3 mo | ||||||
| Showered at least once a day | 131 (16) | 23 (21) | 108 (16) | 1.04 | .97, 1.11 | .32 |
| Shared make-up with others | 88 (12) | 13 (13) | 75 (11) | 1.01 | .93, 1.10 | .82 |
| Shared bar soap with others | 496 (63) | 67 (63) | 429 (63) | 1.00 | .95, 1.05 | .96 |
| Shared clothes with others without washing | 51 (6) | 8 (7) | 43 (6) | 1.03 | .94, 1.13 | .53 |
| Shared towels with others | 389 (48) | 55 (50) | 334 (48) | 1.02 | .97, 1.07 | .42 |
| Wore clothes more than once without washing | 400 (50) | 58 (53) | 342 (49) | 1.02 | .97, 1.07 | .42 |
| Hand-washing frequency after using the bathroom | ||||||
| Mean ± SD | 2.7 ± 0.62 | 2.7 ± 0.76 | 2.8 ± 0.60 | 0.97 | .92, 1.01 | .20 |
| Median (range) | 3 (0–3) | 3 (0–3) | 3 (0–3) | … | … | … |
| Household cleaning scalec | ||||||
| Mean ± SD | 17 ± 7 | 17 ± 8 | 17 ± 7 | 0.99 | .99, 1.00 | .17 |
| Median (range) | 18 (0–34) | 18 (0–29) | 18 (0–34) | … | … | … |
| Use of a gym | 87 (12) | 10 (10) | 77 (12) | 0.98 | .92, 1.05 | .59 |
| Participation in contact sports | 201 (25) | 33 (30) | 168 (24) | 1.04 | .98, 1.10 | .23 |
| Goes to day care | 30 (16) | 3 (10) | 27 (17) | 0.92 | .82, 1.02 | .10 |
| Use of public facilitiesd | 224 (28) | 32 (29) | 192 (28) | 1.00 | .96, 1.06 | .86 |
| Strain Specific Factors | ||||||
| Infecting strain categorization by the CDC case definition | ||||||
| CA-MRSA | 268 (33) | 39 (35) | 229 (33) | 0.95 | .88, 1.01 | .12 |
| HA-MRSA | 258 (32) | 52 (47) | 206 (29) | Ref | … | … |
| CA-MSSA | 100 (12) | 8 (7) | 92 (13) | 0.89 | .82, .97 | .008 |
| HA-MSSA | 186 (23) | 12 (11) | 174 (25) | 0.88 | .82, .94 | .003 |
| ST8-IV-PVL infection strain type | 442 (54) | 87 (78) | 355 (51) | 1.13 | 1.08, 1.19 | <.001e |
| PVL presence | 622 (77) | 95 (86) | 527 (75) | 1.07 | 1.01, 1.13 | .03 |
| SCCmec type IV | 503 (62) | 89 (80) | 414 (59) | 1.10 | 1.05, 1.16 | <.001 |
Statistically significant relationships are bolded; NA indicates cannot be calculated due to zero cell.
Abbreviations: CA-MRSA, community-associated methicillin-resistant Staphylococcus aureus; CA-MSSA, community-associated methicillin-susceptible S. aureus; CDC, Centers for Disease Control and Prevention; CI, confidence interval; HA-MRSA, healthcare-associated methicillin-resistant S. aureus; HA-MSSA, healthcare-associated methicillin-susceptible S. aureus; HIV, human immunodeficiency virus; OR, odds ratio; PVL, Panton-Valentine leukocidin; Ref, reference group; SCCmec type, staphylococcal cassette chromosome mec type; TMP-SMX, trimethoprim-sulfamethoxazole.
Variable significant in multivariable analysis (OR, 2.01 [1.31–3.08]; P = .001).
Variable significant in multivariable analysis (OR, 3.5 [1.3–9.5]; P = .01).
Household cleaning is a measure of the frequency of cleaning for common household items with higher values representing more frequent cleaning.
Use of public facilities is defined as use of a publicly available gym, locker room, shower, swimming pool, sauna, or Jacuzzi.
Variable significant in multivariable analysis (OR, 3.0 [1.7–5.3]; P = .0002).
