Abstract
TM0077 from Thermotoga maritima is a member of the carbohydrate esterase family 7 and is active on a variety of acetylated compounds, including cephalosporin C. TM0077 esterase activity is confined to short-chain acyl esters (C2-C3), and is optimal around 100°C and pH 7.5. The positional specificity of TM0077 was investigated using 4-nitrophenyl-β-D-xylopyranoside monoacetates as substrates in a β-xylosidase-coupled assay. TM0077 hydrolyzes acetate at positions 2, 3 and 4 with equal efficiency. No activity was detected on xylan or acetylated xylan, which implies that TM0077 is an acetyl esterase and not an acetyl xylan esterase as currently annotated. Selenomethionine-substituted and native structures of TM0077 were determined at 2.1 Å and 2.5 Å resolution, respectively, revealing a classic α/β-hydrolase fold. TM0077 assembles into a doughnut-shaped hexamer with small tunnels on either side leading to an inner cavity, which contains the six catalytic centers. Structures of TM0077 with covalently bound phenylmethylsulfonyl fluoride (PMSF) and paraoxon were determined to 2.4 Å and 2.1 Å, respectively, and confirmed that both inhibitors bind covalently to the catalytic serine (Ser188). Upon binding of inhibitor, the catalytic serine adopts an altered conformation, as observed in other esterase and lipases, and supports a previously proposed catalytic mechanism in which this Ser hydroxyl rotation prevents reversal of the reaction and allows access of a water molecule for completion of the reaction.
Keywords: Acetyl esterase, Thermotoga maritima, crystal structure, α/β hydrolase, inhibitor, serine rotation
INTRODUCTION
Thermotoga maritima is a hyperthermophilic bacterium that grows optimally at 80°C and is able to metabolize a variety of simple and complex carbohydrates, including glucose, sucrose, starch, cellulose, and xylan 1. Its carbohydrate utilization potential was confirmed by analysis of its sequenced genome 2. The xylan degrading pathway of T. maritima has been studied using microarrays 2–4, and several genes encoding transporters, xylanases, and a β-xylosidase have been identified. Among the enzymes with a differential expression pattern in the microarray was a predicted acetyl xylan esterase (locus tag TM0077, axeA) 3,5. Depending on the source, the xylan backbone may contain a varying degree of acetylated xylose residues. Therefore, in addition to xylanases and xylosidases, the complete degradation of xylan requires esterases/deacetylases 6.
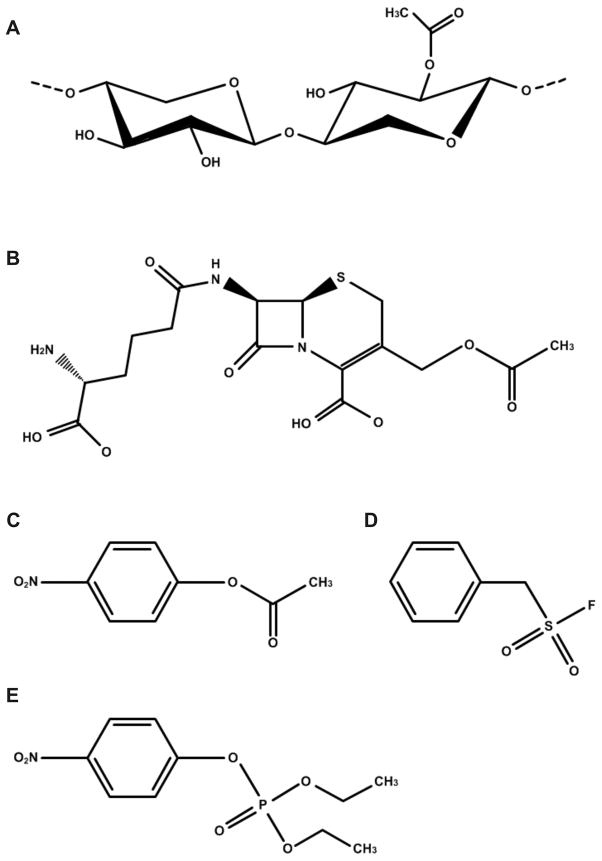
Presently, esterases and deacetylases that are active on carbohydrate substrates have been classified into 16 families by Henrissat and coworkers (Carbohydrate-Active enZymes Server (CAZy)) 7. According to this classification, the predicted acetyl xylan esterase from T. maritima would be a member of family 7 of the carbohydrate esterases (CE7). In addition to the acetyl xylan esterase activity, enzymes in the CE7 family are rather unusual in that they display a high specific activity towards the antibiotic cephalosporin C [(Fig. 1(a-b)] 8. Cephalosporins belong to the β-lactam class of antibiotics, which also includes penicillin, and affect bacterial cell growth by inhibiting the penicillin-binding-protein that cross-links peptide glycans required for cell wall formation 9. The production of deacetylated cephalosporins is of great interest because these compounds are valuable building blocks for the production of semi-synthetic β-lactam antibiotics10,11.
Figure 1.
Substrates and inhibitors of the CE7 family of enzymes. Structures of (A) acetylated xylooligosaccharide, (B) cephalosporin C, (C) p-nitrophenyl-acetate, (D) phenylmethylsulfonyl fluoride (PMSF), and (E) paraoxon.
To explore the catalytic capacity of the predicted acetyl xylan esterase from T. maritima and gain a better insight into the structure and function of the family 7 carbohydrate esterases, TM0077 was expressed and purified, and three-dimensional structures of the native enzyme and its complexes with phenylmethylsulfonyl fluoride (PMSF) and paraoxon inhibitors, were determined by x-ray crystallography. Furthermore, the enzyme was functionally characterized, and various biochemical properties including the positional specificity of the esterase were investigated.
MATERIALS AND METHODS
Gene cloning
TM0077 was selected as part of the Joint Center for Structural Genomics (JCSG) effort on complete structural coverage of the T. maritima soluble proteome as a large-scale center for high-throughput structure determination funded under the NIHGMS Protein Structure Initiative (PSI) 12. The gene encoding TM0077 (GenBank: AAD35171.1, GI:4980565; SwissProt: Q9WXT2) was amplified by polymerase chain reaction (PCR) from genomic DNA using PfuTurbo DNA polymerase (Stratagene) and primers corresponding to the predicted 5′ and 3′ ends. The PCR product was cloned into plasmid pMH1, which encodes an expression and purification tag (MGSDKIHHHHHH) at the amino terminus of the protein. The cloning junctions were confirmed by DNA sequencing.
TM0077-SeMet protein production and purification
Protein production was performed in a selenomethionine-containing medium using the Escherichia coli methionine auxotrophic strain DL41. Expression was induced by the addition of 0.15% L-arabinose. At the end of fermentation, cells were harvested and subjected to one freeze/thaw cycle, and subsequently sonicated in Lysis Buffer [50 mM Tris pH 7.9, 50 mM NaCl, 1 mM MgCl2, 0.25 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 1 mg/ml lysozyme] and the lysate was centrifuged at 3,400 × g for one hour. The soluble fraction was applied to nickel-chelating resin (GE Healthcare) pre-equilibrated with Equilibration Buffer [50 mM potassium phosphate pH 7.8, 300 mM NaCl, 10% (v/v) glycerol, 0.25 mM TCEP] containing 20 mM imidazole. The resin was washed with Equilibration Buffer containing 40 mM imidazole, and the protein was eluted with Elution Buffer [20 mM Tris pH 7.9, 300 mM imidazole, 10% (v/v) glycerol, 0.25 mM TCEP]. The eluate was buffer exchanged with Buffer Q [20 mM Tris pH 7.9, 5% (v/v) glycerol, 0.25 mM TCEP] containing 50 mM NaCl and applied to a RESOURCE Q column (GE Healthcare) pre-equilibrated with the same buffer. The protein was eluted using a linear gradient of 50 to 500 mM NaCl in Buffer Q and purified further with a HiLoad 16/60 Superdex 200 column (GE Healthcare), using Crystallization Buffer [20 mM Tris pH 7.9, 150 mM NaCl, 0.25 mM TCEP] as the mobile phase. For crystallization trials, the peak Superdex 200 fractions were concentrated to ~15 mg/mL by centrifugal ultrafiltration (Millipore). Molecular weight and oligomeric state of TM0077 were determined using a 1 cm × 30 cm Superdex 200 column (GE Healthcare) coupled with miniDAWN static light scattering (SEC/SLS) and Optilab differential refractive index detectors (Wyatt Technology). The mobile phase consisted of 20 mM Tris pH 8.0, 150 mM NaCl, and 0.02% (w/v) sodium azide.
Native TM0077 production and purification
For protein production, E. coli DL41 cells were grown in LB medium for 8 hours (an OD600 well above 2.0 was reached). Subsequently, the culture was induced by adding 0.15% L-arabinose and incubated another 16 hours at 37°C. Cells were harvested by centrifugation at 10,000 × g for 20 min. The cell pellet was resuspended in 30 ml of Lysis Buffer 2 [50 mM Tris-HCl pH 8.0, 50 mM NaCl, 10 mM imidazole, 0.25 mM TCEP]. The cells were disrupted by two passages through a French press at 110 MPa. The crude cell extract was treated with DNAse I at room temperature for 30 min and subsequently centrifuged at 43,000 × g for 30 min in order to remove cell debris. The supernatant was heated at 70°C for 25 min and then centrifuged to remove the precipitated proteins. The supernatant was filtered and loaded onto a nickel-chelating column packed with 20 ml of Ni-NTA His-Bind Resin (Novagen) and equilibrated in 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 2% (v/v) glycerol, and 0.25 mM TCEP. The column was washed with 20 mM imidazole in the same buffer, and proteins were subsequently eluted with a linear gradient of 20–500 mM imidazole in the same buffer. Fractions containing esterase activity were pooled and loaded onto a HiPrep Desalting column (GE Healthcare) equilibrated with 20 mM Tris-HCl pH 8.0, 150 mM NaCl, and 0.25 mM TCEP. The homogeneity of the protein was checked by SDS-PAGE, and activity staining of the SDS-PAGE gel was performed using α-napthyl acetate, as described previously 13. The protein concentration was determined at 280 nm using a NanoDrop ND-1000 Spectrophotometer.
