1. Introduction
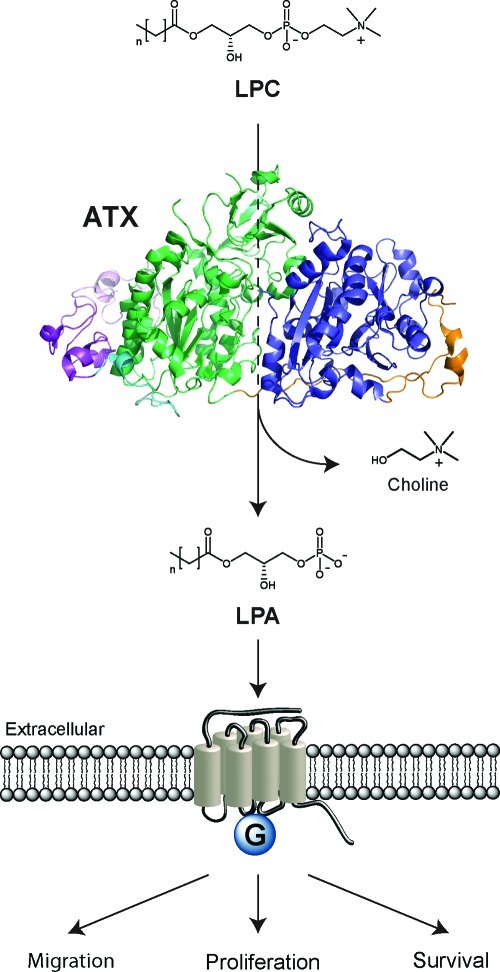
In 1992, autotaxin (ATX or ENPP2) was isolated as an autocrine motility factor from melanoma cells.1 The ∼120 kDa glycoprotein ATX belongs to a small family named ectonucleotide pyrophosphatase and phosphodiesterase (ENPP), which consists of seven family members.2 ATX is the only ENPP family member with lysophospholipase D (lysoPLD) activity and is responsible for the hydrolysis of lysophosphatidylcholine (LPC) to produce the bioactive lipid lysophosphatidic acid (LPA) (Scheme 1).3,4 LPA acts on specific G-protein-coupled receptors and thereby stimulates the migration, proliferation, and survival of many cell types.5,6 ATX is produced in various tissues and is the major LPA-producing enzyme in the circulation. After biosynthesis by ATX, LPA is subject to degradation by membrane-bound lipid phosphate phosphatases (LPPs).7,8
Scheme 1. ATX Is Responsible for Hydrolyzing LPC into LPA in an Extracellular Environment.
This reaction is catalyzed by a threonine residue and two zinc ions present in the ATX active site. LPA activates specific G-protein-coupled receptors stimulating migration, proliferation, and survival of cells. ATX is displayed as a cartoon representation of an ATX crystal structure (PDB ID 2XR9) with the SMB domains in purple, the PDE domain in green, and the nuclease-like domain in blue.
ATX is essential for vascular development9,10 and is found overexpressed in various human cancers.11 Forced overexpression of ATX or individual LPA receptors promotes tumor progression in mouse models,12−15 while LPA receptor deficiency protects from colon carcinogenesis.16 In addition to its role in cancer, ATX–LPA signaling has been implicated in lymphocyte homing and (chronic) inflammation,17 fibrotic diseases,18,19 thrombosis,20 and cholestatic pruritus.21 Given its role in human disease, the ATX–LPA axis is an interesting target for therapy that deserves significant attention. The fact that ATX is an extracellular enzyme makes it even more attractive as a drug target.