Colonizing Strain Types of Household Contacts
Of the 350 households, the presence of >1 S. aureus strain circulating among household contacts was common. Including the index patient’s infecting isolate, 35% (123/350) of households had a single strain type identified, 33% (117/350) had 2 strain types, 20% (40/350) had 3 strain types, 7% (24/350) had 4 strain types, and 5% (16/350) had ≥5 strain types identified.
The genetic backgrounds of S. aureus identified among household members are summarized in Table 2. The most common genetic background was USA300 MRSA, which represented 30% of isolates. USA300 MRSA was found to colonize 16% of CA-MRSA, 20% of HA-MRSA, 16% of CA-MSSA, and 12% of HA-MSSA household members.
Fourteen percent (111/812) of household members had at least 1 colonizing strain type concordant with the index patient’s infecting isolate and 39% (317/812) of their colonizing isolate types were discordant. In households with an index MRSA infection, 17% (91/526) of contacts carried a concordant strain type compared with 7% (20/286) among household contacts of an MSSA index infection and 20% (87/442) of households with a USA300 MRSA index infection.
For household contacts, bivariate associations between hypothesized risk factors for colonization and strain type concordant with the index patient’s infecting strain type are described in Table 4. In multivariable analysis, significant predictors of strain type concordant with the index patient’s infecting strain type included previous skin infection (OR, 2.01 [1.31–3.08]; P = .001), cephalexin use in the past 12 months (OR, 3.5 [1.3–9.5]; P = .01), and the index patient having a USA300 infection strain type (OR, 3.0 [1.7–5.3]; P <.001).
DISCUSSION
Our investigation of 350 households with S. aureus infection in Los Angeles and Chicago comprising 1162 persons demonstrated the prevalence of S. aureus colonization among index patients and their household members is high, yet colonizing strain types are surprisingly diverse, complex, and frequently not concordant with the infecting isolate.
Our investigation yielded several notable findings. First, our data suggest that the USA300 MRSA genetic background appears to spread more easily in households than other genetic backgrounds. An index infection with the USA300 MRSA genetic background was an independent predictor of concordant strain type colonization in another household member. These findings may explain USA300 MRSA’s emergence in communities [53, 54] and infection clusters in households, jails, military barracks, and sports teams [54–57]. Of note, USA300 MRSA was highly prevalent and found in ≥1 persons in 80% of households with an index HA-MRSA infection and 26% of households with an HA-MSSA infection.
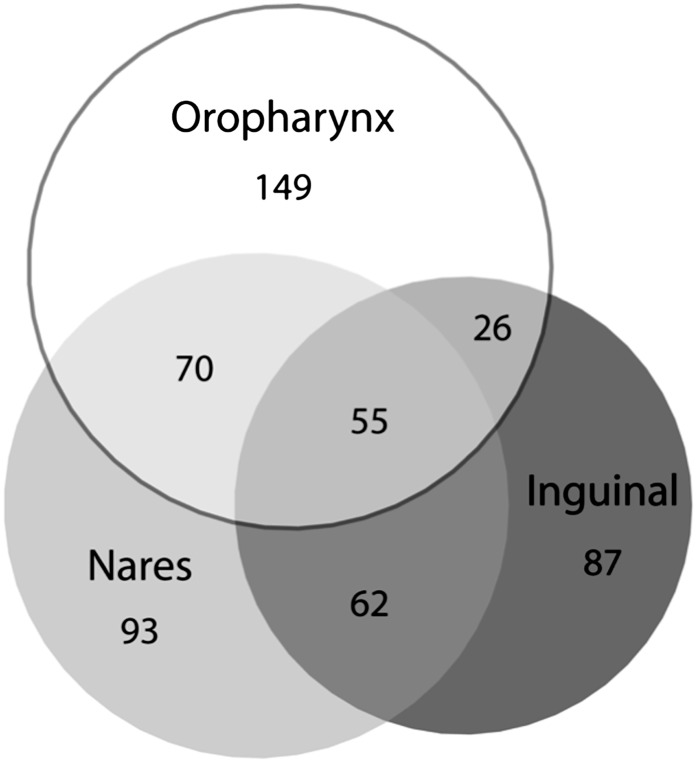
Second, S. aureus colonization of household members was very common (50%) and higher than the 20%–35% prevalence of nasal colonization commonly cited [58–60]. These findings probably reflect that we surveyed 3 body sites and used enrichment broth culture. Similar to studies in other populations [29–32], assessing additional anatomic sites for S. aureus colonization revealed a higher prevalence of S. aureus colonization. Notably, a nares-only culture survey in our population of household contacts would have missed 48% of S. aureus–colonized persons (Figure 1) and 51% of MRSA-colonized persons. These findings suggest that development of successful decolonization regimens to prevent infection should consider agents that eradicate skin and oropharyngeal colonization.
