Abstract
Background:
Prostate cancer (PrCa) is one of the most common cancers affecting men but its aetiology is poorly understood. Family history of PrCa, particularly at a young age, is a strong risk factor. There have been previous reports of increased PrCa risk in male BRCA1 mutation carriers in female breast cancer families, but there is a controversy as to whether this risk is substantiated. We sought to evaluate the role of germline BRCA1 mutations in PrCa predisposition by performing a candidate gene study in a large UK population sample set.
Methods:
We screened 913 cases aged 36–86 years for germline BRCA1 mutation, with the study enriched for cases with an early age of onset. We analysed the entire coding region of the BRCA1 gene using Sanger sequencing. Multiplex ligation-dependent probe amplification was also used to assess the frequency of large rearrangements in 460 cases.
Results:
We identified 4 deleterious mutations and 45 unclassified variants (UV). The frequency of deleterious BRCA1 mutation in this study is 0.45% three of the mutation carriers were affected at age ⩽65 years and one developed PrCa at 69 years. Using previously estimated population carrier frequencies, deleterious BRCA1 mutations confer a relative risk of PrCa of ∼3.75-fold, (95% confidence interval 1.02–9.6) translating to a 8.6% cumulative risk by age 65.
Conclusion
This study shows evidence for an increased risk of PrCa in men who harbour germline mutations in BRCA1. This could have a significant impact on possible screening strategies and targeted treatments.
Keywords: prostate cancer, BRCA1 gene, mutation screening, cancer risk
Prostate cancer (PrCa) is one of the most common cancers affecting men worldwide; in the United Kingdom, PrCa incidence has overtaken lung cancer with 37 000 cases and 10 000 deaths reported in the latest statistics since 2008. Worldwide there were >900 000 cases diagnosed and ∼250 000 deaths reported in 2008 (CRUK CancerStats August 2011). However, the underlying aetiology is complex and poorly understood compared with other common complex diseases. The only well-established risk factors are age (60% of men diagnosed with PrCa are >70 years old), family history (first-degree relatives have an ∼2-fold risk compared with the general population) and ethnic background (men of African descent have an increased risk of PrCa). Recently, common genetic variants (minor allele frequency >5%) that individually account for a moderate increase in PrCa risk (relative risk (RR) <2) were identified by the initial PrCa genome-wide association studies (GWAS; Eeles et al, 2008; Gudmundsson et al, 2008; Thomas et al, 2008), these and subsequent follow-up studies have now found >40 PrCa risk alleles (reviewed in Goh et al, 2012). Predisposition genes with a higher RR have proven more difficult to identify.
Studies on twins were instrumental in providing evidence for the existence of a substantial genetic component in PrCa risk, with identical twins being ∼5 times more likely to be concordant with PrCa compared with non-identical twins (Grönberg et al, 1994; Page et al, 1997). A large Scandinavian twin study into the causes of cancer reported that genetic factors accounted for 42% of the PrCa risk in their sample set and concluded that both rare highly penetrant variants and common polymorphisms with low penetrance contribute to PrCa risk (Lichtenstein et al, 2000).
Deleterious germline mutations in BRCA1 and BRCA2 increase the risk of breast and ovarian cancer in women, and co-aggregation of PrCa and breast cancer was initially established in epidemiological studies of breast cancer families (Thompson and Easton, 2001, 2002). For BRCA1 mutation carriers the Breast Cancer Linkage Consortium reported an increase in PrCa risk in men aged <65 years with a RR of 1.82 (95% confidence interval (CI) 1.01–3.29, P=0.05), but no risk increase was seen for men ⩾65 years. For BRCA2 mutation carriers the study reported an increase in PrCa RR of 4.65 (95% CI 3.48–6.22) rising to 7.33 (95% CI 4.66–11.52) for men <65 years (The Breast Cancer Linkage Consortium, 1999). The frequency and risk estimates for BRCA2 mutation carriers was addressed by several studies, most comprehensively it was recently evaluated in a large cohort by our group (Kote-Jarai et al, 2011), which reported an ∼8.6-fold increased risk of PrCa for BRCA2 mutation carriers.
