Abstract
Background:
Brain tumour stem cells (BTSCs) are a small population of cancer cells that exhibit self-renewal, multi-drug resistance, and recurrence properties. We have shown earlier that peroxisome proliferator-activated receptor gamma (PPARγ) agonists inhibit the expansion of BTSCs in T98G and U87MG glioma. In this study, we analysed the influence of PPARγ agonists on the expression of stemness and differentiation genes in BTSCs.
Methods:
The BTSCs were isolated from T98G and DB29 glioma cells, and cultured in neurobasal medium with epidermal growth factor+basic fibroblast growth factor. Proliferation was measured by WST-1 (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2 H-5-tetrazolio]-1,3-benzene disulphonate) and 3H thymidine uptake assays, and gene expression was analysed by quantitative reverse--transcription PCR and Taqman array. The expression of CD133, SRY box 2, and nanog homeobox (Nanog) was also evaluated by western blotting, immunostaining, and flow cytometry.
Results:
We found that PPARγ agonists, ciglitazone and 15-deoxy-Δ12,14-ProstaglandinJ2, inhibited cell viability and proliferation of T98G- and DB29-BTSCs. The PPARγ agonists reduced the expansion of CD133+ BTSCs and altered the expression of stemness and differentiation genes. They also inhibited Sox2 while enhancing Nanog expression in BTSCs.
Conclusion:
These findings highlight that PPARγ agonists inhibit BTSC proliferation in association with altered expression of Sox2, Nanog, and other stemness genes. Therefore, targeting stemness genes in BTSCs could be a novel strategy in the treatment of glioblastoma.
Keywords: glioblastoma, brain tumour stem cells, PPARγ agonist, stemness genes, Sox2, Nanog
Brain tumours are among the most devastating cancers that remain resistant to therapy and pose major health problems throughout the world. There are numerous types of brain tumours identified in humans with varying biological and clinical outcomes. Glioblastoma is one of the most frequent primary malignant brain tumours in adults. It is estimated that median survival is generally less than 1 year, and most patients die within 2 years from the time of diagnosis (Deorah et al, 2006). Standard therapy includes surgical resection followed by radiation and chemotherapy, which have limited efficacy and significant side effects (Peacock and Lesser, 2006). Multi-drug resistance and fast recurrence are some of the key challenges in combating brain tumours. Thus, further investigations are needed to develop novel therapeutic strategies for the treatment of brain tumours in patients. Cancer stem cells (CSCs) are a small population of cancer cells that exhibit characteristics of typical stem cells, such as asymmetric division, self-renewal, proliferation, and tumour-initiation capabilities. The CSCs were first identified in acute myeloid leukaemia (Bonnet and Dick, 1997), and then in breast (Al-Hajj et al, 2003), prostate (Patrawala et al, 2006), liver (Yang et al, 2008), and skin cancer (Schatton et al, 2008). Earlier studies have also identified putative brain tumour stem cells (BTSCs) on the basis of their ability to form tumour spheres and the expression of neural stem cell marker CD133 (Singh et al, 2003; Singh et al, 2004). The failure to cure cancer has been attributed to the fact that conventional therapies target rapidly growing tumour cells, which respond transiently, whereas sparing CSCs (Liu et al, 2006). Thus, optimal treatment for glioblastoma requires targeting the self-renewal capabilities of BTSCs, which are responsible for tumour recurrence and progression in patients.
Two important growth factors commonly used to maintain the self-renewal of embryonic stem cells (ESCs), neural stem cells (NSCs), and BTSCs are epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF; Wells, 1999). These growth factors induce a complex network of multiple signalling pathways, leading to the expression of stemness gene clusters associated with the self-renewal of stem cells. Stemness genes encode for a group of proteins that have critical roles in maintaining the pluripotency of ESCs (Takahashi and Yamanaka, 2006; Yu et al, 2007). Ectopic expression of SRY (sex determining region Y)-box 2 (Sox2) in conjunction with the octamer-binding transcription factor 4, also known as POU5F1 (POU domain, class 5, transcription factor 1; Oct4), nanog homeobox (Nanog), and Lin28 was sufficient to induce reprogramming of human fibroblasts into induced pluripotent stem cells with all the essential characteristics of ESCs (Yu et al, 2007; Takahashi et al, 2007). The SRY box 2 (Sox2) is an SRY-related high-mobility group box transcription factor that is expressed in the ESCs, NSCs, and trophoblast stem cells, but not in their differentiated derivatives (Avilion et al, 2003; D'Amour and Gage, 2003; Catena et al, 2004; Episkopou, 2005). Gene targeting experiments have shown that the Sox2 expression is required to maintain pluripotency of stem cells during early embryonic development (Avilion et al, 2003). More importantly, constitutive expression of Sox2 maintains stemness and inhibits neuronal differentiation (Graham et al, 2003). Nanog is a homeobox protein, which in combination with Oct4 and Sox2, constitutes a core transcription complex that maintains the self-renewal and stemness characteristics of human ESCs (Boyer et al, 2005). Interestingly, earlier studies have demonstrated the expression of many stemness genes, including Sox2 and Nanog, in human glioma-derived BTSCs (Kappadakunnel et al, 2010), suggesting their significance to the self-renewal and tumourigenicity of glioblastoma.
Peroxisome proliferator-activated receptor (PPAR) is a family of ligand-dependent nuclear receptor transcriptional factors that regulate physiological processes ranging from development to reproduction, metabolism, cell proliferation, differentiation, apoptosis, and homeostasis. Three known subtypes of the PPAR family are PPARα, PPARγ, and PPARδ (Kliewer et al, 1992). The PPARγ is abundantly expressed in many cell types, where it regulates lipid metabolism, glucose homeostasis, tumour progression, and inflammation. Polyunsaturated fatty acids, eicosanoids, prostaglandins, and linoleic acid have been identified as endogenous ligands for PPARγ, whereas thiazolidinedione class of compounds function as high-affinity synthetic agonists for PPARγ (Rubenstrunk et al, 2007). Upon activation with specific ligands, PPARγ forms a heterodimer complex with retinoid X receptor, which then mediates the target gene expression (Schwartz et al, 1998). Because of its physiological significance, PPARγ has been explored as a therapeutic modality for various diseases (Blumberg and Evans, 1998). Earlier studies have shown that PPARγ agonists induce growth arrest and apoptosis in glioma cells in culture, suggesting their use in the treatment of brain tumours (Papi et al, 2009). In a recent report, we showed that PPARγ agonists 15-deoxy-Δ12,14-ProstaglandinJ2 (15d-PGJ2) and ciglitazone inhibit growth and expansion of CD133+ BTSCs in culture (Chearwae and Bright, 2008), but the mechanism by which PPARγ agonists regulate BTSCs is not known.
In this study, we investigated the molecular mechanisms by which PPARγ agonists regulate BTSCs in culture. We found that PPARγ agonists alter the expression of various stemness and differentiation genes, suggesting the significance of PPARγ in the treatment of glioblastoma.
Materials and methods
Glioma cell culture
Human glioblastoma cell line T98G was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). DB29 primary human glioma cells were established in our laboratory by culturing surgically removed brain tumour tissue samples obtained from the tissue repository at Methodist Research Institute with an IRB approved protocol. Glioma cells were cultured in Dulbecco’s modified Eagle medium (DMEM) with 10% FBS, 1 mℳ sodium pyruvate, 100 U ml−1 penicillin G, 100 μg ml−1 streptomycin, 2 mℳ glutamine, 1 mℳ MEM non-essential amino acids, and 50 μℳ 2β-mercaptoethanol in 5% CO2 incubator at 37 °C. The cells were dissociated using 0.25% trypsin with 0.05 mℳ EDTA solution and sub-cultured once in 3–5 days (Chearwae and Bright, 2008; Chakraborty et al, 2011).