Figure 1.
Overlap of nares, oropharynx, and inguinal colonization among the 542 Staphylococcus aureus–colonized subjects from our total population of 1162 persons of households of persons with a recent S. aureus skin infection. Each circle size is proportional to the amount of S. aureus detected at that given anatomic site. Of note, nares-only surveys would have missed 48% of S. aureus–colonized persons.
Third, only 74% (91/122) of household contacts with MRSA colonization were colonized with the same MRSA genetic background as the index patient. The unexpectedly high prevalence of discordant MRSA genetic backgrounds colonizing contacts (26%) suggests that merely surveying for MRSA colonization in household contacts of persons with a skin infection would overestimate the spread of the index patients’ MRSA clones within the household and that many households in our study had >1 S. aureus or even MRSA strain type circulating.
Fourth, S. aureus colonization among index patients (40%) was less common than among household contacts (50%). Households were visited on average 18 days after the infection culture was obtained. Thus, this finding probably reflects index patients’ recent antibiotic treatment that may have eradicated S. aureus colonization.
Finally, our data demonstrate that the distribution of pathogenic strain types differs from that of strain types colonizing index patients and their household contacts. For example, USA300 MRSA genetic background caused 53% of infections but comprised just 29% of colonizing strain types (P < .001). Conversely, sequence type 30 MSSA caused just 1% of infections but was responsible for 7.4% of colonizing isolates. These findings suggest that some genetic backgrounds are unlikely causes of disease and that others, such as USA300 MRSA, have high pathogenic potential. If true, decolonization strategies may need to be refocused to avoid inadvertently eliminating less pathogenic S. aureus strains and disrupting commensal flora. Alternatively, these findings may stem from decolonization of patients’ infecting USA300 strain and subsequent recolonization with less pathogenic strains.
Compared with other investigations of household S. aureus spread, it should be noted that 3 European investigations and 1 American investigation found that all or all but 1 of the MRSA strain types colonizing household members were identical to those infecting the index patient [17, 18, 21, 24]. Perhaps this lack of strong concordance is due to the higher level of endemic MRSA colonization and the differences in the genetic backgrounds of prevalent MRSA clones that exist in the United States compared with many European countries. In the US study noted above, results may have differed from ours because the study was conducted before the emergence of CA-MRSA. Interestingly, 1 smaller US study conducted in an area of endemic CA-MRSA, like ours, found discordance in about half of isolates colonizing household members [19].
There are strengths to our study. First, to our knowledge, our investigation is the largest detailed survey of household contacts of patients with S. aureus skin infections. Unlike many previous investigations of household colonization that did not survey multiple body sites [17, 19, 20, 22, 23, 25, 26], we assessed 3 body sites for colonization and undertook a detailed epidemiologic survey. Second, our study was performed at 2 urban sites in the United States that have different racial and ethnic population distributions. Third, we performed genotyping of isolates. A previous large investigation of HA-MRSA spread to household contacts found that the rate of spread to contacts was 20% [23]. However, in the absence of strain typing, this number may be an overestimation. Furthermore, the purported risk factors for MRSA transmission in this prior investigation would be biased by the lack of strain typing. Finally, other previous investigations did not examine epidemiologic factors or were likely underpowered to detect significant relationships [15–21].
There are limitations to our study. First, our survey may not have detected some colonized individuals because some studies have found S. aureus colonization on the wrists, rectum, axilla, and vagina [30, 61, 62]. However, the yields of these additional sites may have been low [30, 61, 62]. Second, our study is cross sectional and could not determine directionality of strain transfer. Third, our population may not be representative of other populations. Study subjects came from populations of relatively low socioeconomic status in the United States where CA-MRSA infections are epidemic. Fourth, the number of comparisons and tests performed for statistical analyses may increase the likelihood of type 1 error.