Crystallization
Crystals of selenomethionine-substituted TM0077 were obtained by hanging drop vapor diffusion against a 250 μl crystallization solution consisting of 20% (w/v) PEG-3000, 0.1 M HEPES pH 7.5, 0.2 M NaCl. Drops consisted of 0.5 μl protein and 0.5 μl crystallization solution. Native TM0077 was crystallized using nanodrop vapor diffusion techniques against a crystallization solution consisting of 0.2 M calcium acetate hydrate, 20% (w/v) PEG 3350, pH 7.3 at 20°C. Protein was concentrated to 22.8 mg/ml. Drops consisted of 100 nl protein and 100 nl of crystallization solution and a 60 μl reservoir of crystallization solution. Crystals of TM0077 in complex with inhibitors PMSF or paraoxon were obtained at 4°C in the same conditions with the same reagents as the native crystals. PMSF or paraoxon were added in a molar ratio of 1:3 (protein:inhibitor).
Data collection
For cryoprotection, the TM0077-SeMet crystal was transferred to crystallization solution supplemented with 15% (v/v) glycerol. The crystal was mounted in a cryoloop and subsequently flash-cooled in liquid nitrogen. X-ray data were collected at 100 K on beamline BL9-2 at the Stanford Synchrotron Radiation Lightsource (SSRL, Menlo Park, CA) using a Quantum 4 CCD detector (ADSC). A TM0077-SeMet MAD data set was collected to 2.1 Å resolution and the data were indexed in monoclinic space group P21, with unit cell parameters a = 152.6 Å, b = 131.0 Å, and c = 157.8 Å, and β=118.9°, and 12 molecules in the asymmetric unit. Data were indexed and integrated with DENZO and then scaled with SCALEPACK 14.
Native TM0077, TM0077-PMSF complex (TM0077-PMS) and TM0077-paraoxon complex (TM0077-DEP) crystals were transferred to crystallization solution supplemented with 10% (v/v) ethylene glycol and flash-cooled to 100K. Data were collected at beamline 5.0.3 of the Advanced Light Source (ALS, Berkeley, CA) and processed with the HKL2000 package 14. The native data set was collected to 2.5 Å resolution, and TM0077-PMS and TM0077-DEP data sets were collected to 2.4 and 2.1 Å, respectively. All data were indexed in orthorhombic space group P212121, with unit cell parameters approximately a=103Å b=104Å c=221Å (See Table 1), and six molecules in the asymmetric unit. Data reduction and refinement statistics for TM0077-SeMet, TM0077-Native, TM0077-PMS and TM0077-DEP are summarized in Table I.
Table I.
Summary of crystal parameters, data collection, and refinement statistics
| TM0077-SeMet | TM0077-Native | TM0077-PMS | TM0077-DEP | ||
|---|---|---|---|---|---|
| Space group | P 21 | P 21 21 21 | P 21 21 21 | P 21 21 21 | |
| Unit cell parameters | a=152.64Å b=130.95Å c=157.82Å β=118.90° |
a=103.46Å b=103.79Å c=221.02Å |
a=103.57Å b=104.50Å c=221.61Å |
a=103.80Å b=104.43Å c=221.64Å |
|
| Data collection | λ1 MAD-Se | λ2 MAD-Se | |||
| Wavelength (Å) | 0.9791 | 0.9183 | 0.9765 | 0.9765 | 0.9765 |
| Resolution range (Å) | 29.6 – 2.10 | 29.6 – 2.10 | 48.8 – 2.50 | 49.0 – 2.40 | 49.0 – 2.12 |
| No. observations | 1,119,236 | 1,100,249 | 1,222,016 | 765,546 | 989,949 |
| No. unique reflections | 293,140 | 291,757 | 83,045 | 94,681 | 123,070 |
| Completeness (%) | 93.0 (61.8)a | 92.6 (60.8) | 100 (100) | 100 (100) | 89.8 (53.5) |
| Mean I/σ(I) | 9.1 (2.4)a | 9.6 (2.2) | 14.4 (2.9) | 11.5 (3.4) | 15.3 (2.2) |
| Rmerge on I (%) | 12.3 (52.5) a | 11.9 (57.9) | 20.7 (109.7) c | 18.0 (67.4) | 9.5 (51.9) |
| Rmeas on I (%) | 14.3 (62.2) a | 13.9 (68.7) | 21.4 (113.6) | 19.2 (71.9) | 10.2 (60.2) |
| Rpim on I (%) | 7.2 (32.7) a | 7.1 (36.2) | 5.5 (29.2) | 6.7 (24.9) | 3.5 (29.2) |
| Highest resolution shell (Å) | 2.15 – 2.10 | 2.15 – 2.10 | 2.56 – 2.50 | 2.46 – 2.40 | 2.18 – 2.12 |
| Model and refinement statistics | |||||
| Resolution range (Å) | 29.6 – 2.10 | 48.8 – 2.50 | 49.0–2.40 | 49.0 – 2.12 | |
| No. reflections (total) | 293,097 b | 83,045 | 94,680 | 122,994 | |
| No. reflections (test) | 14,726 | 4,200 | 4,742 | 6,188 | |
| Completeness (% total) | 92.8 | 100.0 | 100.0 | 89.8 | |
| Data set used in refinement | λ1 MAD-Se | ||||
| Cutoff criteria | |F| > 0 | |F| > 0 | |F| > 0 | |F| > 0 | |
| Rcryst | 0.186 | 0.167 | 0.160 | 0.167 | |
| Rfree | 0.223 | 0.212 | 0.208 | 0.205 | |
| Stereochemical parameters | |||||
| Restraints (RMSD observed) | |||||
| Bond angle (°) | 1.48 | 1.47 | 1.53 | 1.44 | |
| Bond length (Å) | 0.018 | 0.017 | 0.017 | 0.015 | |
| Av. isotropic B-value (Å2) | 27.9 | 24.7 | 19.4 | 19.6 | |
| ESU based on Rfree | 0.17 | 0.25 | 0.22 | 0.18 | |
| Water molecules/other solvent molecules | 2,464/1 | 507/24 | 946/17 | 987/23 | |
| PDB ID | 1vlq | 3m81 | 3m82 | 3m83 | |
Highest resolution shell
Rmerge=ΣhklΣi|Ii(hkl)-<I(hkl)>|/ΣhklΣiIi(hkl), Rmeas(redundancy-independent Rmerge)=Σhkl[Nhkl/(Nhkl-1)]1/2Σi|Ii(hkl)-<I(hkl)>|/ΣhklΣiIi(hkl), and Rpim(precision-indicating Rmerge)=Σhkl[1/(Nhkl-1)]1/2Σi|Ii(hkl)-<I(hkl)>|/ΣhklΣiIi(hkl) 67–69.
Rcryst = Σ| |Fobs|-|Fcalc| |/Σ|Fobs| where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rcryst, but for 5.0 % of the total reflections chosen at random and omitted from refinement.
Typically, the number of unique reflections used in refinement is slightly less than the total number that were integrated and scaled. Reflections are excluded due to systematic absences, negative intensities, and rounding errors in the resolution limits and cell parameters.
Rmerge of the highest resolution shell is high due to high redundancy (14.7). However, the completeness and mean I/σ of the highest resolution shell are reasonable, and these data were included in the refinement.
Structure solution and refinement
The TM0077-SeMet structure was solved by MAD phasing method using a two-wavelength MAD dataset. At the time of the initial data collection (2001), the structure determination of Se-MAD TM0077 posed a significant challenge to crystallographic programs, which were still under active development. As a result, modifications were made in various structure determination and refinement programs to achieve success. For initial phasing, SHELXD 15 was used to find candidate SeMet substructure sites. Attempts to complete phasing were unsuccessful due to the translational non-crystallographic symmetry (NCS) (not recognized initially). Self-consistent sets (partial sets) were found using the CCP4 program PROFESSS 16 and additional SeMet sites were found by SHELXD, and added to these partial sets. The SHARP 17 run did not complete initially; however, updates of SHARP and ARP/wARP eventually helped to resolve issues and an initial trace was obtained by ARP/wARP. The structure was then refined with BUSTER 18 using tight NCS restraints to an Rcryst and Rfree of 18.6% and 22.3%, respectively. Model building was performed using O 19 and the structure was refined using Refmac5 20. Refinement statistics are summarized in Table I. The final model contains 12 protein molecules (chains A-L) in the asymmetric unit each consisting of residues 2-323. The MolProbity 21 Ramachandran plot analysis showed that 97.4% of all residues are in favored regions with a single outlier, Gln120 of chain B, which is supported by unambiguous electron density. Ramachandran outlier Gln120 of chain B of TM0077-SeMet is due to crystal packing with chain C. The backbone carbonyl oxygens of Gln120 and Gly119 of chain B makes hydrogen bonds with the backbone nitrogen of Gln140 of chain C (3.19 and 3.11 Å, respectively).