However, most BRCA1 mutations reported in PrCa cases to date were identified through familial breast/ovarian cancer studies and PrCa-specific studies restricted to specific populations and assessment of founder mutations (Kirchhoff et al, 2004; Cybulski et al, 2008). Therefore, these studies are likely to be underpowered to assess accurately a BRCA1 mutation carrier’s risk of PrCa. Here, we report the first large-scale (913 cases) prostate centric study, using direct Sanger sequencing and multiplex ligation-dependent probe amplification (MLPA) to provide a comprehensive assessment of germline BRCA1 genetic variation across the entire coding sequence and proximal promoter region of this gene in PrCa cases.
Materials and methods
Samples
A series of men with PrCa were recruited from the UK Genetic Prostate Cancer Study (UKGPCS) as reported previously (Eeles et al, 1997). The majority (546, 83.4%) of patients with a known diagnosis method (655/913) had clinically presenting (not prostate-specific antigen (PSA) screened) disease. Case selection for this study was based primarily on age at diagnosis of ⩽65 years (821 cases; age range 36–65 years); with a further cohort aged >65 years (92 cases; age range 66–88 years) with a family history of one or more first-degree relatives with PrCa (Table 1).
Table 1. Age range and family history of PrCa samples screened for BRCA1 germline mutation.
| Age range (years) | Number of samples | % PrCa family history a | % Br/Ov family history a | % Samples deceased | Number of carriers | % Carriers |
|---|---|---|---|---|---|---|
| 36–55 | 327 | 37.3 | 26.0 | 13.1 | 1 | 0.3 |
| 56–65 | 494 | 53.0 | 28.7 | 12.8 | 2 | 0.4 |
| 66–88 | 92 | 100.0 | 31.5 | 22.8 | 1 | 1 |
| Total | 913 | 4 |
Abbreviations: BrCa=breast cancer; OvCa = ovarian cancer; PrCa = prostate cancer. Family history of (BrCa) and (OvCa) is also shown.
In first- and second-degree relatives.
Mutation detection
Germline DNA was obtained from peripheral blood samples and extracted as reported previously (Edwards et al, 1997). A total of 29 primer pairs were designed using primer3 and exonprimer (Rozen and Skaletsky, 2000; http://ihg.gsf.de/ihg/ExonPrimer.html) to cover all exons, intron/exon junctions, proximal promoter and exon 1a/b region of the full-length NM_007294.3 mRNA transcript; primer sequences available on request. Using Qiagen multiplex PCR kit 206145 (Qiagen, Hilden, Germany), the 29 primer pairs were grouped into four multiplexs (3 × 7 multiplexes and 1 × 6 multiplex) and two singleton reactions. The amplicons were sequenced on an ABI3730 Genetic Analyzer using a 1/16th BigDye v3 protocol (Applied Biosystems, Foster City, CA, USA). Deleterious mutations were confirmed by Sanger sequencing using a different aliquot from stock DNA of each sample. Multiplex ligation-dependent probe amplification was performed on a subset of 460 samples using a modified protocol of the SALSA MLPA kit P002-C2 (MRC-Holland, Amsterdam, The Netherlands).
Sequence analysis
Mutation surveyor 3.97 (Softgenetics, State College, PA, USA) was used to analyse the sequencing data, using a region of interest covering 500 bp of the near promoter exons 1–3, 5–24 and also including ±20 bp per exon to screen the intron/exon boundaries. Results were exported to Human Genome Variation Society nomenclature and converted to hg19 genomic coordinates using Mutalyzer 2.0 β-8, (http://www.mutalyzer.nl/) to allow variant annotation using ANNOVAR (Wildeman et al, 2008; Wang et al, 2010). Variants were designated as ‘novel’ if they were not described in dbSNP132, Breast cancer Information Core (BIC) or the Leiden Open Variation Database as of July 2011 (Szabo et al, 2000; Sherry et al, 2001; Fokkema et al, 2011).
Statistical methods
Estimates for BRCA1 prevalence in the general population were derived from a study where a large population series and a set of high-risk breast cancer families from the United Kingdom were analysed incorporating the effect of BRCA1, BRAC2 and other genes to account for residual family history (Antoniou et al, 2002). Confidence intervals for the prevalence of BRCA1 mutations among PrCa cases and hence the CIs on RR were computed using an exact binomial procedure. Age-specific cumulative risks of PrCa to age 65 in BRCA1 mutation carriers were computed based on the RR estimated from this study and age-specific pooled incidence rates from five cancer registries in England for the year 2002 as reported in Ferlay et al (2007).
