Abstract
Mouse plasmacytoma (PCT) can develop within 45 days when induced by a v-abl/myc replication-deficient retrovirus. This fast-onset PCT development is always associated with trisomy of cytoband E2 of mouse chromosome 11 (11E2). Trisomy of 11E2 was identified as the sole aberration in all fast-onset mouse PCTs in [T38HxBALB/c]N congenic mice, with a reciprocal translocation between chromosome X and 11 (rcpT(X;11)) (Genes Cancer 2010;1:847–858). Using this mouse model, we have now examined the overall and individual telomere lengths in fast-onset PCTs compared with normal B cells using two-dimensional and three-dimensional quantitative fluorescent in situ hybridization of telomeres. We found fast-onset PCTs to have a significantly different three-dimensional telomere profile, compared with primary B cells of wild-type littermates with and without rcpT(X;11) (P < .0001 and P = .006, respectively). Our data also indicate for primary PCT cells, from the above mouse strain, that the translocation chromosome carrying 11E2 is the only chromosome with telomere lengthening (P = 4 x 10-16). This trend is not seen for T(X;11) in primary B cells of control [T38HxBALB/c]N mice with the rcpT(X;11). This finding supports the concept of individual telomere lengthening of chromosomes that are functionally important for the tumorigenic process.
Introduction
Telomeres are DNA-protein structures at the ends of mammalian linear chromosomes. Telomeres shorten with each cycle of cell division [1,2], and once they reach a critically short length, in a primary cell, this cell will normally undergo senescence [3–5]. Therefore, rapidly dividing cells, such as stem and tumor cells, or cells surviving “crisis,” often activate telomere lengthening mechanisms to ensure their replicative potential [6–9]. Approximately 85% of tumor cells use telomerase, a reverse transcriptase with an RNA template, to add telomeric repeats to the 3′ end of parental DNA [10,11]. However, not all tumor cells with elevated telomerase have long telomeres [12]. Approximately 15% of tumor cells use the alternative lengthening of telomeres mechanisms to lengthen telomeres through cycles of homologous recombination [13–16].
Telomere regulation plays a critical role in genome instability and tumorigenesis [17–19]. Critically short telomeres and uncapped telomeres can trigger a series of events that lead to genomic instability and include dynamic chromosome structure and number abnormalities [20–22]. Dysfunctional telomeres can cause the fusion of sister chromatids, or the fusion of neighboring chromosomes, leading to the formation of anaphase bridges [23]. These bridges break as the centromeres are pulled apart (so-called breakage-bridge-fusion cycles), which results in unbalanced translocations and terminal deletions in the daughter cells [8]. The repetitive nature of such aberrant genome remodeling cycles creates ongoing rearrangements, aneuploidy and polyploidy, all of which are commonly found in tumor cells, especially in tumors with complex karyotypes and high levels of cell-to-cell heterogeneity [24–28]. However, these cycles will end if a new telomere is acquired, usually through the cell's activation of telomere lengthening pathways.
Most cases reported in the literature show mean telomere shortening [29–31], but few studies have measured individual telomere lengths. Quantitation of telomere length using a telomeric peptide nucleic acid (PNA) probe was done for the first time in 1996 by Lansdorp et al. [32]. Chromosome-specific features were proposed to control telomere length because particular telomere lengths were associated with specific chromosomes [33]. Telomeres on specific chromosome arms were also found to have interallelic differences, through the use of STELA, a polymerase chain reaction-based technique [34,35]. Chromosome-specific telomere length changes have been identified in esophageal cancer [36], chronic myeloid leukemia [37,38], and breast cancer [39]. These findings in tumor cells have led to the hypothesis that there may be chromosome-specific protective factors leading to lengthening of certain telomeres on chromosomes “key” to the pathogenesis of the cancer [37,38]; however, the connection between a chromosome known to be critical for tumorigenesis and uniquely lengthened telomeres has yet to be made.
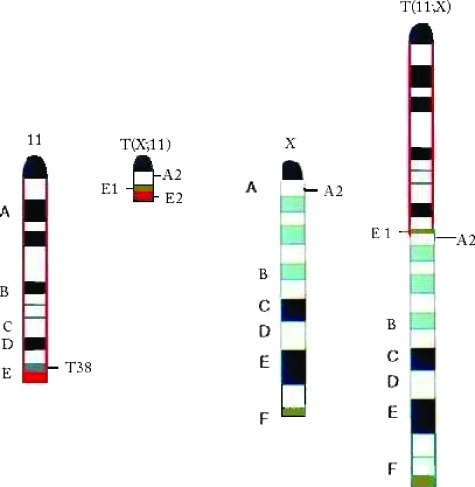
Mouse plasmacytoma (PCT) is a B-cell lineage tumor and the mouse model for human Burkitt lymphoma because they both result from c-myc activating chromosomal translocations [40] and are cytogenetically identical. Fast-onset mouse PCTs, which can develop within 45 days, are induced with v-abl/myc, which generates constitutive retroviral c-Myc expression in infected preB lymphocytes [41]. A characteristic of these tumors is the presence of chromosome 11 aberrations, which are the only chromosomal change in fast-onset PCTs [41]. The generation of a mouse model with a reciprocal translocation (rcpT) between chromosomes 11 and X, congenic [T38HxBALB/c]N with rcpT(X;11) mice, allowed us to narrow down the affected region on chromosome 11. This rcpT generates a very small T(X;11) translocation chromosome with only cytoband E2 and part of E1 (Figure 1). The chromosome 11 aberration found in all fast-onset PCTs was determined to be trisomy of the 11E2 cytoband, which contains all the genes necessary for the accelerated tumorigenesis of these tumors [42].