The native TM0077 structure, TM0077-PMS and TM0077-DEP structures were solved by molecular replacement using PHASER 22,23 with the TM0077-SeMet hexamer coordinates (pdb: 1vlq; A-F chains) as a search model. One hexamer was successfully located and the structure was further refined with Refmac5 20 using tight NCS restraints to an Rcryst and Rfree of 16.7% and 21.2% (native TM0077), 16.0% and 20.8% (TM0077-PMS) and 16.7% and 20.5% (TM0077-DEP), respectively. Iterative cycles of refinement and building were performed with Refmac5, Phenix 24,25 and Coot 26. All other crystallographic manipulations were carried out with the CCP4 package 16. Refinement statistics are summarized in Table I. The final model of native TM0077 contains residues 3-324 (chains A, B, C, D and F) and 3-323 (chain E) in the asymmetric unit. Analysis of main-chain torsion angles using MolProbity 21 showed that 97.8% of the residues are in favored regions of the Ramachandran plot with 0.2% outliers (Asn302 of chains B, C and D), which are supported by unambiguous electron density. The final model of TM0077-PMS contains residues 3-323 for all chains in the asymmetric unit with 97.5% of the residues in favored regions with 0.2% outliers (Asn302 of chains A, B, D and F). The final model of TM0077-DEP contains residues 0-324 for (chains A, B, C and F) and 0-323 (chains D and E) in the asymmetric unit, respectively, with 97.6% of the residues in favored region of the Ramachandran plot with 0.2% outliers (Asn302 of B, C, D and F chains). Ramachandran outlier Asn302 in the TM0077-Native, TM0077-PMS and TM0077-DEP structures is a neighbor to the catalytic triad residue His303 and may reflect a slightly different state for these structures compared to the Se-Met structure.
Structure validation and deposition
The quality of the crystal structure was analyzed using the JCSG Quality Control server (http://smb.slac.stanford.edu/jcsg/QC). This server processes the coordinates and data through a variety of validation tools including AutoDepInputTool 27 MolProbity 21, WHATIF 5.0 28, RESOLVE 29, MOLEMAN2 30 as well as several in-house scripts, and summarizes the results. Protein quaternary structure analysis were performed using the PISA server 30. Figures were prepared with PyMOL (DeLano Scientific) 31. RMSD values were calculated using the ProCKSI-Server 32. The structural data have been deposited in the RCSB Protein Data Bank (PDB) with accession codes 1vlq for TM0077-SeMet, 3m81 for TM0077-native, 3m83 for TM0077-DEP and 3m82 for TM0077-PMS.
Enzyme assays
Esterase activity was measured using p-nitrophenyl esters as described previously 13. Briefly, the standard assay consisted of activity measurements with 0.2 mM p-nitrophenyl acetate as substrate in 50 mM citrate-phosphate (pH 6) at 70°C. The p-nitrophenol liberated was measured continuously at 405 nm on a Hitachi U-2001 spectrophotometer with a temperature-controlled cuvette holder. Extinction coefficients of p-nitrophenol were determined prior to each measurement. Kinetic parameters were determined by direct fitting the data, obtained from multiple measurements, to the Michaelis–Menten curve (Tablecurve 2d, version 5.0).
The effect of pH on esterase activity was studied in the pH range from 5 to 10. The buffers used were 50 mM citrate-phosphate (pH 5–8) and 50 mM CAPS (3-(cyclohexylamino) 1-propanesulphonic acid) (pH 9.5–10). The pH of the buffers was set at room temperature, and temperature corrections were made using their temperature coefficients: −0.0028 pH/°C for citrate-phosphate buffer and −0.018 pH/°C for CAPS buffer. The effect of temperature on esterase activity was studied in the range of 40–100°C using 0.2 mM p-nitrophenyl acetate as substrate. Enzyme thermostability was determined by incubating the enzyme in a 50 mM Tris-HCl, 150 mM NaCl (pH 7.8) buffer at 90°C and 100°C for various time intervals. Residual activity was determined in the standard assay.
Inhibition kinetics of PMSF and paraoxon were determined as described for the acetylcholinesterase from electric eel 33. All experiments were performed at 70°C in 50 mM citrate-phosphate (pH 6) buffer and 0.2 mM p-nitrophenyl acetate as substrate. The kinetic constants for the inhibition of TM0077 with PMSF and paraoxon were measured in the concentration range of 1.0–10.0 mM and 0.2–1.0 mM, respectively.
Deacetylase activity was determined using high-performance liquid chromatography (HPLC) by measuring the amount of acetic acid released from the substrates cephalosporin C, 7-aminocephalosporanic acid, glucose-pentaacetate and acetylated xylan. Xylan was acetylated by the method described by Johnson 34. The reaction mixture contained 0.9 ml of substrate solution (dissolved in 50 mM Tris-HCl, pH 7.5) and 0.1 ml of enzyme solution, and was incubated at 37°C for various time intervals (0–10 min). The reaction was stopped by adding 0.2 ml of stop solution (100 mN H2SO4 and 30 mM crotonate) and placing the sample on ice. The conditions for HPLC were as follows: column, KC811 Shodex; detection, RI and UV detectors; solvent, 3 mN H2SO4; flow rate, 1.5 ml/min; temperature, 30°C; internal standard, crotonate. One unit of enzyme activity was defined as the amount of enzyme that releases one μmol of acetic acid per minute.
Activity on xylan was measured quantitatively using DMSO-extracted xylan (1% polysaccharide solution in 0.1 M sodium phosphate buffer pH 6) at 60°C 35. Xylan will precipitate as a consequence of deacetylation, resulting in a rapid turbidity of the solution.
Positional specificity assay
The positional specificity of TM0077 was investigated using an enzyme-coupled assay on monoacetylated 4-nitrophenyl β-D-xylopyranosides (pNP-Xyl) as described 36. The β-xylosidase XloA (locus tag: TM0076) from T. maritima was cloned into the vector pET24d in frame with a C-terminal His6-tag. The enzyme was expressed and purified as described above for native TM0077. Activity of XloA was confirmed by measuring the release of p-nitrophenol at 405 nm from the substrate 4-nitrophenyl β-D-xylopyranoside.
The enzyme-coupled assay was performed at 60°C in a total volume of 125 μl, which contained 0.1 M sodium phosphate (pH 6 or 7), 2-O-, 3-O-, or 4-O-acetyl pNP-Xyl, the β-xylosidase XloA, and TM0077. Stable 50x-concentrated stock solutions of the substrates were prepared in DMSO. The reaction was started by addition of 2.5 μl of a stock solution to a preheated reaction mixture consisting of phosphate buffer, auxiliary β-xylosidase XloA in excess, and TM0077. The reaction was terminated by addition of 800 μl of a 2% solution of Na2CO3. Liberated p-nitrophenol was determined at 405 nm against substrate and enzyme blanks. A short incubation time for activity determination was used to suppress acetyl migration on the xylopyranosyl-ring, which is significant at pH 6 or 7 37. The kinetic constants were determined at pH 7 and 60°C with reaction times of 2 or 5 minutes.
RESULTS and DISCUSSION
In silico analysis
TM0077 consists of 325 amino acids with a calculated molecular mass of 37 kDa. Sequence analysis, using the SignalP 3.0 server, revealed that TM0077 has no predicted signal sequence and is, therefore, believed to be an intracellular enzyme. Analysis of the gene organization indicates that the TM0077 gene co-localizes with genes encoding a xylanase (TM0070) 38, ABC transporter components (TM0071-TM0075), and a β-xylosidase (TM0076) 39. BLAST-P analysis showed that TM0077 has highest similarity to putative acetyl esterases, acetyl xylan esterases and cephalosporin C deacetylases. Among the BLAST results, a predicted acetyl xylan esterase-related protein from T. maritima (locus tag: TM0435) was also identified. TM0077 was compared with other members of the CE7 family using structure-based, multi-sequence alignment and the putative catalytic triad, Ser188, Asp274, and His303, was identified from conservation throughout the analyzed sequences. The putative nucleophilic serine (Ser188) is located within a conserved pentapeptide consensus sequence, Gly-Xaa-Ser-Gln-Gly, typical of this family. Previously, a signature sequence motif, [RGQ]-(x:~70)-[GxSQG]-(x:~115)-[HE] (where x indicates any amino acid), had been suggested for the CE7 family based on an aminoacid alignment of 12 sequences 40. In an updated alignment consisting now of 50 sequences, we observed many sequences that have this signature motif, but it is not conserved throughout the entire family (See Supporting Information and Fig. S1 for the multi-sequence alignment).
Overall structure
The crystal structure of seleno-methionine incorporated TM0077 (TM0077-SeMet) was determined to 2.1 Å resolution by multi-wavelength anomalous dispersion (MAD) (Table I) with twelve molecules per asymmetric unit. A native apo structure (TM0077-Native) was determined in a different space group (see Methods) to 2.5 Å by molecular replacement, using TM0077-SeMet as a search model, with six molecules in the asymmetric unit (Table I). Each monomer of the native hexamer contained a calcium ion (see below) bound by Lys22, Glu26, and Asp25 via a bridging water molecule. Superposition of the TM0077-SeMet and the TM0077-Native structures gave a root-mean-square difference (RMSD) of 0.12 Å over 321 Cα atoms, which indicates that these structures are nearly identical as expected.