Brain tumour stem cell culture
To induce the growth and expansion of BTSCs as gliospheres, we adopted a culture condition established in our laboratory (Chearwae and Bright, 2008; Chakraborty et al, 2011). Briefly, T98G and DB29 glioma cells were cultured in neurobasal medium (NBM) supplemented with B27 (Invitrogen, Madison, WI, USA) in the presence of 10 ng ml−1 EGF (PeproTech, Rocky Hill, NJ, USA) and bFGF (R&D Systems, Minneapolis, MN, USA). The BTSCs (5 × 104 ml per well) were cultured in 12-well plates in 5% CO2 incubator at 37 oC, and 5 to 7-day-old gliospheres were used for the experiments.
Proliferation assay
The effect of PPARγ agonists on the proliferation of glioma and BTSCs was measured by WST-1 (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2 H-5-tetrazolio]-1,3-benzene disulphonate); Roche, Indianapolis, IN, USA) and 3H thymidine uptake assays. Briefly, T98G and BD29 glioma cells were cultured in 96-well tissue-culture plates (1 × 104/200 μl per well) in DMEM in the presence of 10% FBS (Invitrogen) and 1% penicillin–streptomycin in 5% CO2 incubator at 37 °C. T98G- and DB29-BTSCs were obtained by dissociating 5 to 7-day-old gliospheres generated as above by treatment with Accutase (Sigma Chemicals, St Louis, MO, USA) for 15 min at 37 oC. The BTSCs were cultured in 96-well tissue culture plates (1 × 104/200 μl per well) in NBM in the presence of B27 and 10 ng ml−1 EGF+bFGF in 5% CO2 incubator at 37 oC. Different concentrations of PPARγ agonists, ciglitazone (CalBiochem, San Diego, CA, USA) or 15d-PGJ2 (Sigma Chemicals), were added at the initiation of culture and compared with the DMSO-treated control. The WST-1 reagent (10 μl per well) was added at 46 h, and the OD was measured at 450 nm after 48 h using a titre-plate reader (Alpha Diagnostics, San Antonio, TX, USA). 3H thymidine (0.5 μ Ci per well) was added at 24 h, and cells were harvested after 48 h using a Tomtec 96 harvester (Tomtec, Hamden, CT, USA). The amount of 3H thymidine uptake was measured using Wallac Microbeta liquid scintillation counter (Perkin Elmer, Fremont, CA, USA).
Quantitative reverse-transcription PCR
The effect of PPARγ agonists on the expression of stemness and differentiation genes in BTSCs was determined by quantitative reverse-transcription PCR (qRT–PCR) technique. Briefly, T98G- and DB29-BTSCs were cultured in NBM with 10 ng ml−1 EGF and bFGF with different concentrations of ciglitazone or 15d-PGJ2 at 37 oC for 24 h. Total RNA was extracted using RNeasy kit according to the manufacturer’s instructions (Qiagen, Valencia, CA, USA). Equal amount of RNA was reverse transcribed into cDNA, and qRT−PCR was performed using 384-well TaqMan Low-Density Human Stem Cell Gene Array Card in 7900 HT fast Real-time PCR system (Applied Biosystems, Foster City, CA, USA). To examine the mRNA expression of Sox2 and Nanog, RNA samples were reverse transcribed into cDNA, using random primer and master mix from TaqMan reverse transcription kit (Applied Biosystems, Branchburg, NJ, USA). The cDNA samples were then amplified in TaqMan Universal Master Mix with optimised concentrations of Sox2 and Nanog primer sets in a standard optical 96-well reaction plate. Results were analysed using Prism 7900 relative quantification (ΔΔCt) study software (Applied Biosystems, Foster City, CA, USA). Heat map was constructed using the DataAssist software (Applied Biosystems, Foster City, CA, USA). Box plot, scatter plot, and Venn diagrams were generated using GraphPad Prism 5.0 software (GraphPad, La Jolla, CA, USA).
SDS–PAGE and western blot analysis
The effect of PPARγ agonists on the expression of proteins in BTSCs was determined by SDS–PAGE and western blotting. Briefly, T98G- and DB29-BTSCs were cultured in NBM with 10 ng ml−1 EGF+bFGF in the presence of different concentrations of ciglitazone or 15d-PGJ2 at 37 oC for 24 h. Cells were washed in ice-cold PBS, and whole-cell lysates were prepared by boiling in lysis buffer (0.2 M Tris−HCl pH 6.8, 0.8 μg ml−1 SDS, 4% glycerol, 0.588 ℳ β-mercaptoethanol, 0.05 ℳ EDTA, 8 μg ml−1 bromophenol blue) for 5 min. Total protein was resolved on 10% SDS–PAGE using a mini-gel apparatus (BioRad, Hercules, CA, USA) and transferred to a nylon/PVDF membrane (Millipore, Bedford, MA, USA) using a Novablot transfer system (Pharmacia, Piscataway, NJ, USA). The residual binding sites in the membrane were blocked by incubating with PBST (PBS and 0.1% Tween 20) containing 5% non-fat dry milk powder for 1 h. The blots were incubated with rabbit anti-Sox2, rabbit anti-Nanog, goat anti-glial fibrillary acidic protein (GFAP), rabbit anti-chondroitin sulphate proteoglycan 4 (NG2), mouse anti-β-III Tubulin, mouse anti-CD133, or mouse anti-β-actin antibody (1 : 500, Santa Cruz Biotechnology, Santa Cruz, CA, USA) in PBST containing 1% non-fat milk powder at room temperature for 1 h. Membranes were then washed in PBST and incubated with corresponding peroxidase-conjugated anti-IgG antibody (1 : 2500, Santa Cruz Biotechnology) in PBS containing 1% milk powder for 1 h. The blots were then developed using Super-signal West-pico chemiluminescence reagent (Thermo Scientific, Rockford, IL, USA).
Immunostaining and flow cytometry
The effect of PPARγ agonists on the expansion of CD133+, Sox2+ or Nanog+ BTSCs was analysed by immunostaining and flow cytometry. T98G- and DB29-BTSCs were cultured in 12-well tissue-culture plates in NBM with B27 and 10 ng ml−1 EGF+bFGF in the absence or presence of 10 μℳ ciglitazone or 15d-PGJ2 in 5% CO2 incubator at 37 °C. After 24 or 48 h, BTSCs were dissociated using Accutase for 15 min at 37 oC and suspended in PBS containing 0.1% BSA. Cells were washed, fixed, and permeabilised using BD Cytofix/Cytoperm solution (BD Bioscience Pharmingen, San Diego, CA, USA) at room temperature for 15 min. The BTSCs were washed and incubated with PE-conjugated anti-CD133 antibody (1 : 10, Miltenyi Biotec, Auburn, CA, USA) in PBS with 0.1% BSA for 1 h. Cells were also stained with anti-Nanog or anti-Sox2 antibody (1 : 100, Santa Cruz Biotechnology) in BD Perm/Wash buffer at 4 °C for 1 h. Cells were then washed and incubated with DyLight 633-conjugated secondary antibody (1 : 200, Thermo Scientific) at 4 oC for 30 min. DyLight 633 (1 : 200) and PE-conjugated mouse IgG1 antibody (1 : 10, Miltenyi Biotec) were used as isotype controls. The percentage of CD133+, Sox2+, or Nanog+ BTSCs was analysed using FACS Calibur Flow Cytometer with CellQuest software (BD Biosciences, San Jose, CA, USA).
For immunostaining, T98G and DB29 gliospheres were fixed in 1% paraformaldehyde, washed, and stained with PE-conjugated anti-CD133 antibody (1 : 10, Miltenyi Biotec) in PBS with 0.1% BSA at 4 °C for 1 h. Gliospheres were also stained with anti-Sox2 and anti-Nanog antibody (1 : 100, Santa Cruz Biotechnology) in BD Perm/Wash buffer at 4 °C for 30 min, followed by incubation with DyLight 594-conjugated secondary antibody (1 : 2000, Thermo Scientific) at 4 oC for 30 min. DyLight 594-conjugated secondary antibody (1 : 2000) and PE-conjugated mouse IgG1 antibody (1 : 10, Miltenyi Biotec) were used as isotype controls. Gliospheres were mounted on a glass slide using Vectashield mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA) and photographed using Leica Leitz DMRB fluorescent microscope (Leica Microsystems, Buffalo Grove, IL, USA).