Figure 1.
Graphical illustration of the chromosomal constitution of chromosomes 11 in the (BALB/cxT38H) F1 N backcross generation mouse. Chromosome (Chr) 11 with the breakpoint T38H in the telomeric cytoband 11E1 (brown), whereas the breakpoint in Chr X is located in the centromeric A2 band. The cytoband 11E2 is red. The reciprocally translocated T(11;X) chromosome resulted from the fusion of the ABCD bands of Chr 11 proximal to the T38H breakpoint with the centromeric A2 band of Chr X. The T(X;11) chromosome was generated by the translocation of the X-derived A2 subband onto the 11E1 cytoband of Chr 11. The final, definitive version of this figure has been published in Genes Cancer. 2010; 1(8):847–858. SAGE Publications Ltd. All rights reserved. © http://online.sagepub.com.
We examined for the first time in mouse PCT, whether telomere dysfunction has a role in this tumor's development. We analyzed both the three-dimensional (3D) telomere organization and potential two-dimensional (2D) telomere length changes in the [T38HxBALB/c]N mouse model, which could have been difficult because of the long telomeres in inbred mice [29,43,44]. However, we present the consistent finding, in both 2D and 3D experiments, of significantly shorter telomeres in the fast-onset PCT cells compared with the control [T38HxBALB/c]N mice, with or without the rcpT(X;11) translocation. Our unique rcpT mouse model allows us to study the metaphase telomere length of the T(X;11) translocation chromosome for chromosome-specific telomere length changes that may indicate a protective mechanism. We find the T(X;11) translocation chromosome, which carries the 11E2 band, critical for fast-onset PCT development [42], has significantly longer telomeres in the fast-onset PCT compared with the wild-type rcpT(X:11) mice. Therefore, we demonstrate, for the first time, a connection between a chromosome key to a tumor's development and chromosome-specific telomere lengthening. This finding may provide significant support for a chromosome-specific protective mechanism used by tumor cells.
Materials and Methods
Cell Harvest, Culture, and Fixation
Primary lymphocytes were harvested from the spleens of congenic [T38HxBALB/c]N wild-type and [T38HxBALB/c]N with rcpT (X;11) mice [42]. Primary PCT cells were harvested from the ascites of fast-onset PCT mice [45]. All procedures were done in accordance with Animal Protocol 11-019 as approved by Central Animal Care Services, University of Manitoba (Winnipeg, Manitoba, Canada). For metaphase preparation of wild-type [T38HxBALB/c]N mice, with or without rcpT(X;11), lymphocytes were cultured short term (48–72 hours) in RPMI 1640 with 10% fetal bovine serum, 1% l-glutamine, 1% sodium pyruvate, 1% penicillin-streptomycin, and 0.1% β-mercaptoethanol (Invitrogen/Gibco, Burlington, Ontario, Canada) with lipopolysaccharide at a concentration of 10 µg/ml at 37°C in a humidified atmosphere and 5% CO2. Cells were maintained at a density of around 106 cells/ml.
2D Chromosome and 3D Nuclei Fixation
Primary cells from the mice or cell culture were spun down at 200g for 10 minutes at room temperature. For 3D fixation [24,46,47], the pellet was resuspended in 5 ml of 0.075 M KCl for 10 minutes at room temperature and then overlaid with 1 ml of fresh 3:1 methanol-to-acetic acid fixative and inverted gently. Twice more, the cells were spun down at 200g for 10 minutes at room temperature and resuspended in 2 ml of fixative. Thirty microliters of this solution was gently placed on a slide, and the remainder was stored at -20°C. For 2D fixation, the initial pellet was resuspended in 5 ml of 0.075 ml for 30 minutes at room temperature, spun down at 200g for 10 minutes at room temperature and then the pellet underwent drop fixation with 3:1 methanol-to-acetic acid fixative [48]. For statistical significance, at least 20 metaphases with nonoverlapping chromosomes and 30 nuclei were examined in three independent experiments for each mouse type [24,27].
Telomere Quantitative Fluorescence In Situ Hybridization
Telomere quantitative fluorescence in situ hybridization (Q-FISH) was performed on both 2D metaphases and 3D interphase nuclei with a PNA probe for telomeres purchased from DAKO (Glostrup, Denmark). In brief for the 2D telomere hybridization, the slides were fixed in fresh 3.7% formaldehyde/1x PBS, washed in 1x PBS, followed by a pepsin/HCl treatment and a second fixation. After ethanol dehydrations, the telomere PNA probe was applied, sealed onto the slide with rubber cement, and underwent a 3-minute denaturation at 80°C and 2-hour hybridization at 30°C using a Hybrite (Vysis; Abbott Diagnostics, Des Plains, IL). Then the slides were washed in 70% formamide/10 mM Tris at pH 7.4, 1x PBS, 0.1x SSC at 55°C, and 2x SSC/0.05% Tween 20. The cells were then counterstained with 4′6′-diamidino-2-phenylindole (DAPI), dehydrated, and mounted in Vectashield (Vector Laboratories, Burlington, Ontario, Canada). For the 3D telomere hybridization, briefly, the slides were fixed in 3.7% formaldehyde/1x PBS, incubated in 0.5% Triton X, followed by an hour incubation in glycerol and freeze-thaw treatment with liquid nitrogen. After 1x PBS washed and a HCl incubation, the slides were equilibrated in 70% formamide/2x SSC at pH 7.0. Then the slides underwent the same denaturation and hybridization, subsequent washes, staining, and mounting procedure as in the 2D fixation protocol.