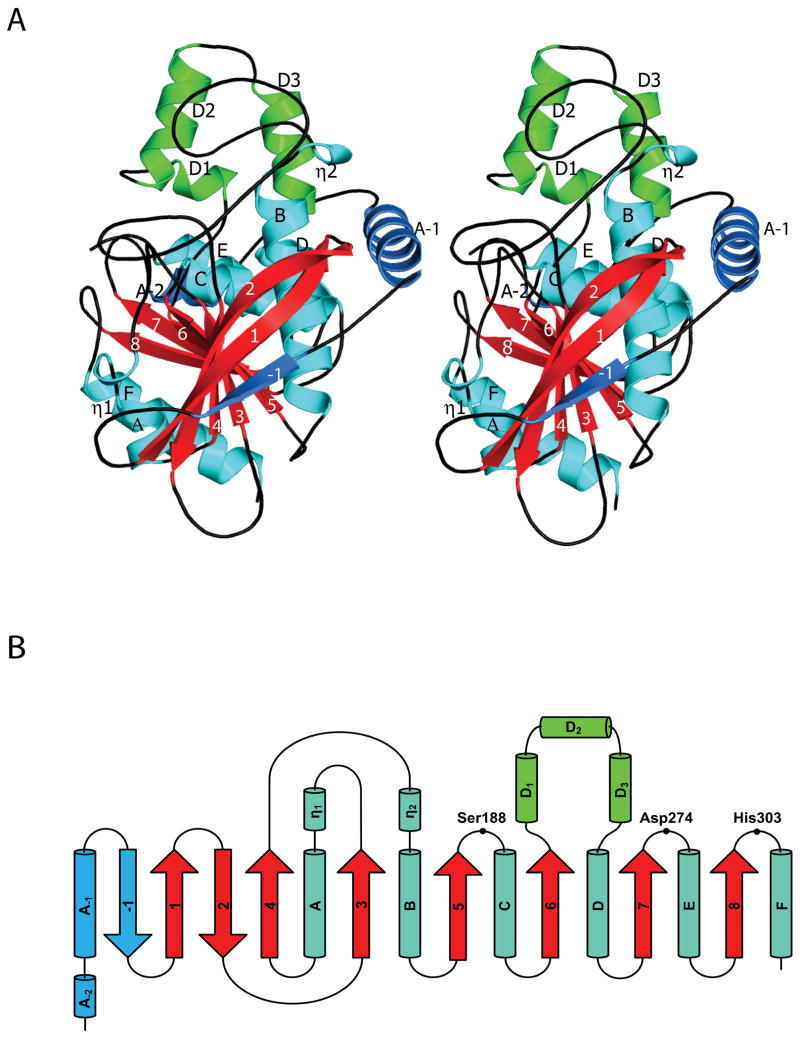
In general, the TM0077 structure resembles the canonical α/β-hydrolase fold, which consists of a central, twisted, eight-stranded β-sheet surrounded by α-helices on both sides, with β2 antiparallel to the other strands. TM0077 deviates slightly from the canonical α/β-hydrolase fold at two locations: a three-helix insertion after strand β6 and an extension of the N-terminus (Fig. 2). Insertions after β6 or β7 are common for α/β-hydrolases and are proposed to help shape the substrate-binding site 41. The N-terminus is extended by two helices (αA-1 and αA-2) and an antiparallel β-strand (β-1) that aligns with the other eight β-strands (β1-β8) and extends the central β-sheet. This nine-stranded β-sheet is highly twisted, and β-1 and β8 at the extreme edges are rotated approximately 130° relative to each other. Helices αA and αB both contain a short 310-helix segment at their N-terminus. Helices αA-1, αA-2, αB, αC, αD, αD1, αD2, αD3, αE, and the 310-helix η2 are located on one side of the central β-sheet, and helices αA, αF and the 310-helix η1 are on the other side.
Figure 2.
Overall fold and topology of TM0077. (A) Stereo view of a TM0077 protomer. The β-strands are labeled numerically (-1 to 8) with the core strands in red, α-helices are labeled alphabetically (A-2 to F) and 310-helices are labeled with an Eta (η1 and η2) with the core helices in cyan. The three-helix insertion after β6 is colored green and the N-terminal extension is colored sky blue. The figure was generated using Pymol 31. (B) Topology diagram of TM0077, with the helices displayed as cylinders and the strands displayed as arrows following the color and label scheme of (A). The location of residues forming the catalytic triad is also indicated.
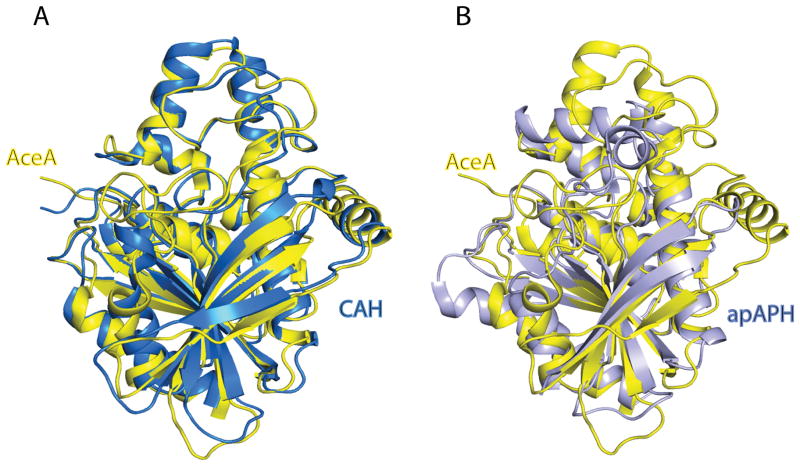
A structural similarity search was performed using the program DALI 42. Monomer A of the TM0077-SeMet structure was used as a search model and similarity was found with cephalosporin C deacetylases, acetyl xylan esterases, acylamino-releasing enzymes, dipeptidyl peptidases and some esterases and lipases. TM0077 is structurally most similar to cephalosporin C deacetylase (CAH) from B. subtilis (PDB: 1ods) 40, acetyl xylan esterase (AXE) from B. pumilus (PDB: 3fvr and 2xlb), acetyl xylan esterase (AXE1) from Thermoanaerobacterium sp. JW/SL YS485 (PDB: 3fcy), and acylpeptide hydrolase/esterase apAPH from Aeropyrum pernix K1 (PDB: 1ve6) 43. The sequence identity between TM0077 and CAH is 41% and the two structures align with a Z-score of 46 and an RMSD of 1.5 Å over 312 Cα atoms. The sequence identity with apAPH is 17% with a Z-score of 23.3 and an RMSD of 2.3 Å over 230 Cα atoms. Superpositions of TM0077 with CAH and with apAPH are shown in Fig. 3.
Figure 3.
Structural superposition of TM0077 with structurally related esterases. Superposition of TM0077 (yellow) with (A) the cephalosporin C deacetylase (CAH) from B. subtilis (PDB: 1ods; blue) 40 and (B) the α/β-hydrolase domain of the acylpeptide hydrolase/esterase apAPH from A. pernix K1 (PDB: 1ve6; grey) 43.
Quaternary structure
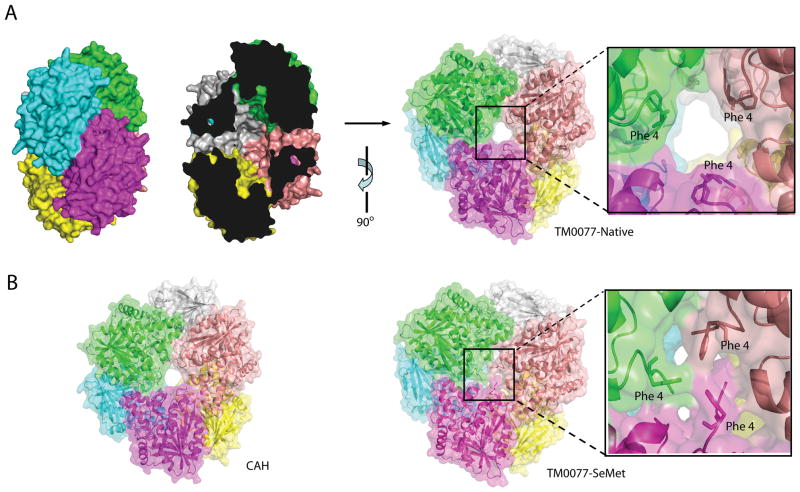
The crystal structure of TM0077-SeMet contains two hexamers in the asymmetric unit that are related by a non-crystallographic two-fold axis. Each hexamer contains a dimer of trimers with a back-to-back arrangement (Fig. 4). The apo and the complex crystals contained one hexamer in the asymmetric unit. Crystallographic packing analysis using PISA (EBI) 44 indicated that the relevant physiological oligomeric state of TM0077 is a hexamer, which was confirmed by size exclusion chromatography coupled with static light scattering. Further analyses of the hexameric assembly indicated that two main interfaces play an essential role in complex formation. The first interface between subunit A and B (green and cyan in Fig. 4) (identical to C/D and E/F) is stabilized by seven hydrogen bonds on average and has a buried surface area of 1024 Å2 contributed by each chain. The second interface between A and F (green and purple in Fig. 4) (B/C and D/E) is stabilized by 17 hydrogen bonds on average with a buried surface area of 1079 Å2 contributed by each chain However, a multiple sequence alignment of TM0077 with other CE7 esterases showed that the residues involved in these two main interfaces are not conserved. Other secondary interfaces bury around 514 Å2 contributed by each chain. The hexamer has a total buried surface area of 18,860 Å2, which is approximately 30% of the total surface area. Approximately 3,143 Å2 per monomer is, therefore, buried upon complex formation.
Figure 4.
TM0077 oligomeric assembly. (A) Surface representation of the biological unit of the TM0077-Native hexamer with each monomer in a different color (left). The “cross section” shows the entrances on either side of the assembly and the internal cavity (center), and a 90° rotated view of the TM0077-Native hexamer, with a close-up view of the open central hole (right). (B) Surface representation of the biological hexamer unit of CAH from B. subtilis 40 (left) and the TM0077-SeMet hexamer with a close-up view of the blocked central hole (right).