Statistical analysis
The data was analysed by one-way ANOVA using the GraphPad Prism 5.0 software. The values are mean±s.d./s.e.m., and the significance is expressed as *P<0.05, **P<0.01, and ***P<0.001 in the figures.
Results
Peroxisome proliferator-activated receptor gamma agonists inhibit the proliferation of glioma and BTSCs in culture
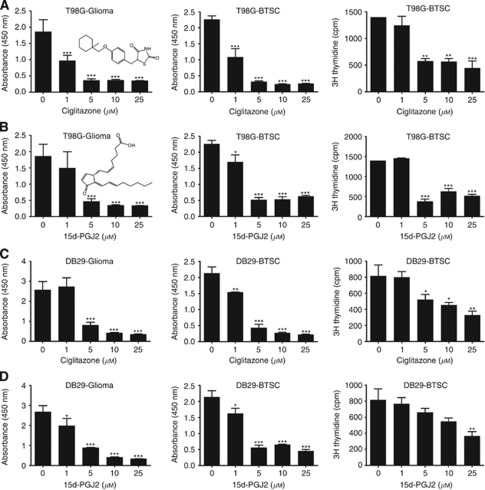
To investigate the use of PPARγ agonists for the treatment of glioblastoma, we first examined the effect of PPARγ agonists on the viability of glioma cells in culture. As shown in Figure 1A and B, addition of a PPARγ agonist, ciglitazone or 15d-PGJ2, resulted in a dose-dependent decrease in T98G glioma viability, reaching a significant effect at 5 μℳ. Similarly, PPARγ agonists also induced a dose-dependent inhibition of cell viability in DB29 primary human glioma in culture (Figure 1C and D). Both T98G and DB29 glioma cells showed comparable inhibitory response to ciglitazone and 15d-PGJ2 in culture.
Figure 1.
Inhibition of glioma and BTSC proliferation by PPARγ agonists. T98G (A and B) and DB29 (C and D) glioma and BTSCs were cultured in DMEM or NBM in the presence of ciglitazone (A and C) or 15d-PGJ2 (B and D). Cell viability and proliferation were determined using WST−1 and 3H thymidine-uptake assays. Chemical structures (inserts) of ciglitazone and 15d-PGJ2 were created using ChemDraw software (CambridgeSoft, Cambridge, MA, USA). Values are mean of triplicates (±s.d.) and *P<0.05, **P<0.01 and ***P<0.001 were considered significant. The figure is a representative of three independent experiments.
To determine the potential use of PPARγ agonists for BTSC-targeted therapy in glioblastoma, we then examined the proliferation of BTSCs using the WST-1 assay. We found that in vitro culture of BTSCs isolated from T98G and DB29 glioma grew and expanded in NBM supplemented with EGF+bFGF. Interestingly, addition of 1, 5, 10, and 25 μℳ ciglitazone or 15d-PGJ2 resulted in a dose-dependent inhibition of proliferation of both T98G- and DB29-BTSCs in culture (Figure 1). To confirm the results, we then examined the proliferation of BTSCs using the 3H thymidine uptake assay. As shown in Figure 1, addition of 1, 5, 10, and 25 μℳ PPARγ agonists resulted in a dose-dependent inhibition of proliferation of both T98G- and DB29-BTSCs. These findings highlight the use of PPARγ agonists for BTSC-targeted therapy in glioblastoma.
Peroxisome proliferator-activated receptor gamma agonists inhibit CD133+ BTSC expansion and promote differentiation
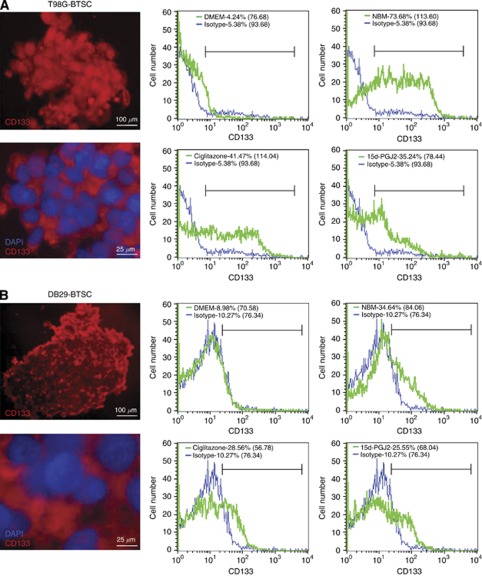
To determine the effect of PPARγ agonists on BTSCs, we examined the expression of CD133, an important stemness marker, in T98G- and DB29-BTSCs. We found that in vitro treatment with PPARγ agonists significantly reduced the percentage of CD133+ cells (Figure 2A and B). The T98G glioma cells cultured in DMEM showed 4.24% CD133+ cells with a mean fluorescent intensity (MFI) of 76.68, whereas T98G-BTSCs cultured in NBM showed 73.68% CD133+ cells with an MFI of 113.60 (Figure 2A). The percentage of CD133+ cells in T98G-BTSCs decreased to 41.47 and 35.24% with an MFI of 114.04 and 78.44 following treatment with ciglitazone and 15d-PGJ2, respectively. Similarly, DB29 glioma cultured in DMEM showed 8.98% CD133+ cells with an MFI of 70.58, whereas DB29-BTSCs cultured in NBM showed 34.64% CD133+ cells with an MFI of 84.06 (Figure 2B). Treatment of DB29-BTSCs with ciglitazone and 15d-PGJ2 also reduced the percentage of CD133+ cells to 28.56 and 25.55% with an MFI of 56.78 and 68.04, respectively.
Figure 2.
Inhibition of CD133 expression in BTSCs by PPARγ agonists. T98G (A) and DB29 (B) glioma and BTSCs were cultured in DMEM or NBM in the presence of ciglitazone or 15d-PGJ2 for 48 h. Cells were dissociated and stained with PE-conjugated anti-CD133 antibody or isotype control. The percentage of CD133+ cells and mean fluorescent intensities (MFI) in parentheses were calculated using CellQuest software. Gliospheres were also stained with PE-conjugated anti-CD133 antibody or isotype control with DAPI and photographed ( × 100 or × 400) under a fluorescent microscope. Scale bars represent 100 and 25 μm. The figure is a representative of four independent experiments.
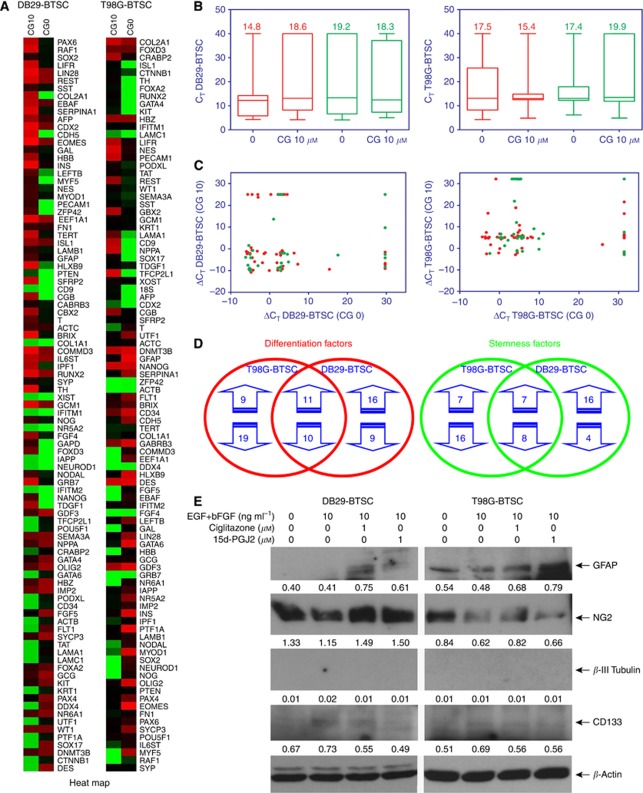
Western blot analyses showed that gliospheres express detectable levels of CD133 that decreased after treatment with PPARγ agonists in culture (Figure 3). To examine if the inhibition of stemness marker CD133 was associated with an increase in the expression of differentiation markers, we treated T98G- and DB29-BTSCs with ciglitazone or 15d-PGJ2 for 24 h, and analysed their protein expression by western blot. As shown in Figure 3E, we found that T98G glioma cells expressed detectable level of astrocyte marker GFAP that decreased in T98G-BTSCs and increased after treatment with 10 μℳ ciglitazone or 15d-PGJ2. DB29 glioma and BTSCs showed low expression of GFAP that also increased after treatment with ciglitazone or 15d-PGJ2 (Figure 3E). We have also found that both T98G and DB29 glioma cells expressed early oligodendrocyte marker NG2 that decreased in BTSCs and increased after treatment with PPARγ agonists, but more so in DB29-BTSCs than T98G-BTSCs (Figure 3E). The expression of neuronal marker β-III Tubulin was not detected in T98G- and DB29-BTSCs cultured in the absence or presence of PPARγ agonists. These results suggest that PPARγ agonists induce glial differentiation of BTSCs in glioma.