In summary, we found that S. aureus colonization was very common among household contacts of persons with acute S. aureus infection. Given the strain type diversity among MSSA and MRSA isolates found in household members, development of successful household S. aureus screening and decolonization programs infections may not be a simple task. It is plausible that decolonization will eradicate less pathogenic strain types, leaving the person vulnerable to recolonization with more pathogenic strain types, such as USA300 MRSA. The complex nature of colonization we observed may prompt rethinking of MRSA prevention strategies. The implications of eliminating S. aureus strain types uncommonly associated with disease are unclear and require further study.
Notes
Acknowlegments.
We thank Everly Macario, Mellie Badar, Ramiro Correa, Sallie Chui, and Grace Tagudar for assistance with survey development, collection of study data, and processing of laboratory specimens. We also thank the staff of the Clinical Microbiology Laboratory at Harbor-UCLA Medical Center for their assistance with specimen collection. Finally, we thank all of the patients and their families for their participation and for allowing us to visit their homes for this study.
Financial support.
This work was supported by a grant from the National Institutes of Health (R01 AI067584-01A1).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Herold BC, Immergluck LC, Maranan MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998;279:593–8. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 2.Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–74. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 3.Seybold U, Kourbatova EV, Johnson JG, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care–associated blood stream infections. Clin Infect Dis. 2006;42:647–56. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 4.Park SH, Park C, Yoo JH, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated bloodstream infections in Korea. Infect Control Hosp Epidemiol. 2009;30:146–55. doi: 10.1086/593953. [DOI] [PubMed] [Google Scholar]
- 5.Maree C, Daum RS, Boyle-Vavra S, Matayoshi K, Miller LG. Community-associated methicillin-resistant Staphylococcus aureus strains causing healthcare-associated infections. Emerg Infect Dis. 2007;13:236–42. doi: 10.3201/eid1302.060781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorwitz RJ, Jernigan DB, Powers JH, Jernigan JA Participants in the CDC-Convened Experts’ Meeting on Management of MRSA in the Community. Strategies for clinical management of MRSA in the community: summary of an experts’ meeting convened by the Centers for Disease Control and Prevention. 2006. Available at: http://cdc.gov/ncidod/dhqp/pdf/ar/CAMRSA_ExpMtgStrategies.pdf. Accessed 22 May 2006. [Google Scholar]
- 7.Miller LG, Kaplan SL. Staphylococcus aureus: a community pathogen. Infect Dis Clin North Am. 2009;23:35–52. doi: 10.1016/j.idc.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Mulligan ME, Murray-Leisure KA, Ribner BS, et al. Methicillin-resistant Staphylococcus aureus: a consensus review of the microbiology, pathogenesis, and epidemiology with implications for prevention and management. Am J Med. 1993;94:313–28. doi: 10.1016/0002-9343(93)90063-u. [DOI] [PubMed] [Google Scholar]
- 9.Moreno F, Crisp C, Jorgensen JH, Patterson JE. Methicillin-resistant Staphylococcus aureus as a community organism. Clin Infect Dis. 1995;21:1308–12. doi: 10.1093/clinids/21.5.1308. [DOI] [PubMed] [Google Scholar]
- 10.Frenay HM, Vandenbroucke-Grauls CM, Molkenboer MJ, Verhoef J. Long-term carriage, and transmission of methicillin-resistant Staphylococcus aureus after discharge from hospital. J Hosp Infect. 1992;22:207–15. doi: 10.1016/0195-6701(92)90045-n. [DOI] [PubMed] [Google Scholar]
- 11.Adcock PM, Pastor P, Medley F, Patterson JE, Murphy TV. Methicillin-resistant Staphylococcus aureus in two child care centers. J Infect Dis. 1998;178:577–80. doi: 10.1086/517478. [DOI] [PubMed] [Google Scholar]