In 3D, very short telomeres are defined as signals at a relative fluorescent intensity from 0 to 10,000 and short telomeres are defined as signals at a relative fluorescent intensity from 10,000 to 20,000. In 2D, very short telomeres are defined as signals at a relative fluorescent intensity from 0 to 30,000 and short telomeres are defined as signals at a relative fluorescent intensity from 30,000 to 50,000.
Image Acquisition
2D and 3D imaging and acquisition were performed using an AxioImager Z2 microscope (Carl Zeiss, Toronto, Canada), an AxioCam HR charge-coupled device (Carl Zeiss), and Zeiss AxioVision 4.8 software (Carl Zeiss). A 63x/1.4 oil objective lens (Carl Zeiss) was used with a DAPI filter, for detection of nuclear DNA staining, and a cyanine 3 filter, for detection of the telomere PNA probe signals. All metaphases images were taken at the same exposure time, and tricolor beads were used for standardization; the same protocol was used for all imaging of all interphase nuclei. For 3D imaging, 80 z stacks at 200 nm each, with x, y = 102 nm, z = 200 nm were acquired. The acquired images were deconvolved using the constrained iterative algorithm [49]. To quantitatively analyze the metaphase telomere signals, Case Data Manager 4.0 software (Applied Spectral Imaging, Migdal HaEmek, Israel) for PC was used. TeloView software [50] was used to quantitatively analyze the 3D interphase telomere signals.
Statistical Analysis
Statistical analysis was done to compare the percentage of interphase telomeric signals at a certain intensity, using bins at an interval of 1000, from 0 to 225,000. An analysis was also done on the percentage of metaphase telomere signals at a certain intensity, using bins at an interval of 5000, from 0 to 195,000. Using a χ2 test, the telomere signal distribution was compared between the fast-onset PCT and the wild-type mice, with and without rcpT(X;11), and between the two types of wild-type mice. One threshold was set at the 50th percentile or median. P < .01 is considered significant.
Statistical analysis was also done to compare the telomere length of translocation chromosome T(X;11) to the telomere length of all other chromosomes. This was done using a Kolmogorov-Smirnov test in both the fast-onset PCT and the [T38HxBALB/c]N with rcpT(X;11) wild-type mice. P < .001 is considered significant.
Results
3D Telomere Organization
To investigate the 3D telomere organization in wild-type mice, with or without rcpT(X;11), and in fast-onset PCTs of the same mice, 3D Q-FISH was done with a telomere-PNA probe. This approach preserves the 3D nuclear architecture of the cells and specifically labels interphase telomeres in the whole cell population. After acquisition and constrained iterative deconvolution, all 3D interphase nuclei were analyzed with TeloView to determine the number of telomeric signals per cell and their intensity. The signal intensity correlates to the length of the telomere [51].
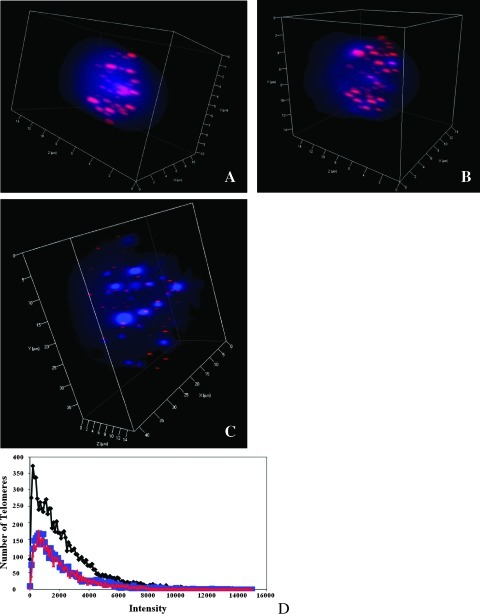
Figure 2 (A–C) shows a representative example of the 3D telomere organization in an interphase nucleus from the [T38HxBALB/c]N wild-type and [T38HxBALB/c]N with rcpT(X;11) control mice and the fast-onset PCT mouse. The telomeric signals were visibly smaller in the fast-onset PCT nuclei than in controls. The results from three independent experiments for each mouse type were combined, and the telomeric profiles of the three mice were compared. Figure 2D is a scatterplot that illustrates the number of telomere signals at a specific intensity. The distribution of telomeres is shown for intensities in the range of 0 to 160,000. As expected, 3D nuclei of the wild-type mice, whether carrying the rcpT(X;11) or not, have a similar telomere profile and are not significantly different, P = .3 (as determined by a χ2 test with P < .01 considered significant). The fast-onset PCT nuclei have many more low-intensity or very short telomeres (Materials and Methods), and their telomere profile is significantly different from both wild-type mice with or without rcpT(X;11) (P < .0001 and P = .006, respectively).