Figure 3.
Alteration of differentiation and stemness genes in BTSCs by PPARγ agonists. T98G- and DB29-BTSCs were cultured in NMB in the presence of 10 μℳ ciglitazone. Total RNA was extracted after 24 h, and the gene expression profile determined by qRT−PCR using TaqMan low-density optical 384-well human stem cell gene panel. Gene expression in DB29- and T98G-BTSCs was normalised to 18S and GAPDH, respectively. The heat map (A) was constructed using DataAssist software. Box plots (B), scatter plots (C), and Venn diagrams (D) were generated using GraphPad Prism 5.0 software. The differentiation genes and stemness genes are shown in red and green, respectively. Lines in the box plots represent the median and the numbers represent the mean. (E) Total cell lysates were prepared from T98G and DB29 glioma, and BTSCs treated with 10 μℳ ciglitazone or 15d-PGJ2 for 24 h, and the expression of GFAP, NG2, β−III Tubulin, CD133, and β-actin was determined by western blot. Numbers represent the relative quantities of protein calculated using β-actin as internal control. The figure is a representative of two independent experiments.
Peroxisome proliferator-activated receptor gamma agonists alter the expression of differentiation genes in BTSCs
To further define the molecular mechanisms of PPARγ agonists, we then examined the effect of ciglitazone on the expression of 50 differentiation genes in DB29- and T98G-BTSCs using the TaqMan Low-Density Human Stem Cell Gene Array Card (384 wells). As shown in Figure 3A, the heat map shows significant changes in the gene expression profile of DB29- and T98G-BTSCs after treatment with ciglitazone (Figure 3A). Box-plot and scatter-plot analyses also showed significant alteration in the expression of various differentiation genes in DB29- and T98G-BTSCs after treatment with ciglitazone in culture (Figure 3B and C). As presented in Table 1, DB29-BTSCs showed a⩾100-fold increase in the transcription of 10- and 1 to 100-fold increase in 14 differentiation genes, whereas 3 differentiation genes went from ND to D (not detected to detected). DB29-BTSCs also showed a 1 to 10-fold decrease in 4 and 10 to 100-fold decrease in 5, whereas 11 differentiation genes went from D to ND. The T98G-BTSCs showed⩾100-fold increase in the expression of 4 and a 1 to 100-fold increase in 7, whereas 9 differentiation factors went from ND to D after treatment with ciglitazone. The T98G-BTSCs also showed a 1 to 100-fold decrease in 12 and a⩾100-fold decrease in 11, whereas 6 differentiation genes went from D to ND. We have also found that three differentiation genes (COL1A1, IAPP, and NEUROD1) were not detected (ND to ND) before and after treatment with ciglitazone in DB29-BTSCs, whereas DDX4 is the only differentiation gene not detected before and after treatment with ciglitazone in T98G-BTSCs (Table 1).
Table 1. Expression of differentiation factors in PPARγ agonist treated-BTSCs.
| Symbol | Gene name | DB29-BTSC | T98G-BTSC |
|---|---|---|---|
| CDH5 | Cadherin 5 type 2 | ND–D | 0.01 |
| PECAM1 | Platelet/endothelial cell adhesion molecule | ND–D | 100.2 |
| MYF5 | Myogenic factor 5 | ND–D | DND |
| COL2A1 | Collagen type II alpha 1 | 3.3 × 1015 | 3.917 |
| CGB | Chorionic gonadotropin beta polypeptide | 1.5 × 1014 | 47.2 |
| HLXB9 | Motor neuron and pancreas homeobox 1 | 2.9 × 107 | 0.004 |
| IPF1 | Pancreatic and duodenal homeobox 1 | 2.1 × 105 | 0.02 |
| SERPINA1 | Serpin peptidase inhibitor clade A | 5700 | 0.018 |
| TH | Tyrosine hydroxylase | 3800 | 9.0 × 1012 |
| PAX6 | Paired box 6 | 2600 | 0.405 |
| INS | Insulin | 948.6 | 0.005 |
| HBB | Haemoglobin beta | 319.9 | D–ND |
| NES | Nestin | 307.0 | 129.8 |
| EOMES | Eomesodermin | 64.65 | 0.001 |
| T | Brachyury homologue | 54.75 | D–ND |
| CDX2 | Caudal type homeobox 2 | 46.92 | ND–D |
| MYOD1 | Myogenic differentiation 1 | 29.23 | D–ND |
| GCM1 | Glial cells missing homologue 1 | 27.06 | 2.017 |
| LAMB1 | Laminin beta 1 | 26.43 | 0.519 |
| AFP | Alpha fetoprotein | 23.42 | ND–D |
| SST | Somatostatin | 22.68 | 5.876 |
| GFAP | Glial fibrillary acidic protein | 19.27 | 0.003 |
| RUNX2 | Runt-related transcription factor 2 | 16.49 | ND–D |
| SYP | Synaptophysin | 16.008 | 0.83 |
| ISL1 | Insulin gene enhancer, ISL LIM homeobox 1 | 4.074 | ND–D |
| ACTC1 | Actin alpha cardiac muscle 1 | 1.736 | 0.421 |
| FN1 | Fibronectin 1 | 1.597 | 0.155 |
| WT1 | Wilms tumour 1 | 0.761 | 9.887 |
| FOXA2 | Forkhead box A2 | 0.23 | ND–D |
| NPPA | Natriuretic peptide A | 0.14 | ND–D |
| OLIG2 | Oligodendrocyte lineage transcription factor 2 | 0.118 | 0.006 |
| HBZ | Haemoglobin zeta | 0.086 | D–ND |
| GCG | Glucagon | 0.066 | 0.886 |
| GATA4 | GATA-binding protein 4 | 0.066 | ND–D |
| SYCP3 | Synaptonemal complex protein 3 | 0.037 | 0.039 |
| PAX4 | Paired box 4 | 0.025 | 0.073 |
| CD34 | CD34 molecule | D–ND | 0.067 |
| DDX4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 | D–ND | ND–ND |
| DES | Desmin | D–ND | 0.626 |
| FLT1 | Vascular endothelial growth factor receptor 1 | D–ND | 0.068 |
| GATA6 | GATA-binding protein 6 | D–ND | 0.001 |
| KRT1 | Keratin 1 | D–ND | 1.547 |
| LAMA1 | Laminin alpha 1 | D–ND | 1.9 × 107 |
| LAMC1 | Laminin gamma 1 | D–ND | ND–D |
| PTF1A | Pancreas-specific transcription factor, 1a | D–ND | 0.002 |
| SOX17 | SRY (sex determining region Y)-box 17 | D–ND | ND–D |
| TAT | Tyrosine aminotransferase | D–ND | 5.015 |
| COL1A1 | Collagen type I alpha 1 | ND–ND | 0.105 |
| IAPP | Islet amyloid polypeptide | ND–ND | 0.151 |
| NEUROD1 | Neurogenic differentiation 1 | ND–ND | D–ND |
Abbreviations: bFGF=basic fibroblast growth factor; BTSCs=brain tumour stem cells; EGF=epidermal growth factor; PPARγ=peroxisome proliferator-activated receptor gamma; qRT-PCR=quantitative reverse-transcription PCR. T98G and DB29-BTSCs were cultured in NMB with EGF+bFGF in the presence of 10 μℳ ciglitazone. Total RNA was extracted after 24 h and the gene expression profile was determined by qRT–PCR using TaqMan optical 384-well human stem cell gene panel. Gene expression was normalised to 18S and GAPDH in DB29-BTSCs and T98G-BTSCs, respectively. The fold change (RQ) in the differentiation gene expression profile compared with DMSO control was calculated and presented in the table. The genes were arranged according to the expression levels in DB29-BTSCs from high to low. D=detected; ND=not detected. The data is a representative of two independent experiments.