The TM0077 hexamer has a doughnut-shape when viewed from the side, with the six active sites located in the interior of the complex, where they line an oval-shaped cavity [Fig. 4(a)]. This cavity is accessible via two entrances, one on each side of the flat hexamer. Each of these entrances is approximately 13 Å wide and connects to a short tunnel or pore spanning approximately 10 Å to reach the inner cavity. Interestingly, in the TM0077-SeMet hexamer, the entrance to the internal cavity is blocked by three phenylalanine residues (Phe4), one for each of three monomers that compose half of the hexamer [Fig. 4(b)]. Residue Phe4 is located in the mobile N-terminus (high B-values), which may indicate some flexibility or multiple conformations.
Calcium ions were identified, by the electron density and coordination geometry, supported by their presence in the crystallization reagents, in the native TM0077, TM0077-PMS and TM0077-DEP structures, but not in the TM0077-SeMet structure. The SeMet protein was crystallized without any calcium in the crystallization reagents. In each subunit of the hexamer, one calcium ion is located at the N-terminal region of helix αA-1, and is coordinated by the backbone carbonyl of Lys22 and the Glu26 carboxylate. The remainder of the calcium coordination sphere is filled with waters from a neighboring solvent channel present in all molecules in the asymmetric unit. The Asp25 carboxylate contributes to the calcium binding via one of the coordinating water molecules. Another calcium ion is bound in a crystal packing interface between chain A and chain C′ of a crystallographic symmetry-related hexamer. This calcium is coordinated by the carboxylates of GluA45 and AspA58 from one chain and the carboxylate from Glu C’45 (bidentate coordination) of the symmetry-related chain with three water molecules completing a capped-octahedral coordination sphere. An equivalent calcium binding site is also observed in the crystal packing interface between chains D and B′. No significant increase or reduction of activity of TM0077 was observed in the presence of calcium ions or EDTA. Therefore, it seems that these calcium ions are not important for activity. On the other hand, calcium may help stabilize the structure. No calcium was present in the B. subtilis CAH structure 40; however, Lys22, Glu26 and Ser25 are conserved and may also act as a calcium binding site.
Enzyme activity
The activity of TM0077 was investigated using p-nitrophenol esters with varying acyl-chain length, ranging from C2 to C18. TM0077 is only active on the short-chain p-nitrophenol esters of acetate and propionate and does not hydrolyze esters with acyl chains longer than four carbons. No significant difference was found in the catalytic efficiency (kcat/Km) for the hydrolysis of p-nitrophenyl with acyl chains containing 2 to 3 carbons (Table II) [Fig. 1(c)].
Table II.
Kinetic parameters for hydrolysis of various esters
| Ester | Km (mM) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|
| pNP-Acetate | 0.185 ± 0.026 | 57.5 ± 2.2 | 310.8 ± 45.3 |
| pNP-Propionate | 0.137 ± 0.013 | 41.3 ± 1.1 | 301.5 ± 29.7 |
| 2-O-acetyl pNP-Xyl | 3.6 ± 0.5 | 76.1 ± 19.2 | 21.1 ± 6.1 |
| 3-O-acetyl pNP-Xyl | 4.2 ± 0.4 | 70.1 ± 7.7 | 16.7 ± 2.4 |
| 4-O-acetyl pNP-Xyl | 4.0 ± 0.1 | 78.6 ± 12.9 | 19.7 ± 3.3 |
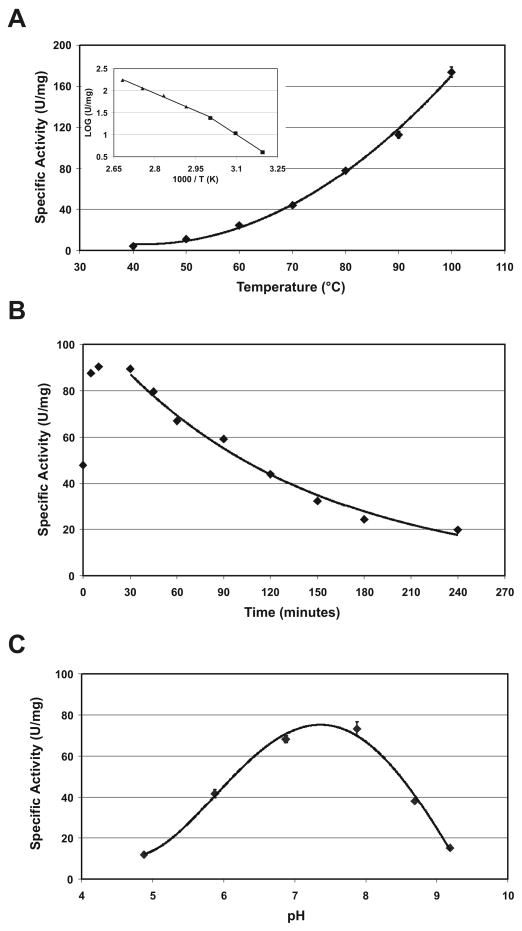
The effect of temperature on activity was studied using p-nitrophenyl acetate as substrate. The esterase activity increased from 40°C upwards until 100°C [Fig. 5(a)]. An Arrhenius analysis resulted in linear plots in the temperature ranges of 40–60°C and 60–100°C [Fig. 5(a); inset], with calculated activation energies for the formation of the enzyme substrate-enzyme complex of 33.7 and 21.9 kJ/mol, respectively. The transition or break in linearity of the Arrhenius plot at 60°C (1000/T (K) = 3.0) could indicate some conformational change of the enzyme. TM0077 is fairly resistant to thermal inactivation. An approximate 50% transient increase in activity is seen during the first 10 to 20 minutes when the enzyme is incubated at 90°C. After 30 minutes, inactivation of function occurs by first order kinetics with a half-life of approximately 120 minutes [Fig. 5(b)]. A transient activation has also been observed for other thermophilic esterases, such from Sulfolobus shibatae 45, and it is believed that a high temperature is needed in order to obtain the optimal conformation for catalysis. TM0077 was not stable at 100°C, resulting in a half-life of less than 5 minutes. However, the optimum temperature and thermal stability of TM0077 are still considerably higher than those reported for other characterized CE7 esterases, including the Thermoanaerobacterium enzyme that has a temperature optimum of 80°C and a half-life of 1h at 75°C 46.
Figure 5.
Effect of temperature and pH on esterase activity. (A) The esterase activity was studied using pNP-C2 as a substrate at temperatures ranging from 40–100°C. The inset shows the temperature dependence as an Arrhenius plot. (B) Thermal stability of TM0077 at 90°C. (C) The effect of pH on esterase activity studied using pNP-C2 as a substrate at pH values in the range of 4.8–9.2.
The effect of pH on activity was measured in the pH range of 4.8 to 9.2 using the substrate p-nitrophenyl acetate. TM0077 displayed maximum activity at approximately pH 7.5 [Fig. 5(c)], which is comparable to other CE7 esterases, such as the acetyl xylan esterases from Thermoanaerobacterium sp. strain JW/SL-YS485 46.
Positional specificity
The positional specificity of TM0077 was tested on three monoacetates of 4-nitrophenyl β-D-xylopyranoside (pNP-Xyl). To determine the enzyme activity, the β-xylosidase XloA 39 (TM0076) from T. maritima is required as an auxiliary enzyme. This thermostable XloA enzyme was, therefore, cloned, heterologously expressed, purified to homogeneity, and its activity was confirmed by measuring release of p-nitrophenol from the substrate pNP-Xyl (data not shown). The β-xylosidase was not active on the three monoacetates of pNP-Xyl. In the XloA-coupled assay, TM0077 hydrolyzed acetate from positions 2, 3 and 4 of pNP-Xyl with similar catalytic efficiency. The results are summarized in Table II.
In addition, TM0077 was investigated for its ability to remove acetyl groups from 7-aminocephalosporanic acid (7-ACA), cephalosporin C, glucose penta-acetate, N-acetyl-D-glucosamine, xylan and acetylated xylan. TM0077 has no activity for acetylated and non-acetylated xylan polymers, indicating that it is, indeed, an acetyl esterase and not an acetyl xylan esterase. As expected for an acetyl esterase, TM0077 displayed high activity on glucose penta-acetate with a turnover number of 2680 s−1. Like other members of CE7, TM0077 was also able to hydrolyze the acetyl groups from both cephalosporin C and 7-ACA with a turnover number of 376 s−1 and 1140 s−1, respectively. TM0077 was not able to hydrolyze the acetyl group from N-acetyl-D-glucosamine, indicating that it is specific for ester bonds and unable to hydrolyze amide bonds.
Inhibitor assays and TM0077 structures complexed with PMSF and paraoxon
PMSF and paraoxon [Fig. 1(d,e)] are competitive irreversible inhibitors of esterases. Inhibition proceeds by the formation of a reversible Michaelis complex, followed by an irreversible step and inhibition can, therefore, be characterized by two parameters: a dissociation constant and a binding rate constant. The inhibition kinetics for paraoxon and PMSF were investigated in the presence of p-nitrophenyl acetate, as described previously 47, and the dissociation and rate constants were 0.5 ± 0.1 mM and 0.13 ± 0.02 s−1 for paraoxon, and 1.1 ± 0.2 mM and 0.020 ± 0.001 s−1 for PMSF, respectively. The acetyl xylan esterase from Bacillus pumilus (BpAxe) has slightly reduced sensitivity to paraoxon (dissociation and rate constants of respectively 5.4 mM and 0.012 s−1), likely due to steric hindrance of two tyrosine residues (Tyr91 and Tyr206) that hamper the binding of paraoxon. Although these residues are essentially conserved in TM0077 (Tyr92 and Phe213), TM0077 is more sensitive to paraoxon than BpAxe48. In comparison to EST2 of Alicyclobacillus acidocaldarius 49 and EstA of T. maritima 47, the TM0077 dissociation constant is slightly higher, but the rate constant is comparable. No significant stimulation or reduction of activity of TM0077 was observed in the presence of divalent metal ions or ethylenediaminetetraacetic acid (EDTA).