Figure 2.
3D telomere organization in interphase nuclei. (A) Nucleus from the [T38HxBALB/c]N wild-type mouse. (B) Nucleus from the [T38HxBALB/c]N with rcpT(X;11) mouse. (C) Nucleus from the fast-onset PCT mouse. (D) Comparison of the telomeric profile of the three mouse types. The intensity correlates to the telomere length. The fast-onset PCT nuclei (black) have significantly more very short telomeres (see Materials and Methods) than the [T38HxBALB/c]N wild-type (red) and the [T38HxBALB/c]N with rcpT(X;11) (blue) mouse (P =.006 and P < .0001, respectively).
2D Telomere Length
To determine the length of individual telomeres on all chromosomes in wild-type mice, with or without rcpT(X;11), and in fast-onset PCTs of the same mice, 2D Q-FISH was done with a telomere-PNA probe. This method hybridizes metaphases with a specific probe for labeling all telomeric signals. After acquisition, all metaphase spreads were analyzed with Case Data Manager 4.0 to determine the intensity of each telomere signal, which correlates to the length of the telomere [51].
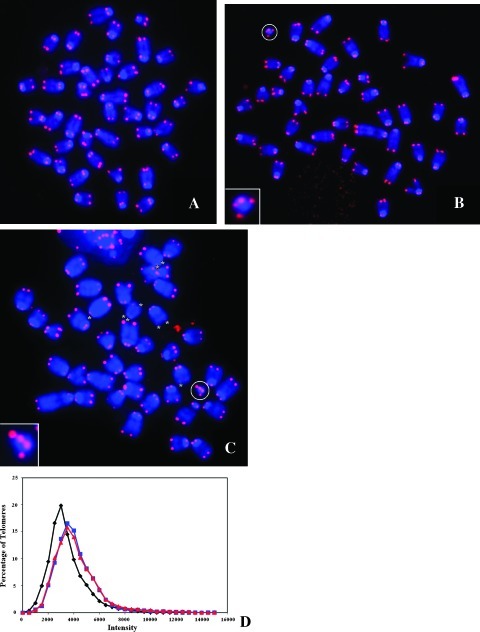
Figure 3 (A–C) illustrates an example of the telomeric signals in a metaphase spread from the [T38HxBALB/c]N wild-type and [T38HxBALB/c]N with rcpT(X;11) control mice and in fast-onset PCTs of [T38HxBALB/c]N mice. As indicated with a circle, the small T(X;11) translocation chromosome is easy to identify based on size alone (see also Figure 1). The results from three independent experiments for each mouse type were combined, and the telomere lengths of the three mice were compared. Figure 3D is a scatterplot that illustrates the percentage of telomere signals at a specific intensity. The distribution of telomeres is shown for intensities in the range of 0 to 160,000. As in the 3D interphase study, chromosomes from wild-type mice, with or without rcpT(X;11), have a similar distribution of telomere lengths and are not significantly different, P = 1.0 (as determined by a χ2 test with P < .01 considered significant). The chromosomes of fast-onset PCTs have more lower intensity or very short telomeres (see Materials and Methods), and their telomere length distribution is significantly different from [T38HxBALB/c]N mice, carrying rcpT(X;11) or not (P < .0001 for both mouse types).
Figure 3.
2D telomere length in metaphase chromosomes. (A) Metaphase spread from the [T38HxBALB/c]N wild-type mouse. (B) Metaphase spread from the [T38HxBALB/c]N with rcpT(X;11) mouse. The translocation chromosome T(X;11) is circled and enlarged. (C) Metaphase spread from the fast-onset PCT mouse. The translocation chromosome T(X;11) is circled and enlarged. Very short telomeres are marked with an asterisk. (D) Comparison of the telomeric length distribution of the three mouse types. The intensity correlates to the telomere length. The fast-onset PCT nuclei (black) have significantly more very short telomeres (see Materials and Methods) than the [T38HxBALB/c]N wild-type (red) and the [T38HxBALB/c]N with rcpT(X;11) (blue) mouse (P < .0001 for both mouse types).
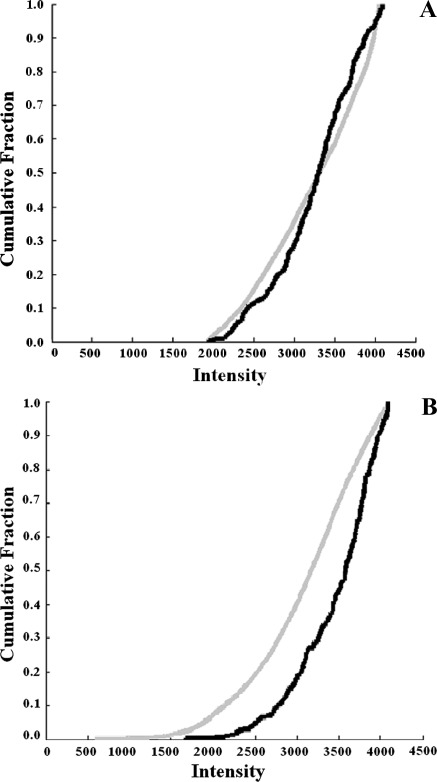
The graphs in Figure 4 (A and B) compare the telomere length of translocation chromosome T(X;11) to the telomere length of the rest of the chromosomes for the [T38HxBALB/c]N with rcpT(X;11) (Figure 4A) and fast-onset PCT mice (Figure 4B). In the tumor-free control mouse, [T38HxBALB/c]N with rcpT(X;11), the telomeres of the T(X;11) translocation chromosome do not have a significantly different length from those of the rest of the chromosomes, P =.006 (as determined by a Kolmogorov-Smirnov test with P < .001 considered significant). However, in the fast-onset PCT mouse, the telomeres of the T(X;11) translocation chromosome, carrying 11E2, are significantly longer than those of the rest of the chromosomes (P = 4 x 10-16).