Further analyses showed that the in vitro treatment with ciglitazone increased the expression of a total of 27 differentiation genes in DB29-BTSCs and 20 differentiation genes in T98G-BTSCs (Figure 3D). Among them, the expression of 11 common differentiation genes (PECAM1, COL2A1, CGB, TH, NES, CDX2, GCM1, AFP, SST, RUNX2, and ISL1) was upregulated by ciglitazone in both DB29- and T98G-BTSCs.
Peroxisome proliferator-activated receptor gamma agonists alter the expression of stemness genes in BTSCs
We also analysed the expression of 40 stemness genes by qRT−PCR using TaqMan Low-Density Human Stem Cell Gene Array Card (384 wells). We found that the in vitro treatment with ciglitazone significantly altered the expression of stemness genes in DB29- and T98G-BTSCs compared with the DMSO-treated control cells (Figure 3A). Box-plot and scatter-plot analyses also showed significant alteration in the expression of various stemness genes in DB29- and T98G-BTSCs after treatment with ciglitazone in culture (Figure 3B and C). Interestingly, DB29-BTSCs showed⩾100-fold increase in the transcription of 9 genes and a 1 to 100-fold increase in 8 stemness genes, whereas 6 stemness genes increased from ND to D levels after treatment with ciglitazone (Table 2). DB29-BTSCs also showed a 1 to 10-fold decrease in the expression of 4 and 10 to 100-fold decrease in 4 stemness genes, whereas 4 stemness genes went from D to ND after treatment with ciglitazone. Similarly, ciglitazone-treated T98G-BTSCs showed a⩾100-fold increase in the transcription of 2 genes and a 1 to 100-fold increase in 10 stemness genes, whereas 3 stemness genes went from ND to D. Moreover, T98G-BTSCs treated with ciglitazone also showed a 1 to 10-fold decrease in the expression of 11 and a 10 to 100-fold decrease in 5 stemness genes, whereas 8 stemness genes went from D to ND. We have also found that five stemness genes (CD9, CRABP2, IFITM1, IFITM2, and NR5A2) were not detected (ND to ND) before and after treatment with ciglitazone in DB29-BTSCs, whereas two stemness genes (GRB7, FGF4) were not detected before and after treatment with ciglitazone in T98G-BTSCs (Table 2).
Table 2. Expression of stemness factors in PPARγ agonist treated-BTSCs.
| Symbol | Gene name | DB29-BTSCs | T98G-BTSCs |
|---|---|---|---|
| SFRP2 | Secreted frizzled-related protein 2 | ND–D | 0.883 |
| TDGF1 | Teratocarcinoma-derived growth factor 1 | ND–D | 4.455 |
| FOXD3 | Forkhead box D3 | ND–D | 1.063 |
| NANOG | Nanog homeobox | ND–D | 0.969 |
| ZFP42 | Zinc finger protein 42 homologue | ND–D | 5.043 |
| PTEN | Phosphatase and tensin homologue | ND–D | 0.73 |
| LEFTB | Left-right determination factor 1 | 1.4 × 106 | 0.064 |
| BRIX | Biogenesis of ribosomes homologue | 7500 | 0.331 |
| TERT | Telomerase reverse transcriptase | 2300 | 0.756 |
| LIFR | Leukaemia inhibitory factor receptor alpha | 2200 | 57.99 |
| GRB7 | Growth factor receptor-bound protein 7 | 2000 | ND–ND |
| FGF4 | Fibroblast growth factor 4 | 1900 | ND–ND |
| GBX2 | Gastrulation brain homeobox 2 | 740.5 | 142.3 |
| XIST | X-inactive specific transcript | 318.1 | 2.099 |
| NODAL | Nodal homologue | 130.8 | D–ND |
| NOG | Noggin | 54.12 | D–ND |
| COMMD3 | COMM domain containing 3 | 37.06 | D–ND |
| GAL | Galanin prepropeptide | 35.94 | D–ND |
| EBAF | Left-right determination factor 2 | 27.29 | D–ND |
| LIN28 | Lin-28 homologue A | 24.44 | 0.028 |
| GABRB3 | Gamma-aminobutyric acid A receptor beta 3 | 16.84 | 0.102 |
| REST | RE1-silencing transcription factor | 13.23 | 1.3 × 104 |
| IL6ST | Interleukin 6 signal transducer | 3.799 | 0.005 |
| SEMA3A | Short basic domain-secreted (semaphorin) 3A | 0.594 | 19.079 |
| DNMT3B | DNA (cytosine-5-)-methyltransferase 3 beta | 0.242 | 0.485 |
| IMP2 | IMP2 inner mitochondrial membrane peptidase-like | 0.207 | 0.128 |
| NR6A1 | Nuclear receptor subfamily 6 group A member 1 | 0.176 | D–ND |
| KIT | V-kit Hardy–Zuckerman 4 feline sarcoma oncogene | 0.081 | ND–D |
| GDF3 | Growth differentiation factor 3 | 0.048 | 0.199 |
| FGF5 | Fibroblast growth factor 5 | 0.008 | D–ND |
| SOX2 | SRY (sex determining region Y)-box 2 | 0.003 | D–ND |
| POU5F1 | POU class 5 homeobox 1 | D–ND | 0.706 |
| PODXL | Podocalyxin-like | D–ND | 42.16 |
| TFCP2L1 | Transcription factor CP2-like 1 | D–ND | ND–D |
| UTF1 | Undifferentiated embryonic cell transcription factor 1 | D–ND | 0.048 |
| CD9 | CD9 molecule | ND–ND | ND–D |
| CRABP2 | Cellular retinoic acid-binding protein 2 | ND–ND | 42.61 |
| IFITM1 | Interferon-induced transmembrane protein 1 | ND–ND | 1.252 |
| IFITM2 | Interferon-induced transmembrane protein 2 | ND–ND | 0.844 |
| NR5A2 | Nuclear receptor subfamily 5 group A member 2 | ND–ND | 0.013 |
Abbreviations: bFGF=basic fibroblast growth factor; BTSCs=brain tumour stem cells; EGF=epidermal growth factor; PPARγ=peroxisome proliferator-activated receptor gamma; qRT-PCR=quantitative reverse transcription PCR. T98G and DB29-BTSCs were cultured in NMB with EGF+bFGF in the presence of 10 μℳ ciglitazone. Total RNA was extracted after 24 h, and the gene expression profile was determined by qRT–PCR using TaqMan optical 384-well human stem cell gene panel. Gene expression was normalised to 18S and GAPDH in DB29-BTSCs and T98G-BTSCs, respectively. The fold change (RQ) in the stemness gene expression profile compared with DMSO control was calculated and presented in the table. The genes were arranged according to the expression levels in DB29-BTSCs from high to low. D=detected; ND=not detected. The data is a representative of two independent experiments.