To obtain more information about inhibitor binding and any possible conformational changes during catalysis, TM0077 was co-crystallized with the inhibitors PMSF and paraoxon and the PMSF (TM0077-PMS) and paraoxon (TM0077-DEP) structures were determined to 2.4 Å and 2.1 Å, respectively (Table I). The electron density map of TM0077 with PMSF showed clear density for the PMSF covalent modification. The fluorine was cleaved from the PMSF molecule during the binding reaction and the phenylmethyl sulfonyl (PMS) moiety is covalently bound to the Oγ atom of Ser188. The native apo and PMS-bound structures superimpose well with RMSD’s of 0.09–0.11 Å over 320–321 Cα atoms. Electron density maps of the paraoxon-bound structure displayed clear density for a diethyl-phosphate moiety covalently bound to the Oγ atom of Ser188. This covalent modification indicates that the p-nitrophenol group of paraoxon was cleaved off during co-crystallization, and a tetrahedral product reminiscent of the first transition state was formed during carboxyl ester hydrolysis. The native apo and paraoxon-bound structures superimpose with RMSD’s of 0.12–0.32 Å over 320–322 Cα atoms. Attempts to obtain co-crystals of TM0077 with cephalosporin C, even at a low temperature of 4°C, were unsuccessful.
Analysis of the active site
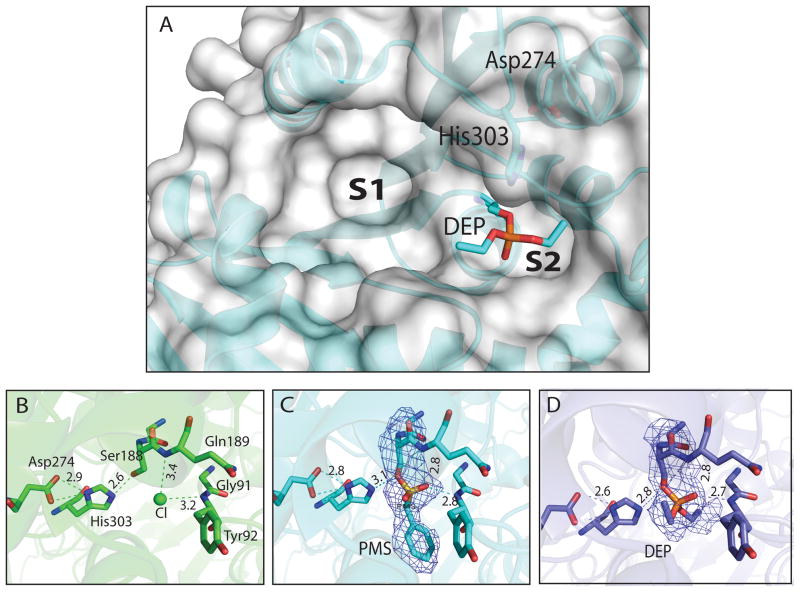
TM0077 has a classic catalytic triad, consisting of Ser188 as the nucleophile, His303 as the proton acceptor/donor, and Asp274 as the acidic residue stabilizing the histidine (Fig. 6). The catalytic serine Ser188 is located within a conserved pentapeptide sequence, Gly-X-Ser-X-Gly (GGSQG), characteristic of esterases and lipases. The positions of Ser188, Asp274, and His303 are consistent with their expected locations in the canonical fold of the α/β-hydrolase family. Ser188 is located at the nucleophile elbow in a sharp turn between β5 and helix αC. The presence of three glycine residues (Gly186, Gly187, and Gly190) in close proximity to Ser188 prevents steric hindrance and facilitates access to the nucleophile elbow. Asp274 and His303 are located in loops between β7 and helix αE, and between β8 and helix αF, respectively. The oxyanion hole is formed by the backbone amide groups of Tyr92 and Gln189. The catalytic triad and oxyanion hole are located in a depression on the surface of TM0077. This ellipsoid pocket (S1), which is approximately 12 Å wide, extends 15 Å from the catalytic serine. A smaller pocket (S2), approximately 5 Å long, extends to the other side of the catalytic serine [Fig. 6(a)]. The volume of both pockets combined (S1 + S2) is 1082 Å3 (CASTp analysis; 50). The substrate-binding pocket is bordered by residues from helices αA and αF, and its base is formed by residues from β-strands 4, 5, 6, and their adjacent C-terminal loops. The overall pocket is hydrophobic, although it does have some polar residues (Gln88, Asp210, and Gln314), which may interact with the substrate.
Figure 6.
TM0077 catalytic site. (A) Surface representation of the TM0077 catalytic site, with His303, Asp274 and the intermediate DEP-modified Ser188 shown as sticks. The two binding pockets are indicated with S1 and S2. (B) Apo TM0077 with a bound chloride ion (green sphere), (C) TM0077 with PMS-modified Ser188 and (D) TM0077 with DEP-modified Ser188. The catalytic residues are shown as sticks, with the hydrogen bonds shown as dashed lines. Carbon atoms are in green (apo), cyan (PMS) or blue (DEP), oxygen atoms in red, sulfur atoms in yellow and phosphate in orange. Electron density omit maps shown for inhibitor modified Ser188 contoured at 1σ show that the PMS and DEP are covalently bonded to Ser188 in (C) and (D), respectively. Distances are shown in Ångströms.
In the native apo structure, the Ser188 hydroxyl makes a hydrogen bond with the imidazole of His303 [Fig. 6(b)]. Extra density was observed near the side chain of Ser188 and was interpreted as a chloride ion based on electron density size and shape as well as the geometry of the interactions with surrounding residues. This chloride ion is bound at the entrance of the oxyanion hole, forming hydrogen bonds with the backbone amides of Tyr92 and Gln189. In the PMSF-bound structure, the phenyl ring of the inhibitor is located in the small active site groove surrounded by hydrophobic residues Tyr92, Trp124, Pro228, Ile276, and His303 [Fig. 6(c)]. The sulfonyl group of PMSF makes hydrogen bonds with the backbone amides of Tyr92 and Gln189. In the paraoxon-bound structure, the diethyl-phosphate (DEP) moiety is stabilized by hydrogen-bonding interactions with the oxyanion hole. One of the two ethyl arms of bound paraoxon points toward the larger pocket in the protein, while the other follows the groove of the small pocket. The two ethyl arms are stabilized by packing against Tyr92, Trp124, Pro228, Ile276, and His303 [Fig. 6(d)].
Two rotamers of the catalytic serine
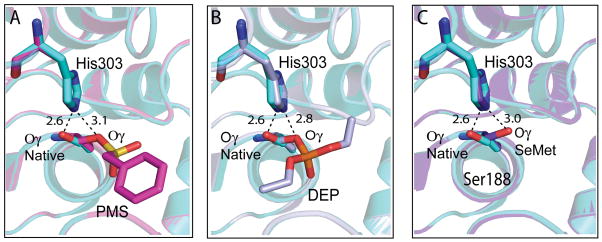
Although no large conformational changes were observed upon binding of PMSF or paraoxon, a different rotamer of the catalytic serine side chain was observed compared to native TM0077 [Fig. 7(a,b)]. Similar changes have been observed in several other esterases and have been shown to play a key role in the catalytic mechanism (see CONCLUSION for more details). In the native structure, the catalytic Ser188 Oγ is in the plane of the imidazole ring of His303, as most commonly observed in the resting state of esterases and lipases 51. The Ser188 Oγ forms a hydrogen bond (2.6 Å) with His303 Nε2. In the PMSF- and paraoxon-bound structures, the conformation of the catalytic serine changes; the Ser188 Oγ rotates about 110°, increasing the distance (3.1 Å and 2.8 Å for PMSF and paraoxon bound structures, respectively) to the His303 imidazole ring. In the TM0077-SeMet structure, the catalytic serine is also rotated over ~110°, with a distance to the imidazole ring of 3.0 Å [Fig. 7(c.)]. A probable explanation for this observation could be the protonation of His303, since TM0077-SeMet was crystallized at a lower pH (pH 4.2) compared to the native TM0077 (pH 7.3). Furthermore, extra electron density was identified in the TM0077-SeMet structure, suggesting a partially occupied acyl intermediate on Ser188. However, as this density is not sufficiently clear and interpretable to fit an acyl intermediate, water molecules were modeled instead. No rearrangements of any other residues in the active site were observed.
Figure 7.
Conformational change of Ser188 Oγ. The Oγ atom of the Ser188 is rotated ~110° between the native apo structure (cyan) and (A) the complexed PMS-modified Ser188 structure (pink), (B) the DEP-modified Ser188 structure (light blue) and (C) the SeMet structure (purple). The different hydrogen bonds made for the Ser Oγ in the native versus complexed structures are shown as dashed black lines with distances in Ångströms.
CONCLUSION
TM0077 from the hyperthermophilic bacterium T. maritima was predicted from its gene sequence to be an acetyl xylan esterase. We have expressed and purified TM0077 and experimentally demonstrated that it has ester-hydrolyzing activity. The TM0077 activity was restricted to short acyl chain esters (C2 and C3) when artificial p-nitrophenyl-esters were used as substrates. In addition, the enzyme has high specific activity on glucose penta-acetate. However, no activity was detected on xylan or acetylated xylan. Thus, TM0077 should be reclassified as an acetyl esterase, and not as an acetyl xylan esterase as currently annotated 52. Furthermore, the lack of any apparent signal sequence suggests that the protein is not secreted. Thus, the predicted intracellular location of TM0077 is compatible with a role other than the deacetylation of extracellular xylan. Based on these results, we conclude that the likely biological function of TM0077 is removal of the remaining acetyl groups from the short, end products of xylan degradation that are imported into the cytoplasm. The resulting deacetylated xylose oligomers are the substrates for a β-xylosidase. This role for TM0077 is in good agreement with the clustering of the TM0077 gene with other genes involved in xylose metabolism. However, it cannot be ruled out that TM0077 may also act on other small, acetylated compounds.