Results
We screened 913 PrCa cases for germline mutations in the BRCA1 gene using direct Sanger sequencing. After quality control (QC) exclusions, data from 886 PrCa cases were analysed. We identified four deleterious mutations (0.45%), of which three were frameshifts and one was a splice site mutation (Table 2). A subset of the PrCa cases (460) was also screened for large-scale rearrangements (LGRs) using MLPA, none were found in our sample set.
Table 2. Patient information for protein-truncating mutation carriers.
| Sample ID | Nucleotide change | Age at diagnosis (years) | Tumour stage | Node stage | Metastases | Final gleason | Years to death | FH of prostate cancer | FH of Br or Ov | FH of other cancers in family |
|---|---|---|---|---|---|---|---|---|---|---|
| PR-1 | c.68_69delAG | 58 | T1a | N0 | M0 | 3+3 | 12 (Alive) | No | Mother Ov | No |
| PR-2 | c.212+1G>T | 57 | T4 | Nx | Mx | 4+4 | 3 | No | Sister Ov, aunt Br, grandmother Br | Mother bladder, grandfather colon |
| PR-3 | c.1954dupA | 51 | Tx | Nx | M0 | — | 11 | No | No | Brother Bladder |
| PR-4 | c.2475delC | 69 | Tx | Nx | Mx | — | 8 | Father | No | No |
Abbreviations: Br=breast cancer; Ov=ovarian cancer. Clinical and family history (FH) data are detailed here if available. Tx, Nx and Mx if status unknown.
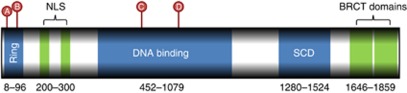
All four deleterious mutations had been previously recorded in BIC (http://research.nhgri.nih.gov/bic/). We also identified 45 missense variants (10 novel), 15 synonymous substitutions (5 novel) and 6 intronic variants (1 novel; Supplementary Table 1). The majority of deleterious mutations occurred in PrCa cases <65 years (3 out of 4) and all mutations are within two distinct functional domains of BRCA1; mutations c.68_69delAG (A) and c.212+1G>T (B) in the N-terminus RING finger domain and c.1954dupA (C) and c.2475delC (D) in the large central DNA-binding domain (Figure 1).
Figure 1.
Schematic diagram of the positions of the deleterious mutations (A–D) found in this study within the BRCA1 transcript. The locations of the functional domains are marked in blue or green.
In addition we have also identified several missense unclassified variants (UV) and a deletion in the proximal promoter region, c.1-1462delC immediately 3′ of a CREB-binding site regulated by methylation (Mancini et al, 1998; Atlas et al, 2001; DiNardo et al, 2001).
The missense UVs identified were annotated by ANNOVAR against dbNSFP normalised (0–1) scores for SIFT, Polyphen2, LRT, MutationTaster and Phylop, (Chun and Fay, 2009; Kumar et al, 2009; Adzhubei et al, 2010; Pollard et al, 2010; Schwarz et al, 2010; Liu et al, 2011). In addition, all non-synonymous SNPs were annotated with Align-GVGD (Mathe et al, 2006; Tavtigian et al, 2008). The intersection of UVs predicted to be ‘probably damaging’ by both SIFT and Polyphen2 selects 13 of the 45 UVs found in the study, of which 11 are in BIC and 2 are novel (Supplementary Table 1). Of the BIC annotated mutations three are classed as ‘of no clinical significance’, the rest as ‘of unknown significance’, whereas 10 of these 13 variants have been classified as ‘of no clinical significance’ using multifactorial likelihood approaches (Vallée et al, 2011). This leaves three variants, H279Q, M1400R and M1783T (Supplementary Table 2) with some potential to be deleterious, although only M1783T is situated in a known functional domain (BRCT repeat 2).
Based on the previously estimated carrier frequency of BRCA1 mutations in the United Kingdom (which estimated the allele frequency of BRCA1 mutation to be 0.006 corresponding to a carrier frequency of 0.0012 (∼1/1000), Antoniou et al, 2002) and the observed prevalence of BRCA1 mutations in our series of PrCa cases of 4/886 (0.45%), we estimate that germline mutations in the BRCA1 gene confer a RR of PrCa of ∼3.75-fold, (95% CI 1.02–9.6).