Figure 4.
Comparison of the length of the T(X;11) translocation chromosome. (A) In the [T38HxBALB/c]N with rcpT(X;11) mouse, telomeres of the T(X;11) translocation chromosome (black) are not significantly longer than those of the other chromosomes (gray) (P = .006). (B) In the fast-onset PCT mouse, the telomeres of the T(X;11) translocation chromosome (black) are significantly longer than the other chromosomes (gray) (P = 4 x 10-16).
Discussion
Although short telomeres are a common finding in many tumor cells, 3D telomere organization and telomere length have not been previously examined in mouse PCT. Our mouse model offers a unique opportunity to study chromosome-specific telomere length regulation because its rcpT places only the genes necessary for accelerated tumorigenesis on a single easily identifiable chromosome, the T(X:11) translocation chromosome [42]. Because we also studied control wild-type mice, with and without the rcpT(X;11) translocation, we are able to make conclusions about tumor-specific telomere changes. Inbred mice can pose a problem for telomere studies because they normally have long telomeres [43,44]. When comparing generations of telomerase knockout mice, significant decreases in telomere size were not seen until generation 6 [29]. However, we present similar results regarding significant alterations in telomere length within the identical mouse strain/generation. Using two complementary approaches, 2D and 3D telomere Q-FISH, we identified significant telomere length changes during fast-onset PCT development.
We found that the 3D interphase nuclei of fast-onset PCT mice had visibly and significantly shorter telomeres compared with the wild-type mice, with or without the rcpT(X;11) translocation (P = .006 and P < .0001, respectively; Figure 2). We also determined that, overall, the telomeres on the metaphase chromosomes of fast-onset PCT mice were significantly shorter than in wild-type mice (P < .0001 for both wild-type mice; Figure 3). With a 3D interphase nuclei study, features of every cell can be analyzed, whereas only metaphases can be examined in a 2D study; therefore, a 3D analysis is more exact, and although it is complemented by the results of a 2D study, it cannot be replaced by solely a 2D analysis. By examining the telomeres of the T(X;11) translocation chromosome compared with all other telomeres, we identified that T(X;11) telomeres were significantly longer than telomeres on all other chromosomes in the fast-onset PCT cells (P = 4 x 10-16) but not significantly different in the tumor-free [T38HxBALB/c]N with rcpT(X;11) cells (P =.006; Figure 4).
It is now apparent that gene expression depends on more than the underlying DNA sequence. Therefore, it is important to study the nuclear architecture of cells, such as telomeres, because this may lead to new diagnostic and treatment options. Multiple studies have identified changes in the 3D telomere organization in disease states [25,46,52–56]. For example, myelodysplastic syndrome and acute myeloid leukemia are two distinct disorders, but part of the same disease spectrum because some cases of myelodysplastic syndrome will transform into acute myeloid leukemia [57]. Unique 3D telomeric signatures have been identified for normal versus tumor cells, which can aid in the diagnosis of patients [56].
Our findings in fast-onset mouse PCTs identify a tumor for which there is a unique 3D telomere profile compared with control cells (Figure 2D). More short telomeres were observed in the fast-onset PCT mouse compared with the control mice. This is consistent with our mouse model because a fast-developing tumor, with 45 days of latency period, exhibits a high rate of cell division and consequently displays enhanced telomere shortening, as we observed in this study.
Telomerase and alternative lengthening of telomeres, the mechanisms by which tumor cells lengthen telomeres to avoid cell senescence from critically short telomeres, are well described. However, chromosome-specific telomere alterations are not as well understood. In 2009, Samassekou et al. [37] identified chromosome arms, in chronic myeloid leukemia, that consistently had longer telomeres (18p, Xp, 1p, and 14p) and ones that consistently had shorter telomeres (20q, 21p, 21q, and 9q) than the rest of the chromosomes. In their most recent study, Samassekou et al. [38] found that Xp and 5p had significantly longer telomeres at a higher frequency in chronic myeloid leukemia than in healthy cells. They propose that the presence of only one X chromosome may drive the cell to protect it from telomere shortening. They also suggest that because the gene encoding the reverse transcriptase component of telomerase is located on 5p, the cell may lengthen its telomeres to regulate the gene. These findings have led them to propose that tumor cells may protect key genes responsible for their tumorigenesis through a chromosome-specific telomere lengthening mechanism.