TM0077 is the first esterase from the CE7 family to be tested for its positional specificity for the deacetylation of 4-nitrophenyl-β-D-xylopyranoside. TM0077 hydrolyzes acetate at the 2, 3 and 4 positions of 4-nitrophenyl-β-D-xylopyranoside with similar efficiency. Conversely, the CtAxe esterase from Clostridium thermocellum in the CE4 family shows a clear preference for hydrolyzing acetate at the 2 position 53, and Penicillium purpurogenum AXE II esterase, a member of the CE5 family, also has a preference for acetate at position 2 54. This lack of preference for a specific position of the acetate group correlates with the relative broad substrate specificity of the CE7 esterases.
Esterases and deacetylases in the CE7 family are unusual in that they are active towards both acetylated xylo-oligosaccharides and the antibiotic cephalosporin C [Fig. 1(a,b)]. Therefore, TM0077 was investigated for activity towards the substrates 7-ACA and cephalosporin C. The activity of TM0077 on these substrates is approximately ten-fold higher than that of the acetyl xylan esterase from B. pumilus 55 or the acetyl esterase from Thermoanaerobacterium sp. strain JW/SL YS485 56. TM0077 has a higher hydrolytic activity on 7-ACA compared to cephalosporin C, as described for other CE7 esterases 40,55,56. Nonetheless, it is unlikely that both compounds are natural substrates, because the stability of these compounds at the optimal growth temperature (80°C) of T. maritima is very low.
Crystal structures of TM0077 in complex with inhibitors PMSF and paraoxon revealed that, upon binding of PMSF or paraoxon, the reaction is trapped at the acylation step via the formation of a covalent tetrahedral reaction product. In the complexed structures, the negatively charged oxygen of the tetrahedral intermediate, derived from the substrate oxyanion, is stabilized by hydrogen bonds with the backbone amide groups of Tyr92 and Gln189. Comparison of the TM0077 complexed structures with the native structure shows that the catalytic serine (Ser188) Oγ rotates about 110°, thereby increasing the distance between Ser188 Oγ and His303 Nε2. Such a conformational change of the catalytic serine has been observed in several other esterases, including Fusarium solani cutinase 57, Penicillium purpurogenum acetyl xylan esterase 51, Rhodococcus sp. strain MB1 cocaine esterase 58, Bacillus subtilis lipase 59, Rhodococcus sp. strain H1 heroine esterase 60, and Aspergillus niger feruloyl esterase 61. The classical model for the catalytic mechanism of esterases consists of a sequential two-step hydrolysis. The first reaction involves nucleophilic attack by the catalytic serine on the substrate carbonyl carbon, resulting in an acyl-enzyme and the liberation of an alcohol. In the second reaction, a water molecule performs a nucleophilic attack on the acyl-enzyme, the acyl-enzyme bond breaks and the carboxylate is released 62. Although the catalytic mechanism of esterases is well established, it is unclear why the initially generated tetrahedral intermediate does not collapse back to the reactant complex during the nucleophilic attack of the substrate. A previously proposed mechanism that would prevent this collapse is the spatial reorganization of the catalytic residues during the initial catalytic step, causing the residues to separate and thereby drive the reaction forward 62–64. The apo and inhibitor bound structures of TM0077, presented herein, support this proposed mechanism. Moreover, in a recent study of the serine protease mechanism, it was suggested that subtle atomic motions of the catalytic serine and histidine residues during the catalytic cycle favor the forward reaction 65. Thus, rotation of Ser188 Oγ of TM0077 may be required to inhibit reversal of the reaction. In addition, such changes may facilitate the access of water to the catalytic histidine so that the second step of the reaction can go to completion.
Deacetyl cephalosporins are valuable building blocks for the production of semisynthetic β-lactam antibiotics. These compounds are derived from cephalosporin C or 7-aminocephalosporanic acid via enzymatic or chemical processes 10. The thermostable TM0077 esterase may be valuable in the preparation of derivatives of β-lactam antibiotics. Recently, the substrate specificity of the acetyl xylan esterase from P. purpurogenum was engineered to accept a range of fatty acid esters of up to 14 carbons compared to its wild-type preference for acetate54. It might also be possible to engineer TM0077 and enable the (de)acetylation of cephalosporins at the C10 position with various acyl chains. Because of its high stability and activity on 7-ACA and cephalosporin C, TM0077 presents an attractive candidate for the production of new semi-synthetic antibiotics.
Supplementary Material
Acknowledgments
We gratefully acknowledge contributions from George Sheldrick for modifications of the SHELXD program, and for Global Phasing Ltd. that made significant improvements in the automation of autoSHARP. We also thank Victor Lamzin for updates of chain docking of the ARP/wARP program, and Gerard Bricogne and Eleanor Dodson for helpful discussion on phasing for the large TM0077-SeMet structure, and Willem J. van Berkel for valuable discussion on the catalytic mechanism of TM0077. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL) and the Advanced Light Source (ALS). The SSRL is a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209), and the National Institute of General Medical Sciences. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231 at Lawrence Berkeley National Laboratory. Genomic DNA from Thermotoga maritima MSB8 (DSM3109) (ATCC #43589D-5) was obtained from the American Type Culture Collection (ATCC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Grant sponsor: NIH Grant numbers U54 GM094586 and U54 GM074898 (Protein Structure Initiative); Grant sponsor: The Graduate School VLAG Wageningen, the Netherlands (ML).
References
- 1.Huber R, Hannig M. Thermotogales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The Prokaryotes: An Evolving Electronic Resource for the Microbiological Community. Vol. 7. New-York: Springer-Verlag; 2006. pp. 899–922. [Google Scholar]
- 2.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, Utterback TR, Malek JA, Linher KD, Garrett MM, Stewart AM, Cotton MD, Pratt MS, Phillips CA, Richardson D, Heidelberg J, Sutton GG, Fleischmann RD, Eisen JA, White O, Salzberg SL, Smith HO, Venter JC, Fraser CM. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 3.Chhabra SR, Shockley KR, Conners SB, Scott KL, Wolfinger RD, Kelly RM. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J Biol Chem. 2003;278:7540–7552. doi: 10.1074/jbc.M211748200. [DOI] [PubMed] [Google Scholar]
- 4.Conners SB, Montero CI, Comfort DA, Shockley KR, Johnson MR, Chhabra SR, Kelly RM. An expression-driven approach to the prediction of carbohydrate transport and utilization regulons in the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol. 2005;187:7267–7282. doi: 10.1128/JB.187.21.7267-7282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.VanFossen AL, Lewis DL, Nichols JD, Kelly RM. Polysaccharide degradation and synthesis by extremely thermophilic anaerobes. Ann N Y Acad Sci. 2008;1125:322–337. doi: 10.1196/annals.1419.017. [DOI] [PubMed] [Google Scholar]
- 6.Biely P, Mackenzie CR, Puls J, Schneider H. Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Bio-Technology. 1986;4:731–733. [Google Scholar]
- 7.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topakas E, Paul C. Microbial xylanolytic carbohydrate esterases. Industrial Enzymes. 2007:83–97. [Google Scholar]
- 9.Weil J, Miramonti J, Ladisch MR. Cephalosporin-C: Mode of action and biosynthetic pathway. Enzyme Microb Technol. 1995;17:85–87. [Google Scholar]
- 10.Barends TRM, Yoshida H, Dijkstra BW. Three-dimensional structures of enzymes useful for beta-lactam antibiotic production. Curr Opin Biotechnol. 2004;15:356–363. doi: 10.1016/j.copbio.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Martínez-Martínez I, Montoro-García S, Lozada-Ramírez JD, Sánchez-Ferrer Á, García-Carmona F. A colorimetric assay for the determination of acetyl xylan esterase or cephalosporin C acetyl esterase activities using 7-amino cephalosporanic acid, cephalosporin C, or acetylated xylan as substrate. Anal Biochem. 2007;369:210–217. doi: 10.1016/j.ab.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 12.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, Stevens RC. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc Natl Acad Sci USA. 2002;99:11664–11669. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levisson M, van der Oost J, Kengen SW. Characterization and structural modeling of a new type of thermostable esterase from Thermotoga maritima. FEBS Journal. 2007;274:2832–2842. doi: 10.1111/j.1742-4658.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 16.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 17.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr Sect D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 18.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Smart O, Vonrhein CTW. BUSTER, version 2.8.0. Cambridge, United Kingdom: Global Phasing Ltd; 2009. [Google Scholar]
- 19.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A Found Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 20.Winn MD, Murshudov GN, Papiz MZ. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 2003;374:300–321. doi: 10.1016/S0076-6879(03)74014-2. [DOI] [PubMed] [Google Scholar]