- 12.Miller LG, Quan C, Shay A, et al. A prospective investigation of outcomes after hospital discharge for endemic, community-acquired methicillin-resistant and -susceptible Staphylococcus aureus skin infection. Clin Infect Dis. 2007;44:483–92. doi: 10.1086/511041. [DOI] [PubMed] [Google Scholar]
- 13.Miller LG, Perdreau-Remington F, Bayer AS, et al. Clinical and epidemiologic characteristics cannot distinguish community-associated methicillin-resistant Staphylococcus aureus infection from methicillin-susceptible S. aureus infection: a prospective investigation. Clin Infect Dis. 2007;44:471–82. doi: 10.1086/511033. [DOI] [PubMed] [Google Scholar]
- 14.Huang YC, Ho CF, Chen CJ, Su LH, Lin TY. Nasal carriage of methicillin-resistant Staphylococcus aureus in household contacts of children with community-acquired diseases in Taiwan. Pediatr Infect Dis J. 2007;26:1066–8. doi: 10.1097/INF.0b013e31813429e8. [DOI] [PubMed] [Google Scholar]
- 15.Hewlett AL, Falk PS, Hughes KS, Mayhall CG. Epidemiology of methicillin-resistant Staphylococcus aureus in a university medical center day care facility. Infect Control Hosp Epidemiol. 2009;30:985–92. doi: 10.1086/605721. [DOI] [PubMed] [Google Scholar]
- 16.Lautenbach E, Tolomeo P, Nachamkin I, Hu B, Zaoutis TE. The impact of household transmission on duration of outpatient colonization with methicillin-resistant Staphylococcus aureus. Epidemiol Infect. 2010;138:683–5. doi: 10.1017/S0950268810000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eveillard M, Martin Y, Hidri N, Boussougant Y, Joly-Guillou ML. Carriage of methicillin-resistant Staphylococcus aureus among hospital employees: prevalence, duration, and transmission to households. Infect Control Hosp Epidemiol. 2004;25:114–20. doi: 10.1086/502360. [DOI] [PubMed] [Google Scholar]
- 18.Mollema FP, Richardus JH, Behrendt M, et al. Transmission of methicillin-resistant Staphylococcus aureus to household contacts. J Clin Microbiol. 2010;48:202–7. doi: 10.1128/JCM.01499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zafar U, Johnson LB, Hanna M, et al. Prevalence of nasal colonization among patients with community-associated methicillin-resistant Staphylococcus aureus infection and their household contacts. Infect Control Hosp Epidemiol. 2007;28:966–9. doi: 10.1086/518965. [DOI] [PubMed] [Google Scholar]
- 20.Huang YC, Su LH, Lin TY. Nasal carriage of methicillin-resistant Staphylococcus aureus in contacts of an adolescent with community-acquired disseminated disease. Pediatr Infect Dis J. 2004;23:919–22. doi: 10.1097/01.inf.0000141745.12941.ef. [DOI] [PubMed] [Google Scholar]
- 21.Johansson PJ, Gustafsson EB, Ringberg H. High prevalence of MRSA in household contacts. Scand J Infect Dis. 2007;39:764–8. doi: 10.1080/00365540701302501. [DOI] [PubMed] [Google Scholar]
- 22.Baggett HC, Hennessy TW, Rudolph K, et al. Community-onset methicillin-resistant Staphylococcus aureus associated with antibiotic use and the cytotoxin Panton-Valentine leukocidin during a furunculosis outbreak in rural Alaska. J Infect Dis. 2004;189:1565–73. doi: 10.1086/383247. [DOI] [PubMed] [Google Scholar]
- 23.Lucet JC, Paoletti X, Demontpion C, et al. Carriage of methicillin-resistant Staphylococcus aureus in home care settings: prevalence, duration, and transmission to household members. Arch Intern Med. 2009;169:1372–8. doi: 10.1001/archinternmed.2009.217. [DOI] [PubMed] [Google Scholar]
- 24.Calfee DP, Durbin LJ, Germanson TP, Toney DM, Smith EB, Farr BM. Spread of methicillin-resistant Staphylococcus aureus (MRSA) among household contacts of individuals with nosocomially acquired MRSA. Infect Control Hosp Epidemiol. 2003;24:422–6. doi: 10.1086/502225. [DOI] [PubMed] [Google Scholar]