Discussion
This study utilised direct Sanger sequencing to screen the complete BRCA1 coding transcript (NM_007294.3) in 913 PrCa patients, the majority of whom were clinically detected (83.4% of cases for which the diagnosis method was reported). This is the largest direct sequencing study of BRCA1 reported so far in PrCa and therefore presents the most comprehensive assessment of the contribution of BRCA1 mutations to PrCa risk.
After QC, 886 PrCa cases were included in our mutation analysis of the sequencing data and four pathogenic mutations were identified (0.45%). In the 460 samples, which were also submitted for MLPA, no LGRs were identified.
Of the four identified pathogenic mutations, c.68_69delAG (BIC 185delAG) is a well-known Ashkenazi-Jewish founder mutation (reported 1979 times in BIC), which has been extensively studied and has a frequency of ∼1% in the general Ashkenazi-Jewish population. From the UK 2001 census, 266 740 people (0.5% UK population) are recorded Jewish as their religion; however, this is believed to be an underestimate due to the non-counting of secular Ashkenazi-Jews and the self-reporting nature of this statistic. A recent meta-analysis on six Ashekenazi-Jewish studies of PrCa (3005 cases and 6834 controls) by Fachal et al showed a non-statistically significant odds ratio 1.8 (95% CI 0.91–3.57; P=0.09) for this particular mutation (Fachal et al, 2011). This correlates with the observation by Al-Mulla et al, 2009, where breast cancer patients with BRCA1-truncating mutations in exon 2 (28/30 mutations assessed were c.68_69delAG) had a significantly higher age of diagnosis compared with carriers of mutations in exons 11, 13 and 20, therefore suggesting protein-truncating mutations in exon 2 have a lower penetrance than would be expected in this gene. The c.68_69delAG mutation has also been found in the non-Ashkenazi-Jewish, Spanish and UK (Yorkshire) populations, with haplotype evidence for this mutation having arisen by independent mutational events in different populations (Neuhausen et al, 1996; Fackenthal and Olopade, 2007). Therefore, without detailed haplotype analysis it cannot be determined which ancestry is relevant in this study. The carrier of this mutation in our study was diagnosed with PrCa at 58 years and following treatment had 12 years disease-free survival.
The c.212+1G>T (BIC IVS5+1G>T) mutation disrupts the splice donor site, allowing read-through into the intron and termination of transcription at codon 64. This mutation has also been observed in a Spanish breast cancer study in a family with breast and bladder cancers (Diez et al, 2010). The patient with this mutation was diagnosed at 57 years and survived for only 3 years after diagnosis; he also has a close relative who was diagnosed with bladder cancer.
The c.1954dupA (BIC 2080insA) mutation is a frame-shift causing insertion, resulting in termination of transcription at codon 672 and has been reported as a Pakistani founder mutation (Liede et al, 2002).
The c.2475delC mutation (BIC 2594delC) is a frame-shift deletion, resulting in termination at codon 845. This mutation appears to be a Scandinavian/Northern European founder mutation from the ethnicity of the study samples recorded in BIC and in two Scandinavian studies (Johannsson et al, 1996; Thomassen et al, 2008).
The incidence of truncating BRCA1 mutations in this study for PrCa cases diagnosed at ⩽65 years (3/802) is 0.37%, and 0.45% in all cases (4/886). Of the four pathogenic mutations identified, only one patient had a family history of PrCa (father, age unavailable), one patient had a family history of bladder cancer (brother, age unavailable) and two patients were from families with first-degree relatives with breast or ovarian cancer (details in Table 2). The small number of mutation carriers does not allow the statistical evaluation of the tumour characteristics in this study; however, we have conducted a clinical assessment of a much larger set of BRCA1 and BRCA2 mutation carriers and non-carrier PrCa cases, which indicates that both BRCA1 and BRCA2 mutations confer a more aggressive PrCa phenotype (Castro et al, submitted).