2. ATX Protein
Alternative splicing of the ATX gene (enpp2) results in three distinct isoforms (α, β, and γ) which are differentially expressed.22,23 ATX β (863 aa) is the best studied isoform and is identical to plasma lysoPLD. ATX β is mainly expressed in peripheral tissues, whereas lower expression levels are observed in the central nervous system. In contrast, the ATX γ (889 aa) isoform is predominantly expressed in the central nervous system. ATX α (915 aa), the original melanoma-derived isoform, exhibits the lowest expression levels in both the central nervous system and peripheral tissues. The ATX α isoform contains a nonspecific protease cleavage site that is not present in the other isoforms.22 All the three ATX isoforms exhibit similar catalytic activities in vitro.22
ATX is produced initially as a pre-proenzyme that has an N-terminal signal peptide required for secretion.24 This signal peptide is removed by a signal peptidase, and ATX is subsequently cleaved by proprotein convertases (PCs) like furin.24 The removal of an N-terminal octapeptide in ATX by PCs is associated with an enhancement of ATX activity.24 The proteolytically processed ATX is secreted and consists of several domains. Starting from its N-terminus, ATX has two somatomedin B (SMB)-like domains, a central catalytic phosphodiesterase (PDE) domain, and an inactive nuclease-like domain, as displayed in Scheme 1. The hydrolytic activity of ATX predominantly originates from a threonine residue and two zinc ions in the ATX active site located in the PDE domain.25 Extending from the ATX active site there is a hydrophobic pocket where the alkyl chain of its lipid substrates binds.26
3. Natural Substrates of ATX
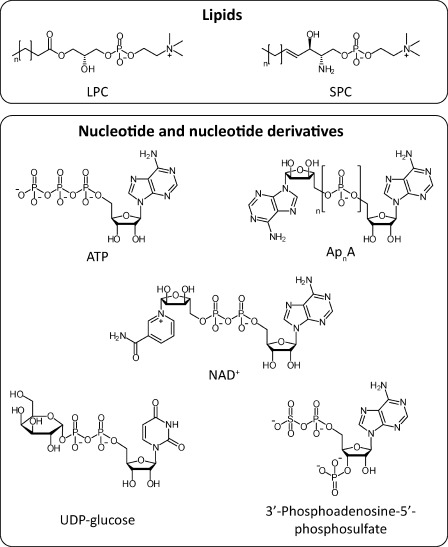
Next to LPC hydrolysis, ATX is also capable of hydrolyzing sphingosylphosphorylcholine (SPC, Figure 1) into sphingosine 1-phosphate (S1P).27 S1P has signaling properties comparable to those of LPA while acting on S1P receptors.28−30 It is however doubtful how relevant the contribution of ATX is to S1P production in vivo. S1P is thought to originate mainly from the phosphorylation of sphingosine by sphingosine kinases, rather than through SPC hydrolysis by ATX.31
Figure 1.
Identified natural substrates of ATX.
Next to recognizing the lipids LPC and SPC as substrates, ATX can also hydrolyze nucleotides, like its family members ENPP1 and ENPP3. In vitro established nucleotide and nucleotide-derived substrates of ATX consist of adenosine-5′-triphosphate (ATP), diadenosine polyphosphates (ApnA), uridine diphosphate glucose (UDP-glucose), nicotinamide adenine dinucleotide (NAD+), and 3′-phosphoadenosine-5′-phosphosulfate (Figure 1).2 The physiological relevance of ATX-mediated hydrolysis of these nucleotide substrates is still unclear.
4. Assays To Study ATX Activity
In the search for inhibitors, appropriate in vitro assays are required to monitor the activity of the enzyme of interest. Over the last 10 years various assays have been developed and used to study the activity of ATX. ATX assays can roughly be divided in two classes depending on the kind of ATX substrate used for the activity measurement. The first class uses the physiological ATX substrate LPC and the second class uses unnatural ATX substrates.