Further analyses showed that the in vitro treatment with ciglitazone resulted in the downregulation of a total of 12 stemness genes in DB29-BTSCs and 24 stemness genes in T98G-BTSCs (Figure 3D). Among them, the expression of eight common stemness genes (DNMT3B, IMP2, NR6A1, GDX4, FGF5, SOX2, POU5F1, and UTF1) was reduced by ciglitazone in DB29- and T98G-BTSCs. Interestingly, Sox2 expression was inhibited by ciglitazone in both DB29- and T98G-BTSCs, whereas Nanog expression was increased in DB29-BTSCs while remaining relatively unchanged in T98G-BTSCs. These results indicate that the PPARγ agonists differentially regulate the expression of a multitude of stemness and differentiation factors in BTSCs.
Peroxisome proliferator-activated receptor gamma agonists alter the expression of Sox2 in BTSCs
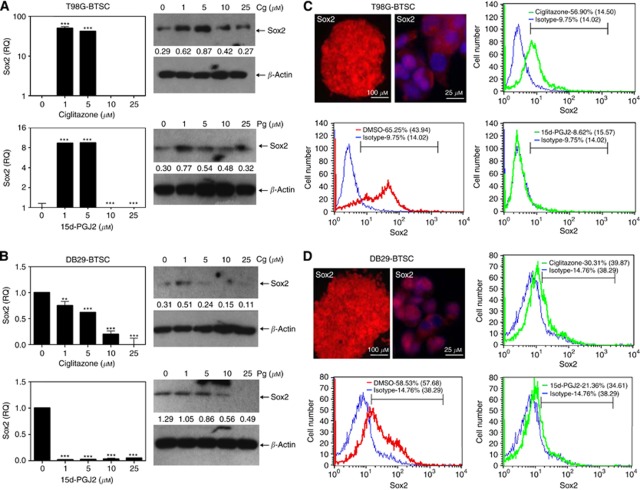
To understand the mechanism by which PPARγ agonists inhibit the growth of BTSCs, we then examined the expression of Sox2 mRNA by qRT−PCR analysis. As shown in Figure 4A, T98G-BTSCs cultured in NBM with EGF+bFGF expressed higher level of Sox2 mRNA following the addition of 1 and 5 μℳ ciglitazone or 15d-PGJ2, but expressed lower levels of Sox2 after treatment with 10 and 25 μℳ PPARγ agonists. Similarly, western blot analysis also showed that the in vitro treatment of T98G-BTSCs with ciglitazone or 15d-PGJ2 induced the expression of Sox2 protein at 1 and 5 μℳ, but inhibited its expression back to the starting level at 10 and 25 μℳ doses. In the case of DB29-BTSCs, qRT−PCR analysis showed that cells cultured in NBM with EGF+bFGF expressed detectable levels of Sox2 mRNA (Figure 4B). However, treatment with ciglitazone or 15d-PGJ2 resulted in a decrease of Sox2 mRNA and protein expression in DB29-BTSCs. These results suggest that PPARγ agonists may modulate the growth of BTSCs by altering the expression of Sox2.
Figure 4.
Modulation of Sox2 expression by PPARγ agonists in BTSCs. T98G-BTSCs (A and C) and DB29-BTSCs (B and D) were cultured in NBM with EGF+bFGF in the presence of 0, 1, 5, 10, and 25 μℳ ciglitazone or 15d-PGJ2 for 24 h. Sox2 mRNA expression was determined by qRT−PCR, and the values are presented as fold change (RQ) compared with DMSO control. Sox2 protein expression was determined by western blot and quantified using β-actin as internal control. T98G (C) and DB29 gliospheres (D) were stained with anti-Sox2 antibody followed by DyLight 594-conjugated second antibody and photographed ( × 100 or × 400) under a fluorescent microscope. Scale bars represent 100 and 25 μm. T98G and DB29 glioma and BTSCs (C and D) were stained with anti-Sox2 antibody followed by DyLight 633-conjugated second antibody or isotype control and analysed by flow cytometry. Histograms show isotype control (blue), DMSO (red), and either ciglitazone or 15d-PGJ2 (green), with mean fluorescent intensities (MFI) in parenthesis. Values are mean of triplicates (±s.e.m.) and **P<0.01 and ***P<0.001 were considered significant. The figure is a representative of three independent experiments.
To examine whether the inhibition of Sox2 expression was associated with a decrease in Sox2+ BTSCs, we then analysed the effect of PPARγ agonists on the percentage of Sox2+ cells in culture. As shown in Figure 4C, immunofluorescence and flow cytometry analyses demonstrated that T98G-BTSCs cultured in NBM with EGF+bFGF generated gliospheres with significant number of Sox2+ cells. Flow cytometry analyses showed that T98G-BTSCs constitute 65% of Sox2+ cells with an MFI of 44 that decreased to 57, and 9% with an MFI of 15 and 16 following treatment with 10 μℳ ciglitazone and 15d-PGJ2, respectively. Similarly, DB29 gliospheres cultured in NBM with EGF+bFGF constitute 59% of Sox2+ BTSCs with an MFI of 58 that decreased to 30 and 21%, with an MFI of 40 and 35 following treatment with ciglitazone and 15d-PGJ2, respectively (Figure 4D). Although both PPARγ agonists inhibited the expansion of Sox2+ BTSCs, treatment with 15d-PGJ2 resulted in a more significant inhibition of Sox2 expression in both T98G- and DB29-BTSCs. These findings suggest that PPARγ agonists modulate the expansion of Sox2+ BTSCs in glioma.
Peroxisome proliferator-activated receptor gamma agonists alter the expression of Nanog in BTSCs
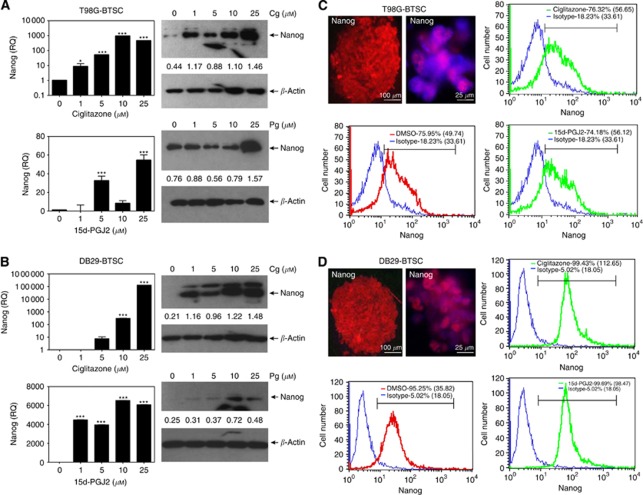
To further examine the effect of PPARγ agonists on BTSCs, we then analysed the expression of Nanog mRNA by qRT−PCR analysis. As shown in Figure 5A, T98G-BTSCs cultured in NBM with EGF+bFGF expressed detectable level of Nanog mRNA, which increased significantly after treatment with ciglitazone or 15d-PGJ2 in culture. Western blot analysis also showed that PPARγ agonists upregulated the Nanog expression in T98G-BTSCs. Additionally, DB29-BTSCs cultured in NBM with EGF+bFGF expressed low levels of Nanog, which was elevated after treatment with 1, 5, 10, and 25 μℳ ciglitazone or 15d-PGJ2 in culture (Figure 5B), suggesting the alteration of Nanog expression by PPARγ agonists in BTSCs.
Figure 5.