Based on the previously estimated carrier frequency of BRCA1 mutations in the United Kingdom, we estimate that germline mutations in the BRCA1 gene confer a RR of PrCa of ∼3.7-fold and this translates to an 8.6% cumulative risk by the age of 65 years. Other studies have estimated higher frequencies of BRCA1 mutation prevalence but these did not allow for the presence of other genetic effects (Whittemore et al, 2004). It is important to emphasise that in addition to the population frequency of BRCA1 mutations various other factors could influence the estimates of RR. Also, if one or more of the missense changes (UVs) were pathogenic, the risk estimates would be higher. On balance our estimate of ∼3.7-fold increased risk for BRCA1 mutation carriers is accurate based on incidence rates in the United Kingdom. Several studies addressed the estimation of PrCa risk by modelling and incorporating various factors such as PSA level (Thompson et al, 2006) or the recently discovered common genetic variants found by GWAS (Macinnis et al, 2011). This latest model also has the capacity to incorporate clinical information and effect of rare genes once this becomes available.
These results suggest that routine testing of early onset PrCa cases for germline BRCA1 mutations would further help to refine the prevalence and risk associated with BRCA1 mutations and may be useful for guiding management options.
Acknowledgments
We acknowledge the NCRN nurses and Consultants for their work in the UKGPCS study. This work was supported by The Prostate Cancer Research Foundation (now Prostate Action) and by Cancer Research UK (grant numbers C5047/A7357, C1287/A10118, C1287/A5260, C5047/A3354, C5047/A10692, C16913/A6135 and C16913/A6835). We also thank the following for funding support: the Institute of Cancer Research and the Everyman Campaign, Prostate Research Campaign UK, the Orchid Cancer Appeal, the National Cancer Research Network, UK and the National Cancer Research Institute, UK. We acknowledge NHS funding to the NIHR Biomedical Research Centre at the Institute of Cancer Research and the Royal Marsden NHS Foundation Trust.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
The authors declare no conflict of interest.
Supplementary Material
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mulla F, Bland JM, Serratt D, Miller J, Chu C, Taylor GT (2009) Age-dependent penetrance of different germline mutations in the BRCA1 gene. J Clin Pathol 62: 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Pharoah PDP, McMullan G, Day NE, Stratton MR, Peto J, Ponder BJ, Easton DF (2002) A comprehensive model for familial breast cancer incorporating BRCA1, BRCA2 and other genes. Br J Cancer 86: 76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlas E, Stramwasser M, Mueller CR (2001) A CREB site in the BRCA1 proximal promoter acts as a constitutive transcriptional element. Oncogene 20: 7110–7114 [DOI] [PubMed] [Google Scholar]
- Chun S, Fay JC (2009) Identification of deleterious mutations within three human genomes. Genome Res 19: 1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Górski B, Gronwald J, Huzarski T, Byrski T, Debniak T, Jakubowska A, Wokołorczyk D, Gliniewicz B, Sikorski A, Stawicka M, Godlewski D, Kwias Z, Antczak A, Krajka K, Lauer W, Sosnowski M, Sikorska-Radek P, Bar K, Klijer R, Romuald Z, Małkiewicz B, Borkowski A, Borkowski T, Szwiec M, Posmyk M, Narod SA, Lubiński J (2008) BRCA1 mutations and prostate cancer in Poland. Eur J Cancer Prev 17: 62–66 [DOI] [PubMed] [Google Scholar]
- Diez O, Gutiérrez-Enríquez S, Balmaña J (2010) Heterogeneous prevalence of recurrent BRCA1 and BRCA2 mutations in Spain according to the geographical area: implications for genetic testing. Fam Cancer 9: 187–191 [DOI] [PubMed] [Google Scholar]
- DiNardo DN, Butcher DT, Robinson DP, Archer TK, Rodenhiser DI (2001) Functional analysis of CpG methylation in the BRCA1 promoter region. Oncogene 20: 5331–5340 [DOI] [PubMed] [Google Scholar]