4.1. LPC-Based Assays
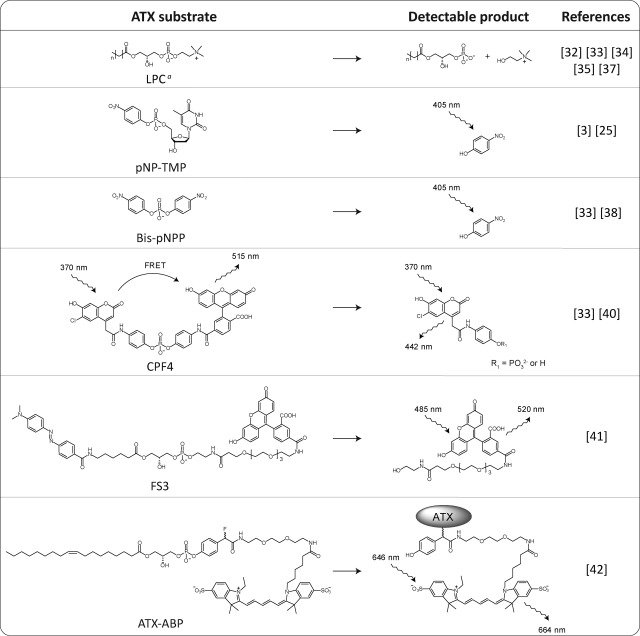
As depicted in Table 1, ATX hydrolyzes LPC into LPA and choline. Both products can be used to measure ATX activity. When a 14C label is introduced in the lipid tail of LPC, ATX activity can be measured by radiometry.32,33 When radiolabeled LPC is hydrolyzed by ATX, the produced LPA contains this radiolabel. After lipid extraction of the ATX–LPC incubation mixture and separation of LPC and LPA using thin layer chromatography (TLC),4 the activity of ATX can be quantified from the 14C-LPA produced. Although this classical method is robust and very sensitive, it is not very suitable for high-throughput screening (HTS).
Table 1. Substrates for ATX-Activity Assays.
LPA can be detected by radiometry or mass spectrometry. Choline can be detected in a two-step enzymatic colorimetric reaction.
Another way to measure the formation of LPA is by using liquid chromatography–tandem mass spectrometry (LC–MS/MS).34,35 From an ATX incubation mixture, LPC and LPA are separated by LC and detected by tandem MS. This method is very sensitive and suitable to detect naturally occurring LPA in biological fluids (i.e., plasma).34−36
ATX activity can also be measured by the detection of choline.4,37 When choline is released from the ATX-mediated LPC hydrolysis, it can be converted by choline oxidase into betaine (trimethylglycine) and hydrogen peroxide. Subsequently, hydrogen peroxide is used by horseradish peroxidase (HRP) to convert a coloring substrate into its oxidized chromophoric state. Different HRP coloring substrates can be used like 2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), homovanillic acid (HVA), or Amplex red. ABTS can be detected using absorbance while HVA and Amplex red are detected by fluorescence.36,38,39 The throughput of this assay is high and suitable for HTS screening.
A danger of the latter assay is that small molecules that do not inhibit ATX may interfere with the readout by inhibiting the enzymes (HRP or choline oxidase) used in the coloring reaction, which will result in false positives. Another possibility that could result in false positives is that reactive compounds tested in this assay can react with the coloring agent or hydrogen peroxide that is generated during that coloring reaction. Incubating molecules that are active in this assay with only choline and subsequently adding the coloring reagents should reveal if these molecules are interfering with the assay readout.38
4.2. Unnatural ATX Substrate-Based Assays
As a member of the ENPP family, ATX also hydrolyzes nucleotides. Therefore, the nucleotide thymidine 5′-monophosphate p-nitrophenyl ester (pNP-TMP) can be used as an ATX substrate (Table 1).3,25 Upon hydrolysis of pNP-TMP by ATX, 4-nitrophenol is released, which has an absorbance at 405 nm and is detectable by colorimetry.
The same product is formed when bis(p-nitrophenyl) phosphate (bis-pNPP) is hydrolyzed by ATX (Table 1).33,38 Bis-pNPP is a cheap ATX substrate that provides a direct readout.
Another artificial ATX substrate is CPF4, which has a similar core structure as bis-pNPP. In CPF4 the two nitro groups of bis-pNPP are replaced by coumarin and fluorescein (Table 1).33,40 When the coumarin donor group in CPF4 is excited, Förster fluorescence resonance energy transfer (FRET) between coumarin and the fluorescein acceptor occurs. After hydrolysis the FRET pair is separated and FRET is lost, providing a very sensitive assay reagent. Although this reagent was originally developed for phosphodiesterase I,40 it provides a very sensitive ATX activity sensor as well.33
FS3 is another synthetic ATX substrate that is based on LPC (Table 1).41 In this substrate a dabcyl moiety, a quencher of fluorescein, is connected via a lipid backbone to fluorescein. The fluorescein moiety in FS3 is quenched by the dabcyl moiety and becomes fluorescent when FS3 is hydrolyzed by ATX.