Modulation of Nanog expression by PPARγ agonists in BTSCs. T98G-BTSCs (A and C) and DB29-BTSCs (B and D) were cultured in NBM with EGF+bFGF in the presence of 0, 1, 5, 10, and 25 μℳ ciglitazone or 15d-PGJ2 for 24 h. Nanog mRNA expression was determined by qRT−PCR, and the values are presented as fold change (RQ) compared with DMSO control. Nanog protein expression was determined by western blot and quantified using β-actin as internal control. T98G (C) and DB29 gliospheres (D) were stained with anti-Nanog antibody followed by DyLight 594-conjugated second antibody and photographed ( × 100 or × 400) under a fluorescent microscope. Scale bars represent 100 and 25 μm. T98G and DB29 glioma and BTSCs (C and D) were stained with anti-Nanog antibody followed by DyLight 633-conjugated second antibody or isotype control and analysed by flow cytometry. Histograms show isotype control (blue), DMSO (red), and either ciglitazone or 15d-PGJ2 (green), with mean fluorescent intensities (MFI) in parenthesis. Values are mean of triplicates (±s.e.m.) and *P<0.05 and ***P<0.001 were considered significant. The figure is a representative of three independent experiments.
To examine whether the induction of Nanog expression was associated with an increase in Nanog+ BTSCs, we then analysed the effect of PPARγ agonists on the percentage of Nanog+ cells in culture. As shown in Figure 5C and D, immunofluorescence staining revealed the presence of Nanog expression in T98G- and DB29-BTSCs. Flow cytometry analyses showed that T98G gliospheres constitute 76% of Nanog+ BTSCs that remained at 76 and 74% after treatment with ciglitazone and 15d-PGJ2, respectively. However, the MFI was slightly changed, as the DMSO-treated T98G-BTSCs showed an MFI of 50 that increased to 57 and 56, following treatment with ciglitazone and 15d-PGJ2, respectively (Figure 5C). DB29 gliospheres constituted 95% of Nanog+ BTSCs that slightly increased to 99 and 100%, following treatment with ciglitazone and 15d-PGJ2, respectively. The DMSO-treated DB29-BTSCs showed an MFI of 36 that significantly increased to 113 and 98, following treatment with ciglitazone and 15d-PGJ2, respectively (Figure 5D). These findings suggest that PPARγ agonists modulate the expansion of Nanog+ BTSCs in glioma.
Discussion
The BTSCs represent a small fraction of self-renewing cancer cells that are resistant to therapy and responsible for brain tumour recurrence. Recent studies have established suitable conditions to expand CD133+ BTSCs as gliospheres in culture (Chearwae and Bright, 2008; Chakraborty et al, 2011). The EGF and bFGF are two important growth factors that are commonly used to maintain the self-renewal and multipotency of BTSCs. Growing evidence suggests that deregulated signalling pathways contribute to the continuous expansion of self-renewing CSCs in tumour (Pardal et al, 2003; Das et al, 2008; Kim and Dirks, 2008). Many glioma patients express elevated levels of EGF receptor, which is undetectable in normal brain (Martens et al, 2008). Likewise, bFGF is induced during reactive gliosis (Finklestein et al, 1988; Frautschy et al, 1991), and its overexpression seems prevalent in glioma (Murphy et al, 1989; Morrison et al, 1994). In this study, we have confirmed our earlier findings that PPARγ agonists inhibit the EGF- and bFGF-induced growth and expansion of BTSCs in culture (Chearwae and Bright, 2008). We have also found that PPARγ agonists inhibit CD133 expression and promote the expression of glial differentiation markers in BTSCs.
We found that the in vitro treatment of BTSCs with ciglitazone significantly altered the expression of various stemness and differentiation genes. One of the eight stemness factors repressed by ciglitazone in both DB29- and T98G-BTSCs is DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B). The DNMT3B has been shown to be elevated in glioma patients compared with normal brain (Kreth et al, 2011). Similarly, growth differentiation factor 3 (GDF3) is another stemness gene repressed by ciglitazone in both DB29- and T98G-BTSCs. This is significant, because an earlier study showed that GDF3 induces the progression of melanoma in mice (Kreth et al, 2011), suggesting that the repression of GDF3 may contribute to the inhibition of BTSC growth by PPARγ agonists. An earlier report also showed that FGF5 is important in the progression of malignant glioma (Allerstorfer et al, 2008). In this study, we found that ciglitazone inhibits FGF5 expression in both DB29- and T98G-BTSCs, suggesting its contribution to the regulation of BTSCs by PPARγ agonists.
Among the 90 genes examined in this study, we focused on the altered expression of Sox2 and Nanog by PPARγ agonists in BTSCs. The Sox2 is a key member of the Sox (SRY-like HMG box) family of transcription factors and is essential in maintaining ESC pluripotency (Bowles et al, 2000; Schepers et al, 2002). Along with Oct3/4, Sox2 regulates transcription of development-related genes and its own transcription via both positive and negative feedback mechanisms (Rizzino, 2009). It has been implicated in the pathogenesis of several cancers, including pulmonary, oesophageal, gastric, breast, pancreatic, and rectal cancer (Li et al, 2004; Sanada et al, 2006; Rodriguez-Pinilla et al, 2007; Chen et al, 2008; Park et al, 2008; Sholl et al, 2010). It is also overexpressed in malignant glioma, whereas minimally expressed in normal tissues (Schmitz et al, 2007). Silencing of Sox2 expression in BTSCs resulted in reduced growth and tumourigenicity in animal models (Gangemi et al, 2009). Earlier studies have shown that a partial reduction or elevation in Sox2 expression can lead to ESC differentiation, suggesting existence of a complex regulatory mechanism (Chew et al, 2005). In this study, we found that PPARγ agonists caused a slight upregulation of Sox2 at lower doses of PPARγ agonists and an inhibition of Sox2 at higher doses in both T98G- and DB29-BTSCs, with an exception being DB29-BTSCs treated with 15d-PGJ2 that showed a reduction of Sox2 expression at all doses. A reduction of Sox2 expression observed in this study suggests that this could be a mechanism by which PPARγ agonists regulate the growth and differentiation of BTSCs in glioblastoma.
Similarly, Nanog is a homeobox transcription factor that has a critical role in maintaining the self-renewal and pluripotency of ESCs during development (Takahashi and Yamanaka, 2006; Takahashi et al, 2007; Yu et al, 2007). Interestingly, earlier studies have demonstrated the presence of Nanog in human glioma-derived BTSCs, suggesting its significance to the self-renewal of BTSCs in glioblastoma (Kappadakunnel et al, 2010). In this study, we found that PPARγ agonists induce Nanog expression in both DB29- and T98G-BTSCs. In ESCs, overexpression of Sox2 can repress Nanog as well as itself through feedback inhibition (Rizzino, 2009). The direct link between Sox2 repression and Nanog upregulation by PPARγ agonists in BTSCs is yet to be determined. However, the differential regulation of Sox2 and Nanog by PPARγ agonists observed in this study suggests a critical role for these stemness factors in modulating growth and differentiation of BTSCs in glioma. Hence, our future studies will explore the mechanisms by which PPARγ agonists regulate differentiation and self-renewal of BTSCs.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