- Edwards SM, Dearnaley DP, Ardern-Jones A, Hamoudi RA, Easton DF, Ford D, Shearer R, Dowe A, Eeles RA. The CRC/BPG UK Familial Prostate Cancer Study Collaborators (1997) No germline mutations in the dimerization domain of MXI1 in prostate cancer clusters. Br J Cancer 76: 992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Dearnaley DP, Ardern-Jones AT, Shearer RJ, Easton DF, Ford D, Edwards SM, Dowe A (1997) Familial prostate cancer: the evidence and the Cancer Research Campaign/British Prostate Group (CRC/BPG) UK Familial Prostate Cancer Study. Br J Urol 79(Suppl 1): 8–14 [DOI] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Giles GG, Olama AAA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF (2008) Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet 40: 316–321 [DOI] [PubMed] [Google Scholar]
- Fachal L, Gómez-Caamaño A, Celeiro-Muñoz C, Peleteiro P, Blanco A, Carballo A, Forteza J, Carracedo A, Vega A (2011) BRCA1 mutations do not increase prostate cancer risk: results from a meta-analysis including new data. Prostate 71(16): 1768–1779 [DOI] [PubMed] [Google Scholar]
- Fackenthal JD, Olopade OI (2007) Breast cancer risk associated with BRCA1 and BRCA2 in diverse populations. Nat Rev Cancer 7: 937–948 [DOI] [PubMed] [Google Scholar]
- Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P (2007) Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol 18: 581–592 [DOI] [PubMed] [Google Scholar]
- Fokkema IFAC, Taschner PEM, Schaafsma GCP, Celli J, Laros JFJ, den Dunnen JT (2011) LOVD v.2.0: the next generation in gene variant databases. Hum Mutat 32: 557–563 [DOI] [PubMed] [Google Scholar]
- Goh CL, Schumacher FR, Easton D, Muir K, Henderson B, Kote-Jarai Z, Eeles RA (2012) Genetic variants associated with predisposition to prostate cancer and potential clinical implications. J Intern Med 271(4): 353–365 [DOI] [PubMed] [Google Scholar]
- Grönberg H, Damber L, Damber JE (1994) Studies of genetic factors in prostate cancer in a twin population. J Urol 152: 1484–1487, , discussion 1487–1489 [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Blondal T, Jakobsdottir M, Stacey SN, Kostic J, Kristinsson KT, Birgisdottir B, Ghosh S, Magnusdottir DN, Thorlacius S, Thorleifsson G, Zheng SL, Sun J, Chang B-L, Elmore JB, Breyer JP, McReynolds KM, Bradley KM, Yaspan BL, Wiklund F, Stattin P, Lindström S, Adami H-O, McDonnell SK, Schaid DJ, Cunningham JM, Wang L, Cerhan JR, Sauver JL, Isaacs SD, Wiley KE, Partin AW, Walsh PC, Polo S, Ruiz-Echarri M, Navarrete S, Fuertes F, Saez B, Godino J, Weijerman PC, Swinkels DW, Aben KK, Witjes JA, Suarez BK, Helfand BT, Frigge ML, Kristjansson K, Ober C, Jonsson E, Einarsson GV, Xu J, Gronberg H, Smith JR, Thibodeau SN, Isaacs WB, Catalona WJ, Mayordomo JI, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K (2008) Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet 40: 281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannsson O, Ostermeyer EA, Håkansson S, Friedman LS, Johansson U, Sellberg G, Brøndum-Nielsen K, Sele V, Olsson H, King MC, Borg A (1996) Founding BRCA1 mutations in hereditary breast and ovarian cancer in southern Sweden. Am J Hum Genet 58: 441–450 [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff T, Kauff ND, Mitra N, Nafa K, Huang H, Palmer C, Gulati T, Wadsworth E, Donat S, Robson ME, Ellis NA, Offit K (2004) BRCA mutations and risk of prostate cancer in Ashkenazi Jews. Clin Cancer Res 10: 2918–2921 [DOI] [PubMed] [Google Scholar]
- Kote-Jarai Z, Leongamornlert DA, Saunders E, Tymrakiewicz M, Castro E, Mahmud N, Guy M, Edwards SM, O’Brien L, Sawyer E, Hall AL, Wilkinson R, Dadaev T, Goh C, Easton DF, Goldgar DE, Eeles RA (2011) BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: implications for genetic testing in prostate cancer patients. Br J Cancer 105: 1230–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K (2000) Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343: 78–85 [DOI] [PubMed] [Google Scholar]