Our study supports this hypothesis and goes one step further to prove its potential validity. In fast-onset PCTs, the T(X;11) translocation chromosome contains all the genes necessary for the tumor's accelerated development in cytoband 11E2 [42], and the trisomy of 11E2 represents the only aberration; therefore, it is probably the most important chromosome to protect. Our findings indicate that the T(X;11) translocation chromosome has significantly longer telomeres than all other chromosomes in the fast-onset PCT cells but not in B cells of the tumor-free [T38HxBALB/c]N mice with rcpT (X:11). Therefore, these findings illustrate a chromosome-specific protective mechanism and suggest a functional tumor-dependent requirement for its selective protection.
Mouse PCT is cytogenetically identical to human Burkitt lymphoma and therefore is an excellent mouse model of the cancer. The genes within mouse chromosome 11E2 are syntenic to rat chromosome 10q32 [58] and human chromosome 17q25 [59]. This region is duplicated, translocated and amplified in many lymphoid and nonlymphoid tumor types such as human acute myeloid leukemia, breast, ovarian, and thyroid cancers [59–62]. Therefore, the chromosome-specific telomere length increase observed on translocation chromosome T(X;11) in fast-onset mouse PCT may also occur on the human chromosome 17 due to the functional relevance of 17q25 for tumor development and warrants further investigation in an independent study.
To examine whether telomere dysfunction has an important role in mouse PCT, we used the techniques of 2D and 3D telomere Q-FISH to study the tumorigenic changes associated with fast-onset mouse PCT in interphase telomere organization and metaphase telomere length. We found selective telomere lengthening and aberrant 3D telomere organization in the fast-onset PCTs. Therefore, we provide significant support for a chromosome-specific protective mechanism that has a functional tumor-dependent requirement for its selective protection.
Acknowledgments
The authors thank Ludger Klewes for his assistance with mouse cell harvest and Mary Cheang for her assistance with statistical analysis.
Abbreviations
- 2D
two-dimensional
- 3D
three-dimensional
- PCT
plasmacytoma
- Q-FISH
quantitative fluorescence in situ hybridization
- rcpT
reciprocal translocation
Footnotes
The authors acknowledge funding support from Canadian Institutes of Health Research.
References
- 1.Olovnikov AM. The immune response and the process of marginotomy in lymphoid cells. Vestn Akad Med Nauk SSSR. 1972;27:85–87. [PubMed] [Google Scholar]
- 2.Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 3.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 4.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 5.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumor suppression: the senescence connection. Nat Rev Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 7.Misri S, Pandita S, Kumar R, Pandita TK. Telomeres, histone code, and DNA damage response. Cytogenet Genome Res. 2008;122:297–307. doi: 10.1159/000167816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lansdorp PM. Telomeres and disease. EMBO J. 2009;28:2532–2540. doi: 10.1038/emboj.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zvereva MI, Shcherbakova DM, Dontsova OA. Telomerase: structure, functions and activity regulation. Biochemistry (Mosc) 2010;75:1563–1583. doi: 10.1134/s0006297910130055. [DOI] [PubMed] [Google Scholar]
- 10.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 11.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 12.Xu L, Blackburn EH. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol Cell. 2007;28:315–327. doi: 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 14.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000;26:447–450. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 15.Muntoni A, Reddel RR. The first molecular details of ALT in human tumor cells. Hum Mol Genet. 2005;14:R191–R196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- 16.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 17.Odagiri E, Kanada N, Jibiki K, Demura R, Aikawa E, Dumura H. Reduction of telomeric length and c-erbB-2 gene amplification in human breast cancer, fibroadenoma, and gynecomastia. Relationship to histologic grade and clinical parameters. Cancer. 1994;73:2978–2984. doi: 10.1002/1097-0142(19940615)73:12<2978::aid-cncr2820731215>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 18.Meeker AK, Hicks JL, Iacobuzio-Donahue CA, Montgomery EA, Westra WH, Chan TY, Ronnett BM, De Marzo AM. Telomere length abnormalities occur early in the initiation of epithelial carcinogenesis. Clin Cancer Res. 2004;10:3317–3326. doi: 10.1158/1078-0432.CCR-0984-03. [DOI] [PubMed] [Google Scholar]