- 25.Faires MC, Tater KC, Weese JS. An investigation of methicillin-resistant Staphylococcus aureus colonization in people and pets in the same household with an infected person or infected pet. J Am Vet Med Assoc. 2009;235:540–3. doi: 10.2460/javma.235.5.540. [DOI] [PubMed] [Google Scholar]
- 26.Nerby JM, Gorwitz R, Lesher L, et al. Risk factors for household transmission of community-associated methicillin-resistant Staphylococcus aureus. Pediatric Infect Dis J. 2011;30:927–32. doi: 10.1097/INF.0b013e31822256c3. [DOI] [PubMed] [Google Scholar]
- 27.Mertz D, Frei R, Periat N, et al. Exclusive Staphylococcus aureus throat carriage: at-risk populations. Arch Intern Med. 2009;169:172–8. doi: 10.1001/archinternmed.2008.536. [DOI] [PubMed] [Google Scholar]
- 28.Ringberg H, Cathrine Petersson A, Walder M, Hugo Johansson PJ. The throat: an important site for MRSA colonization. Scand J Infect Dis. 2006;38:888–93. doi: 10.1080/00365540600740546. [DOI] [PubMed] [Google Scholar]
- 29.Mertz D, Frei R, Jaussi B, et al. Throat swabs are necessary to reliably detect carriers of Staphylococcus aureus. Clin Infect Dis. 2007;45:475–7. doi: 10.1086/520016. [DOI] [PubMed] [Google Scholar]
- 30.Yang ES, Tan J, Eells S, Rieg G, Tagudar G, Miller LG. Body site colonization prevalence in patients with community-associated methicillin-resistant Staphylococcus aureus and other forms of Staphylococcus aureus and skin infections. Clin Microbiol Infect. 2010;16:425–31. doi: 10.1111/j.1469-0691.2009.02836.x. [DOI] [PubMed] [Google Scholar]
- 31.Ide L, Lootens J, Thibo P. The nose is not the only relevant MRSA screening site. Clin Microbiol Infect. 2009;15:1192–3. doi: 10.1111/j.1469-0691.2009.02954.x. [DOI] [PubMed] [Google Scholar]
- 32.Mody L, Kauffman CA, Donabedian S, Zervos M, Bradley SF. Epidemiology of Staphylococcus aureus colonization in nursing home residents. Clin Infect Dis. 2008;46:1368–73. doi: 10.1086/586751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorwitz RJ, Kruszon-Moran D, McAllister SK, et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis. 2008;197:1226–34. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 34.Maree CM, Eells SJ, Tan J, et al. Risk factors for infection and colonization with community-associated methicillin-resistant Staphylococcus aureus in the Los Angeles County jail: a case-control study. Clin Infect Dis. 2010;51:1248–57. doi: 10.1086/657067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graffunder EM, Venezia RA. Risk factors associated with nosocomial methicillin-resistant Staphylococcus aureus (MRSA) infection including previous use of antimicrobials. J Antimicrob Chemother. 2002;49:999–1005. doi: 10.1093/jac/dkf009. [DOI] [PubMed] [Google Scholar]
- 36.Lee NE, Taylor MM, Bancroft E, et al. Risk factors for community-associated methicillin-resistant Staphylococcus aureus skin infections among HIV-positive men who have sex with men. Clin Infect Dis. 2005;40:1529–34. doi: 10.1086/429827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eady EA, Cove JH. Staphylococcal resistance revisited: community-acquired methicillin resistant Staphylococcus aureus—an emerging problem for the management of skin and soft tissue infections. Curr Opin Infect Dis. 2003;16:103–24. doi: 10.1097/00001432-200304000-00007. [DOI] [PubMed] [Google Scholar]
- 38.Chambers HF. The changing epidemiology of Staphylococcus aureus? Emerg Infect Dis. 2001;7:178–82. doi: 10.3201/eid0702.010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salgado CD, Farr BM, Calfee DP. Community-acquired methicillin-resistant Staphylococcus aureus: a meta-analysis of prevalence and risk factors. Clin Infect Dis. 2003;36:131–9. doi: 10.1086/345436. [DOI] [PubMed] [Google Scholar]
- 40.Naimi TS, LeDell KH, Boxrud DJ, et al. Epidemiology and clonality of community-acquired methicillin-resistant Staphylococcus aureus in Minnesota, 1996–1998. Clin Infect Dis. 2001;33:990–6. doi: 10.1086/322693. [DOI] [PubMed] [Google Scholar]