The authors declare no conflict of interest.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100(7): 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allerstorfer S, Sonvilla G, Fischer H, Spiegl-Kreinecker S, Gauglhofer C, Setinek U, Czech T, Marosi C, Buchroithner J, Pichler J, Silye R, Mohr T, Holzmann K, Grasl-Kraupp B, Marian B, Grusch M, Fischer J, Micksche M, Berger W (2008) FGF5 as an oncogenic factor in human glioblastoma multiforme: autocrine and paracrine activities. Oncogene 27(30): 4180–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R (2003) Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17(1): 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Evans RM (1998) Orphan nuclear receptors--new ligands and new possibilities. Genes Dev 12(20): 3149–3155 [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE (1997) Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3(7): 730–737 [DOI] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P (2000) Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol 227(2): 239–255 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122(6): 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catena R, Tiveron C, Ronchi A, Porta S, Ferri A, Tatangelo L, Cavallaro M, Favaro R, Ottolenghi S, Reinbold R, Scholer H, Nicolis SK (2004) Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem 279(40): 41846–41857 [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Kanakasabai S, Bright JJ (2011) Constitutive androstane receptor agonist CITCO inhibits growth and expansion of brain tumour stem cells. Br J Cancer 104(3): 448–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chearwae W, Bright JJ (2008) PPARgamma agonists inhibit growth and expansion of CD133+ brain tumour stem cells. Br J Cancer 99(12): 2044–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y, Shang Y (2008) The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem 283(26): 17969–17978 [DOI] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, Ng HH (2005) Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol 25(14): 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour KA, Gage FH (2003) Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc Natl Acad Sci USA 100(Suppl 1): 11866–11872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Srikanth M, Kessler JA (2008) Cancer stem cells and glioma. Nat Clin Pract Neurol 4(8): 427–435 [DOI] [PubMed] [Google Scholar]
- Deorah S, Lynch CF, Sibenaller ZA, Ryken TC (2006) Trends in brain cancer incidence and survival in the United States: Surveillance, Epidemiology, and End Results Program, 1973 to 2001. Neurosurg Focus 20(4): E1. [DOI] [PubMed] [Google Scholar]
- Episkopou V (2005) SOX2 functions in adult neural stem cells. Trends Neurosci 28(5): 219–221 [DOI] [PubMed] [Google Scholar]
- Finklestein SP, Apostolides PJ, Caday CG, Prosser J, Philips MF, Klagsbrun M (1988) Increased basic fibroblast growth factor (bFGF) immunoreactivity at the site of focal brain wounds. Brain Res 460(2): 253–259 [DOI] [PubMed] [Google Scholar]
- Frautschy SA, Walicke PA, Baird A (1991) Localization of basic fibroblast growth factor and its mRNA after CNS injury. Brain Res 553(2): 291–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G (2009) SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells 27(1): 40–48 [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, Pevny L (2003) SOX2 functions to maintain neural progenitor identity. Neuron 39(5): 749–765 [DOI] [PubMed] [Google Scholar]
- Kappadakunnel M, Eskin A, Dong J, Nelson SF, Mischel PS, Liau LM, Ngheimphu P, Lai A, Cloughesy TF, Goldin J, Pope WB (2010) Stem cell associated gene expression in glioblastoma multiforme: relationship to survival and the subventricular zone. J Neurooncol 96(3): 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CF, Dirks PB (2008) Cancer and stem cell biology: how tightly intertwined? Cell Stem Cell 3(2): 147–150 [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM (1992) Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358(6389): 771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth S, Thon N, Eigenbrod S, Lutz J, Ledderose C, Egensperger R, Tonn JC, Kretzschmar HA, Hinske LC, Kreth FW (2011) O-Methylguanine-DNA methyltransferase (MGMT) mRNA expression predicts outcome in malignant glioma independent of MGMT promoter methylation. PLoS One 6(2): e17156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XL, Eishi Y, Bai YQ, Sakai H, Akiyama Y, Tani M, Takizawa T, Koike M, Yuasa Y (2004) Expression of the SRY-related HMG box protein SOX2 in human gastric carcinoma. Int J Oncol 24(2): 257–263 [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS (2006) Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer 5: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens T, Laabs Y, Gunther HS, Kemming D, Zhu Z, Witte L, Hagel C, Westphal M, Lamszus K (2008) Inhibition of glioblastoma growth in a highly invasive nude mouse model can be achieved by targeting epidermal growth factor receptor but not vascular endothelial growth factor receptor-2. Clin Cancer Res 14(17): 5447–5458 [DOI] [PubMed] [Google Scholar]
- Morrison RS, Yamaguchi F, Saya H, Bruner JM, Yahanda AM, Donehower LA, Berger M (1994) Basic fibroblast growth factor and fibroblast growth factor receptor I are implicated in the growth of human astrocytomas. J Neurooncol 18(3): 207–216 [DOI] [PubMed] [Google Scholar]
- Murphy PR, Myal Y, Sato Y, Sato R, West M, Friesen HG (1989) Elevated expression of basic fibroblast growth factor messenger ribonucleic acid in acoustic neuromas. Mol Endocrinol 3(2): 225–231 [DOI] [PubMed] [Google Scholar]
- Papi A, Tatenhorst L, Terwel D, Hermes M, Kummer MP, Orlandi M, Heneka MT (2009) PPARgamma and RXRgamma ligands act synergistically as potent antineoplastic agents in vitro and in vivo glioma models. J Neurochem 109(6): 1779–1790 [DOI] [PubMed] [Google Scholar]
- Pardal R, Clarke MF, Morrison SJ (2003) Applying the principles of stem-cell biology to cancer. Nat Rev Cancer 3(12): 895–902 [DOI] [PubMed] [Google Scholar]
- Park ET, Gum JR, Kakar S, Kwon SW, Deng G, Kim YS (2008) Aberrant expression of SOX2 upregulates MUC5AC gastric foveolar mucin in mucinous cancers of the colorectum and related lesions. Int J Cancer 122(6): 1253–1260 [DOI] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, Coghlan L, Tang DG (2006) Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene 25(12): 1696–1708 [DOI] [PubMed] [Google Scholar]
- Peacock KH, Lesser GJ (2006) Current therapeutic approaches in patients with brain metastases. Curr Treat Options Oncol 7(6): 479–489 [DOI] [PubMed] [Google Scholar]
- Rizzino A (2009) Sox2 and Oct-3/4: a versatile pair of master regulators that orchestrate the self-renewal and pluripotency of embryonic stem cells. Wiley Interdiscip Rev Syst Biol Med 1(2): 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Pinilla SM, Sarrio D, Moreno-Bueno G, Rodriguez-Gil Y, Martinez MA, Hernandez L, Hardisson D, Reis-Filho JS, Palacios J (2007) Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod Pathol 20(4): 474–481 [DOI] [PubMed] [Google Scholar]
- Rubenstrunk A, Hanf R, Hum DW, Fruchart JC, Staels B (2007) Safety issues and prospects for future generations of PPAR modulators. Biochim Biophys Acta 1771(8): 1065–1081 [DOI] [PubMed] [Google Scholar]
- Sanada Y, Yoshida K, Ohara M, Oeda M, Konishi K, Tsutani Y (2006) Histopathologic evaluation of stepwise progression of pancreatic carcinoma with immunohistochemical analysis of gastric epithelial transcription factor SOX2: comparison of expression patterns between invasive components and cancerous or nonneoplastic intraductal components. Pancreas 32(2): 164–170 [DOI] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, Fuhlbrigge RC, Kupper TS, Sayegh MH, Frank MH (2008) Identification of cells initiating human melanomas. Nature 451(7176): 345–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers GE, Teasdale RD, Koopman P (2002) Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell 3(2): 167–170 [DOI] [PubMed] [Google Scholar]
- Schmitz M, Temme A, Senner V, Ebner R, Schwind S, Stevanovic S, Wehner R, Schackert G, Schackert HK, Fussel M, Bachmann M, Rieber EP, Weigle B (2007) Identification of SOX2 as a novel glioma-associated antigen and potential target for T cell-based immunotherapy. Br J Cancer 96(8): 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Raskin P, Fonseca V, Graveline JF (1998) Effect of troglitazone in insulin-treated patients with type II diabetes mellitus. Troglitazone and Exogenous Insulin Study Group. N Engl J Med 338(13): 861–866 [DOI] [PubMed] [Google Scholar]
- Sholl LM, Long KB, Hornick JL (2010) Sox2 expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl Immunohistochem Mol Morphol 18(1): 55–61 [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB (2003) Identification of a cancer stem cell in human brain tumors. Cancer Res 63(18): 5821–5828 [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB (2004) Identification of human brain tumour initiating cells. Nature 432(7015): 396–401 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131(5): 861–872 [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4): 663–676 [DOI] [PubMed] [Google Scholar]
- Wells A (1999) EGF receptor. Int J Biochem Cell Biol 31(6): 637–643 [DOI] [PubMed] [Google Scholar]
- Yang ZF, Ngai P, Ho DW, Yu WC, Ng MN, Lau CK, Li ML, Tam KH, Lam CT, Poon RT, Fan ST (2008) Identification of local and circulating cancer stem cells in human liver cancer. Hepatology 47(3): 919–928 [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318(5858): 1917–1920 [DOI] [PubMed] [Google Scholar]