- 19.Rampazzo E, Bertorelle R, Serra L, Terrin L, Candiotto C, Pucciarelli S, Del Blanco P, Nitti D, De Rossi A. Relationship between telomere shortening, genetic instability, and site of tumor origin in colorectal cancers. Br J Cancer. 2010;102:1300–1305. doi: 10.1038/sj.bjc.6605644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 21.Gisselsson D, Jonson T, Petersen A, Strombeck B, Dal Cin P, Hoglund M, Mitelman F, Mertens F, Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc Natl Acad Sci USA. 2001;98:12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Murnane JP, Sabatier L. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. Bioessays. 2004;26:1164–1174. doi: 10.1002/bies.20125. [DOI] [PubMed] [Google Scholar]
- 24.Louis SF, Vermolen BJ, Garini Y, Young IT, Guffei A, Lichtensztejn Z, Kuttler F, Chuang TC, Moshir S, Mougey V, et al. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc Natl Acad Sci USA. 2005;102:9613–9618. doi: 10.1073/pnas.0407512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mai S, Garini Y. Oncogenic remodeling of the three-dimensional organization of the interphase nucleus: c-Myc induces telomeric aggregates whose formation precedes chromosomal rearrangements. Cell Cycle. 2005;4:1327–1331. doi: 10.4161/cc.4.10.2082. [DOI] [PubMed] [Google Scholar]
- 26.Mai S. Initiation of telomere-mediated chromosomal rearrangement in cancer. J Cell Biochem. 2010;109:1095–1102. doi: 10.1002/jcb.22501. [DOI] [PubMed] [Google Scholar]
- 27.Guffei A, Sarkar R, Klewes L, Righolt C, Knecht H, Mai S. Dynamic chromosomal rearrangements in Hodgkin's lymphomaare dueto ongoing three-dimensional nuclear remodeling and breakage-bridge-fusion cycles. Haematologica. 2010;95:2038–2046. doi: 10.3324/haematol.2010.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez P, Blasco MA. Telomeric and extratelomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 29.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 30.Rudolph KL, Chang S, Lee HW, Blasco M, Gottlieb GJ, Greider C, DePinho RA. Longevity, stress response, and cancer in aging telomerase-deficient mice. Cell. 1999;96:701–712. doi: 10.1016/s0092-8674(00)80580-2. [DOI] [PubMed] [Google Scholar]
- 31.Willeit P, Willeit J, Brandstatter A, Ehrlenbach S, Mayr A, Gasperi A, Weger S, Oberhollenzer F, Reindl M, Kronenberg F, et al. Cellular aging reflected by leukocyte telomere length predicts advanced atherosclerosis and cardiovascular disease risk. Arterioscler Thromb Vasc Biol. 2010;30:1649–1656. doi: 10.1161/ATVBAHA.110.205492. [DOI] [PubMed] [Google Scholar]
- 32.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum Mol Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 33.Martens UM, Zijlmans JM, Poon SS, Dragowska W, Yui J, Chavez EA, Ward RK, Lansdorp PM. Short telomeres on human chromosome 17p. Nat Genet. 1998;18:76–80. doi: 10.1038/ng0198-018. [DOI] [PubMed] [Google Scholar]
- 34.Baird DM, Rowson J, Wynford-Thomas D, Kipling D. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat Genet. 2003;33:203–207. doi: 10.1038/ng1084. [DOI] [PubMed] [Google Scholar]
- 35.Britt-Compton B, Rowson J, Locke M, Mackenzie I, Kipling D, Baird DM. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans. Hum Mol Genet. 2006;15:725–733. doi: 10.1093/hmg/ddi486. [DOI] [PubMed] [Google Scholar]
- 36.Xing J, Ajani JA, Chen M, Izzo J, Lin J, Chez Z, Gu J, Wu X. Constitutive short telomere length of chromosome 17p and 12q but not 11q and 2p is associated with an increased risk for esophageal cancer. Cancer Prev Res (Phila) 2009;2:459–465. doi: 10.1158/1940-6207.CAPR-08-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samassekou O, Ntwari A, Hebert J, Yan J. Individual telomere lengths in chronic myeloid leukemia. Neoplasia. 2009;11:1146–1154. doi: 10.1593/neo.09836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samassekou O, Li H, Hebert J, Ntwari A, Wang H, Cliché CG, Bouchard E, Huang S, Yan J. Chromosome arm-specific long telomeres: a new clonal event in primary chronic myelogenous leukemia cells. Neoplasia. 2011;13:550–560. doi: 10.1593/neo.11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng YL, Loffredo CA, Shields PG, Selim SM. Chromosome 9 arm-specific telomere length and breast cancer risk. Carcinogenesis. 2009;30:1380–1386. doi: 10.1093/carcin/bgp151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams JM, Gerondakis S, Webb E, Corcoran LM, Cory S. Cellular myc oncogene is altered by chromosome translocation to an immunoglobulin locus in murine plasmacytomas and is rearranged similarly in human Burkitt lymphomas. Proc Natl Acad Sci USA. 1983;80:1982–1986. doi: 10.1073/pnas.80.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiener F, Coleman A, Mock BA, Potter M. Nonrandom chromosomal change (trisomy 11) in murine plasmacytomas induced by an ABL-MYC retrovirus. Cancer Res. 1995;55:1181–1188. [PubMed] [Google Scholar]
- 42.Wiener F, Schmalter AK, Mowat MR, Mai S. Duplication of subcytoband 11E2 of chromosome 11 is regularly associated with accelerated tumor development in v-abl/myc-induced mouse plasmacytomas. Genes Cancer. 2010;1:847–858. doi: 10.1177/1947601910382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hande P, Slijepcevic P, Silver A, Bouffler S, van Buul P, Bryant P, Lansdorp P. Elongated telomeres in SCID mice. Genomics. 1999;56:221–223. doi: 10.1006/geno.1998.5668. [DOI] [PubMed] [Google Scholar]