- 41.Shahin R, Johnson IL, Jamieson F, McGeer A, Tolkin J, Ford-Jones EL. Methicillin-resistant Staphylococcus aureus carriage in a child care center following a case of disease. Toronto Child Care Center Study Group. Arch Pediatr Adolesc Med. 1999;153:864–8. doi: 10.1001/archpedi.153.8.864. [DOI] [PubMed] [Google Scholar]
- 42.McManus R. CDC's Gerberding Warns of Anti-Microbial-Resistant Infections. http://nihrecord.od.nih.gov/newsletters/03_02_2004/story01.htm. Accessed 4 March 2012. [Google Scholar]
- 43.Baggett HC, Hennessy TW, Leman R, et al. An outbreak of community-onset methicillin-resistant Staphylococcus aureus skin infections in southwestern Alaska. Infect Control Hosp Epidemiol. 2003;24:397–402. doi: 10.1086/502221. [DOI] [PubMed] [Google Scholar]
- 44.Macario E, Daum RS, Eells SJ, Bradburn N, Miller LG. Using cognitive interviews to refine a household contacts survey on the epidemiology of community-associated methicillin resistant Staphylococcus aureus. J Infect Prev. 2010;11:44–8. [Google Scholar]
- 45.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. Spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42:792–9. doi: 10.1128/JCM.42.2.792-799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961–7. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyle-Vavra S, Ereshefsky B, Wang CC, Daum RS. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J Clin Microbiol. 2005;43:4719–30. doi: 10.1128/JCM.43.9.4719-4730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 50.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–83. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 51.Minnesota Department of Health. Community-associated methicillin-resistant Staphylococcus aureus in Minnesota. MDH Dis Control Newsl. 2004;32:61–72. [Google Scholar]
- 52.Kleinbaum DG, Klein M. Statistics for biology and health 2002. Springer-Verlag, New York: Logistic regression a self-learning text. [Google Scholar]
- 53.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–87. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller LG, Diep BA. Colonization, fomites, and virulence: rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:742–50. doi: 10.1086/526773. [DOI] [PubMed] [Google Scholar]
- 55.Crum NF, Lee RU, Thornton SA, et al. Fifteen-year study of the changing epidemiology of methicillin-resistant Staphylococcus aureus. Am J Med. 2006;119:943–51. doi: 10.1016/j.amjmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 56.Lu D, Holtom P. Community-acquired methicillin-resistant Staphylococcus aureus, a new player in sports medicine. Curr Sports Med Rep. 2005;4:265–70. doi: 10.1097/01.csmr.0000306220.17928.7c. [DOI] [PubMed] [Google Scholar]
- 57.Centers for Disease Control and Prevention. Methicillin-resistant Staphylococcus aureus infections in correctional facilities—Georgia, California, and Texas, 2001–2003. MMWR Morb Mortal Wkly Rep. 2003;52:992–6. [PubMed] [Google Scholar]
- 58.Williams RE. Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol Rev. 1963;27:56–71. doi: 10.1128/br.27.1.56-71.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casewell MW. The nose: an underestimated source of Staphylococcus aureus causing wound infection. J Hosp Infect. 1998;40(suppl B):S3–11. doi: 10.1016/s0195-6701(98)90199-2. [DOI] [PubMed] [Google Scholar]
- 60.Safdar N, Bradley EA. The risk of infection after nasal colonization with Staphylococcus aureus. Am J Med. 2008;121:310–15. doi: 10.1016/j.amjmed.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 61.Hill RL, Duckworth GJ, Casewell MW. Elimination of nasal carriage of methicillin-resistant Staphylococcus aureus with mupirocin during a hospital outbreak. J Antimicrob Chemother. 1988;22:377–84. doi: 10.1093/jac/22.3.377. [DOI] [PubMed] [Google Scholar]
- 62.Beigi R, Hanrahan J. Staphylococcus aureus and MRSA colonization rates among gravidas admitted to labor and delivery: a pilot study. Infect Dis Obstet Gynecol. 2007;2007:70876. doi: 10.1155/2007/70876. [DOI] [PMC free article] [PubMed] [Google Scholar]