- Liede A, Malik IA, Aziz Z, Rios Pd P de los, Kwan E, Narod SA (2002) Contribution of BRCA1 and BRCA2 mutations to breast and ovarian cancer in Pakistan. Am J Hum Genet 71: 595–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jian X, Boerwinkle E (2011) dbNSFP: a lightweight database of human non-synonymous SNPs and their functional predictions. Hum Mutat 32(8): 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macinnis RJ, Antoniou AC, Eeles R a, Severi G, Al Olama AA, McGuffog L, Kote-Jarai Z, Guy M, O’Brien LT, Hall AL, Wilkinson R a, Sawyer E, Ardern-Jones AT, Dearnaley DP, Horwich A, Khoo VS, Parker CC, Huddart R a, Van As N, McCredie MR, English DR, Giles GG, Hopper JL, Easton DF (2011) A risk prediction algorithm based on family history and common genetic variants: application to prostate cancer with potential clinical impact. Genet Epidemiol 35: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini DN, Rodenhiser DI, Ainsworth PJ, O’Malley FP, Singh SM, Xing W, Archer TK (1998) CpG methylation within the 5′ regulatory region of the BRCA1 gene is tumor specific and includes a putative CREB binding site. Oncogene 16: 1161–1169 [DOI] [PubMed] [Google Scholar]
- Mathe E, Olivier M, Kato S, Ishioka C, Hainaut P, Tavtigian SV (2006) Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res 34: 1317–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhausen SL, Mazoyer S, Friedman L, Stratton M, Offit K, Caligo A, Tomlinson G, Cannon-Albright LA, Bishop T, Kelsell D, Solomon E, Weber B, Couch F, Struewing J, Tonin P, Durocher F, Narod SA, Skolnick MH, Lenoir G, Serova O, Ponder B, Stoppa-Lyonnet D, Easton DF, King MC, Goldgar DE (1996) Haplotype and phenotype analysis of six recurrent BRCA1 mutations in 61 families: results of an international study. Am J Hum Genet 58: 271–280 [PMC free article] [PubMed] [Google Scholar]
- Page WF, Braun MM, Partin AW, Caporaso N, Walsh P (1997) Heredity and prostate cancer: a study of World War II veteran twins. Prostate 33: 240–245 [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rödelsperger C, Schuelke M, Seelow D (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7: 575–576 [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom TM Exonprimer. Available at http://ihg.gsf.de/ihg/ExonPrimer.html
- Szabo C, Masiello A, Ryan JF, Brody LC (2000) The breast cancer information core: database design, structure, and scope. Hum Mutat 16: 123–131 [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Byrnes GB, Goldgar DE, Thomas A (2008) Classification of rare missense substitutions, using risk surfaces, with genetic- and molecular-epidemiology applications. Hum Mutat 29: 1342–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Breast Cancer Linkage Consortium (1999) Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst 91: 1310–1316 [DOI] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, Cancel-Tassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Hoover R, Hayes RB, Hunter DJ, Chanock SJ (2008) Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet 40: 310–315 [DOI] [PubMed] [Google Scholar]
- Thomassen M, Hansen TVO, Borg A, Lianee HT, Wikman F, Pedersen IS, Bisgaard ML, Nielsen FC, Kruse TA, Gerdes A-M (2008) BRCA1 and BRCA2 mutations in Danish families with hereditary breast and/or ovarian cancer. Acta Oncol 47: 772–777 [DOI] [PubMed] [Google Scholar]
- Thompson D, Easton DF (2001) Variation in cancer risks, by mutation position, in BRCA2 mutation carriers. Am J Hum Genet 68: 410–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Easton DF (2002) Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst 94: 1358–1365 [DOI] [PubMed] [Google Scholar]
- Thompson IM, Ankerst DP, Chi C, Goodman PJ, Tangen CM, Lucia MS, Feng Z, Parnes HL, Coltman CA (2006) Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst 98: 529–534 [DOI] [PubMed] [Google Scholar]
- Vallée MP, Francy TC, Judkins MK, Babikyan D, Lesueur F, Gammon A, Goldgar DE, Couch FJ, Tavtigian SV (2011) Classification of missense substitutions in the BRCA genes: a database dedicated to Ex-UVs. Hum Mutat 33(1): 22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore AS, Gong G, John EM, McGuire V, Li FP, Ostrow KL, DiCioccio R, Felberg A, West DW (2004) Prevalence of BRCA1 mutation carriers among U.S. non-hispanic whites. Cancer Epidemiol Biomarkers Prev 13: 2078–2083 [PubMed] [Google Scholar]
- Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PEM (2008) Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat 29: 6–13 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.