- 44.Hemann MT, Greider CW. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res. 2000;28:4474–4478. doi: 10.1093/nar/28.22.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Potter M, Wiener F. Plasmacytomagenesis in mice: model of neoplastic development dependent upon chromosomal translocations. Carcinogenesis. 1992;13:1681–1697. doi: 10.1093/carcin/13.10.1681. [DOI] [PubMed] [Google Scholar]
- 46.Chuang TC, Moshir S, Garini Y, Chuang AY, Young IT, Vermolen B, van den Doel R, Mougey V, Perrin M, Braun M, et al. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004;3:12. doi: 10.1186/1741-7007-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caporali A, Wark L, Vermolen BJ, Garini Y, Mai S. Telomeric aggregates and end-to-end chromosomal fusions require myc box II. Oncogene. 2007;26:1398–1406. doi: 10.1038/sj.onc.1209928. [DOI] [PubMed] [Google Scholar]
- 48.Mai S, Wiener F. The impact of p53 loss on murine plasmacytoma development. Chromosome Res. 2002;10:239–251. doi: 10.1023/a:1015200307448. [DOI] [PubMed] [Google Scholar]
- 49.Schaefer LH, Schuster D, Herz H. Generalized approach for accelerated maximum likelihood based image restoration applied to three-dimensional fluorescence microscopy. J Microsc. 2001;204(pt 2):99–107. doi: 10.1046/j.1365-2818.2001.00949.x. [DOI] [PubMed] [Google Scholar]
- 50.Vermolen BJ, Garini Y, Mai S, Mougey V, Fest T, Chuang TC, Chuang AY, Wark L, Young IT. Characterizing the three-dimensional organization of telomeres. Cytometry A. 2005;67:144–150. doi: 10.1002/cyto.a.20159. [DOI] [PubMed] [Google Scholar]
- 51.Poon SS, Martens UM, Ward RK, Lansdorp PM. Telomere length measurements using digital fluorescence microscopy. Cytometry. 1999;36:267–278. doi: 10.1002/(sici)1097-0320(19990801)36:4<267::aid-cyto1>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 52.Mai S, Garini Y. The significance of telomeric aggregates in the interphase nuclei of tumor cells. J Cell Biochem. 2006;97:904–915. doi: 10.1002/jcb.20760. [DOI] [PubMed] [Google Scholar]
- 53.Gadji M, Fortin D, Tsanaclis AM, Garini Y, Katzir N, Wienburg Y, Yan J, Klewes L, Klonisch T, Drouin R, et al. Three-dimensional nuclear telomere architecture is associated with differential time to progression and overall survival in glioblastoma patients. Neoplasia. 2010;12:183–191. doi: 10.1593/neo.91752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knecht H, Sawan B, Lichtensztejn D, Lemieux B, Wellinger RJ, Mai S. The 3D nuclear organization of telomeres marks the transition from Hodgkin to Reed-Sternberg cells. Leukemia. 2009;23:565–573. doi: 10.1038/leu.2008.314. [DOI] [PubMed] [Google Scholar]
- 55.Knecht H, Bruderlein S, Wegener S, Lichtensztejn D, Lichtensztejn Z, Lemieux B, Moller P, Mai S. 3D nuclear organization of telomeres in the Hodgkin cell lines U-HO1 and U-HO1-PTPN1: PTPN1 expression prevents the formation of very short telomeres including “t-stumps”. BMC Cell Biol. 2010;11:99. doi: 10.1186/1471-2121-11-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klewes L, Hobsch C, Katzir N, Rourke D, Garini Y, Mai S. Novel automated three-dimensional genome scanning based on the nuclear architecture of telomeres. Cytometry A. 2011;79:159–166. doi: 10.1002/cyto.a.21012. [DOI] [PubMed] [Google Scholar]
- 57.Gadji M, Awe JA, Rodriguez P, Klewes L, Kumar R, Houston DS, Falcao RP, de Oliveira FM, Mai S. Three-dimensional nuclear telomeric architecture defines cytopenias of myelodysplastic syndrome and its transformation to acute myeloid leukemia; Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research, 2012 March 31–April 4, Chicago, Illinois; Philadelphia, PA: AACR; 2012. Abstract nr 2044. [Google Scholar]
- 58.Koelsch BU, Rajewsky MF, Kindler-Rohrborn A. A 6-MB contigbased comparative gene and linkage map of the rat schwannoma tumor suppressor region at 10q32.3. Genomics. 2005;85:322–329. doi: 10.1016/j.ygeno.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 59.Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T, Hissong B, Weaver Z, Ried T. The Septin 9 (MSF) gene is amplified and over-expressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63:2179–2187. [PubMed] [Google Scholar]
- 60.Russell SE, McIlhatton MA, Burrows JF, Donaghy PG, Chanduloy S, Petty EM, Kalikin LM, Church SW, McIlroy S, Harkin DP, et al. Isolation and mapping of human septin gene to a region on chromosome 17q, commonly deleted in sporadic epithelial ovarian tumors. Cancer Res. 2000;60:4729–4734. [PubMed] [Google Scholar]
- 61.Langan JE, Cole CG, Huckle EJ, Byrne S, McRonald FE, Rowbottom L, Ellis A, Shaw JM, Leigh IM, Kelsell DP, et al. Novel microsatellite markers and single nucleotide polymorphisms refine the tylosis with oesophageal cancer (TOC) minimal region of 17q25 to 42.5 kb: sequence does not identify the causative gene. Hum Genet. 2004;114:534–540. doi: 10.1007/s00439-004-1100-3. [DOI] [PubMed] [Google Scholar]
- 62.Turhan N, Yurur-Kutlay N, Topcuoglu P, Sayki M, Yuksel M, Gurman G, Tukun A. Translocation (13;17)(q14;q25) as a novel chromosomal abnormality in acute myeloid leukemia-M4. Leuk Res. 2006;30:903–905. doi: 10.1016/j.leukres.2005.12.011. [DOI] [PubMed] [Google Scholar]