- 21.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr Sect D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 23.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 25.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr Sect D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:1833–1839. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- 28.Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger TC. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr Sect D Biol Crystallogr. 2003;59:45–49. doi: 10.1107/S0907444902018048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleywegt GJ. Validation of protein crystal structures. Acta Crystallogr Sect D Biol Crystallogr. 2000;56:249–265. doi: 10.1107/s0907444999016364. [DOI] [PubMed] [Google Scholar]
- 31.DeLano WL. The Pymol molecular graphics system. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 32.Barthel D, Hirst JD, Blazewicz J, Burke EK, Krasnogor N. ProCKSI: a decision support system for Protein (structure) Comparison, Knowledge, Similarity and Information. BMC Bioinformatics. 2007;8:416. doi: 10.1186/1471-2105-8-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forsberg A, Puu G. Kinetics for the inhibition of acetylcholinesterase from the electric eel by some organophosphates and carbamates. Eur J Biochem. 1984;140:153–156. doi: 10.1111/j.1432-1033.1984.tb08079.x. [DOI] [PubMed] [Google Scholar]
- 34.Johnson KG, Harrison BA, Schneider H, Mackenzie CR, Fontana JD. Xylan-hydrolyzing enzymes from Streptomyces spp. Enzyme Microb Technol. 1988;10:403–409. [Google Scholar]
- 35.Biely P, Mackenzie CR, Schneider H. Production of acetyl xylan esterase by Trichoderma reesei and Schizophyllum commune. Can J Microbiol. 1988;34:767–772. [Google Scholar]
- 36.Biely P, Mastihubova M, la Grange DC, van Zyl WH, Prior BA. Enzyme-coupled assay of acetylxylan esterases on monoacetylated 4-nitrophenyl beta-D-xylopyranosides. Anal Biochem. 2004;332:109–115. doi: 10.1016/j.ab.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 37.Mastihubova M, Biely P. Lipase-catalysed preparation of acetates of 4-nitrophenyl beta-D-xylopyranoside and their use in kinetic studies of acetyl migration. Carbohydr Res. 2004;339:1353–1360. doi: 10.1016/j.carres.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 38.Jiang ZQ, Kobayashi A, Ahsan MM, Lite L, Kitaoka M, Hayashi K. Characterization of a thermostable family 10 endo-xylanase (XynB) from Thermotoga maritima that cleaves p-nitrophenyl-beta-D-xyloside. J Biosci Bioeng. 2001;92:423–428. doi: 10.1263/jbb.92.423. [DOI] [PubMed] [Google Scholar]
- 39.Xue YM, Shao WL. Expression and characterization of a thermostable beta-xylosidase from the hyperthermophile, Thermotoga maritima. Biotechnol Lett. 2004;26:1511–1515. doi: 10.1023/B:BILE.0000044454.70768.81. [DOI] [PubMed] [Google Scholar]
- 40.Vincent F, Charnock SJ, Verschueren KHG, Turkenburg JP, Scott DJ, Offen WA, Roberts S, Pell G, Gilbert HJ, Davies GJ, Brannigan JA. Multifunctional xylooligosaccharide/cephalosporin C deacetylase revealed by the hexameric structure of the Bacillus subtilis enzyme at 1. 9 angstrom resolution. J Mol Biol. 2003;330:593–606. doi: 10.1016/s0022-2836(03)00632-6. [DOI] [PubMed] [Google Scholar]
- 41.Nardini M, Dijkstra BW. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 42.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartlam M, Wang G, Yang H, Gao R, Zhao X, Xie G, Cao S, Feng Y, Rao Z. Crystal structure of an acylpeptide hydrolase/esterase from Aeropyrum pernix K1. Structure. 2004;12:1481–1488. doi: 10.1016/j.str.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 44.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Huddleston S, Yallop CA, Charalambous BM. The identification and partial characterisation of a novel inducible extracellular thermostable esterase from the archaeon Sulfolobus shibatae. Biochem Biophys Res Commun. 1995;216:495–500. doi: 10.1006/bbrc.1995.2650. [DOI] [PubMed] [Google Scholar]
- 46.Shao WL, Wiegel J. Purification and characterization of 2 thermostable acetyl xylan esterases from Thermoanaerobacterium sp strain JW/SL-YS485. Appl Environ Microbiol. 1995;61:729–733. doi: 10.1128/aem.61.2.729-733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levisson M, Sun L, Hendriks S, Swinkels P, Akveld T, Bultema JB, Barendregt A, van den Heuvel RH, Dijkstra BW, van der Oost J, Kengen SW. Crystal structure and biochemical properties of a novel thermostable esterase containing an immunoglobulin-like domain. J Mol Biol. 2009;385:949–962. doi: 10.1016/j.jmb.2008.10.075. [DOI] [PubMed] [Google Scholar]
- 48.Montoro-García S, Gil-Ortiz F, García-Carmona F, Polo LM, Rubio VAS-F. The crystal structure of the cephalosporin deacetylating enzyme acetyl xylan esterase bound to paraoxon explains the low sensitivity of this serine hydrolase to organophosphate inactivation. Biochem J. 2011 doi: 10.1042/BJ20101859. [DOI] [PubMed] [Google Scholar]
- 49.Febbraio F, D’Andrea SE, Mandrich L, Merone L, Rossi M, Nucci R, Manco G. Irreversible inhibition of the thermophilic esterase EST2 from Alicyclobacillus acidocaldarius. Extremophiles. 2008;12:719–728. doi: 10.1007/s00792-008-0179-1. [DOI] [PubMed] [Google Scholar]
- 50.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghosh D, Sawicki M, Lala P, Erman M, Pangborn W, Eyzaguirre J, Gutierrez R, Jornvall H, Thiel DJ. Multiple conformations of catalytic serine and histidine in acetylxylan esterase at 0.90 angstrom. J Biol Chem. 2001;276:11159–11166. doi: 10.1074/jbc.M008831200. [DOI] [PubMed] [Google Scholar]
- 52.Williamson G, Kroon PA, Faulds CB. Hairy plant polysaccharides: a close shave with microbial esterases. Microbiology. 1998;144:2011–2023. doi: 10.1099/00221287-144-8-2011. [DOI] [PubMed] [Google Scholar]
- 53.Biely P, Mastihubova M, Puchart V. The vicinal hydroxyl group is prerequisite for metal activation of Clostridium thermocellum acetylxylan esterase. Biochim Biophys Acta. 2007;1770:565–570. doi: 10.1016/j.bbagen.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 54.Colombres M, Garate JA, Lagos CF, Araya-Secchi R, Norambuena P, Quiroz S, Larrondo L, Perez-Acle T, Eyzaguirre J. An eleven amino acid residue deletion expands the substrate specificity of acetyl xylan esterase II (AXE II) from Penicillium purpurogenum. J Comput Aided Mol Des. 2008;22:19–28. doi: 10.1007/s10822-007-9149-4. [DOI] [PubMed] [Google Scholar]
- 55.Degrassi G, Kojic M, Ljubijankic G, Venturi V. The acetyl xylan esterase of Bacillus pumilus belongs to a family of esterases with broad substrate specificity. Microbiology. 2000;146:1585–1591. doi: 10.1099/00221287-146-7-1585. [DOI] [PubMed] [Google Scholar]
- 56.Lorenz WW, Wiegel J. Isolation, analysis, and expression of two genes from Thermoanaerobacterium sp. strain JW/SL YS485: a beta-xylosidase and a novel acetyl xylan esterase with cephalosporin C deacetylase activity. J Bacteriol. 1997;179:5436–5441. doi: 10.1128/jb.179.17.5436-5441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Longhi S, Czjzek M, Lamzin V, Nicolas A, Cambillau C. Atomic resolution (1. 0 angstrom) crystal structure of Fusarium solani cutinase: Stereochemical analysis. J Mol Biol. 1997;268:779–799. doi: 10.1006/jmbi.1997.1000. [DOI] [PubMed] [Google Scholar]
- 58.Larsen NA, Turner JM, Stevens J, Rosser SJ, Basran A, Lerner RA, Bruce NC, Wilson IA. Crystal structure of a bacterial cocaine esterase. Nat Struct Biol. 2002;9:17–21. doi: 10.1038/nsb742. [DOI] [PubMed] [Google Scholar]
- 59.Kawasaki K, Kondo H, Suzuki M, Ohgiya S, Tsuda S. Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1. 3 angstrom resolution. Acta Crystallogr Sect D Biol Crystallogr. 2002;58:1168–1174. doi: 10.1107/s090744490200714x. [DOI] [PubMed] [Google Scholar]
- 60.Zhu X, Larsen NA, Basran A, Bruce NC, Wilson IA. Observation of an arsenic adduct in an acetyl esterase crystal structure. J Biol Chem. 2003;278:2008–2014. doi: 10.1074/jbc.M210103200. [DOI] [PubMed] [Google Scholar]
- 61.McAuley KE, Svendsen A, Patkar SA, Wilson KS. Structure of a feruloyl esterase from Aspergillus niger. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:878–887. doi: 10.1107/S0907444904004937. [DOI] [PubMed] [Google Scholar]
- 62.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4523. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 63.Ash EL, Sudmeier JL, Day RM, Vincent M, Torchilin EV, Haddad KC, Bradshaw EM, Sanford DG, Bachovchin WW. Unusual 1H NMR chemical shifts support (His) Cε1-H···O=C H-bond: Proposal for reaction-driven ring flip mechanism in serine protease catalysis. Proc Natl Acad Sci USA. 2000;97:10371–10376. doi: 10.1073/pnas.97.19.10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bizzozero SA, Dutler H. Stereochemical aspects of peptide hydrolysis catalyzed by serine proteases of the chymotrypsin type. Bioorg Chem. 1981;10:46–62. [Google Scholar]
- 65.Radisky ES, Lee JM, Lu CJK, Koshland DE., Jr Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates. Proc Natl Acad Sci USA. 2006;103:6835–6840. doi: 10.1073/pnas.0601910103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cruickshank DW. Remarks about protein structure precision. Acta Crystallogr Sect D Biol Crystallogr. 1999;55:583–601. doi: 10.1107/s0907444998012645. [DOI] [PubMed] [Google Scholar]
- 67.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 68.Weiss MS, Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J Appl Crystallogr. 1997;30:203–205. [Google Scholar]
- 69.Weiss MS. Global indicators of X-ray data quality. J Appl Crystallogr. 2001;34:130–135. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.