A common advantage of the above-mentioned assays is that they provide a realtime readout, allowing direct kinetic studies of ATX activity.
4.3. ATX Activity-Based Probe
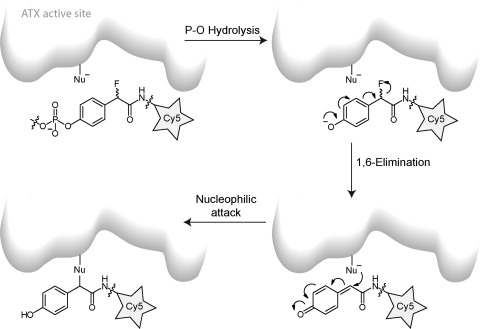
A first generation ATX activity-based probe (ATX-ABP) has recently been reported (Table 1).42 This probe makes use of the activity of ATX to label ATX covalently. Upon the hydrolysis of the phosphodiester bond in ATX-ABP, the released intermediate undergoes an 1,6-elimination of a fluoride atom (Scheme 2). This generates a reactive quinone methide species that traps nearby nucleophiles in the ATX active site, resulting in covalent labeling of ATX with a fluorescent Cy5 dye in an activity-dependent manner. This ATX-ABP is able to label all the three known ATX isoforms.42 In addition, ATX-ABP can label ATX in human plasma; however, an additional affinity-purifying step with an anti-ATX monoclonal antibody is required to make labeled ATX detectable. Further development of these types of probes could turn them into diagnostic reagents to monitor ATX activity in complex samples such as body fluids.
Scheme 2. Labeling Mechanism of ATX with ATX-ABP.
Only one of the potential labeled products resulting from the labeling is shown.
5. Inhibitory Effect of Metal Chelators on ATX Activity
l-Histidine has been reported as the first in vitro “ATX inhibitor” with millimolar IC50 values for ATX using LPC or pNP-TMP as assay substrate.43 It acts by scavenging metal ions in solution, such as zinc, which are essential for ATX activity. In addition, other metal chelating agents such as ethylenediaminetetraacetic acid (EDTA) and 1,10-phenanthroline also have an inhibitory effect on ATX activity.43
6. Lipid and Lipid-Based Inhibitors of ATX
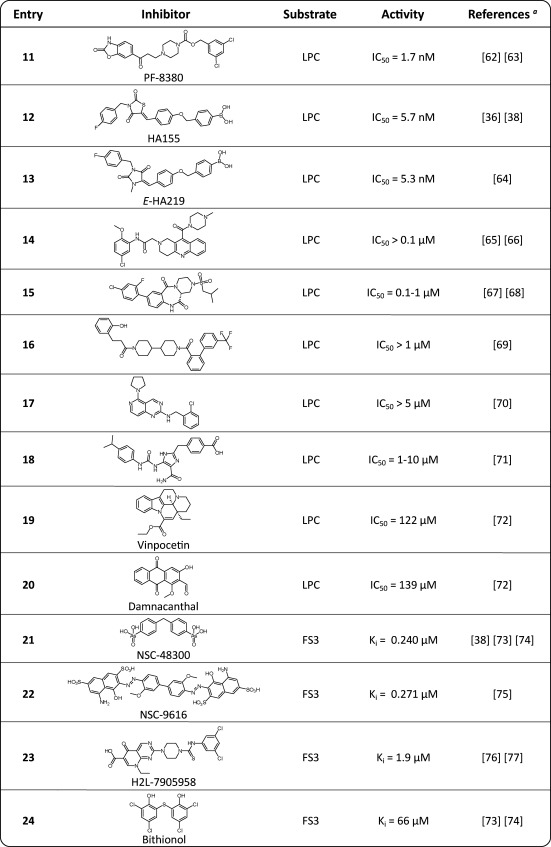
A distinct class of ATX inhibitors is based on lipids. The discovery of product inhibition of ATX by LPA and S1P33 triggered the development of lipid-based ATX inhibitors (Table 2).
Table 2. Lipid and Lipid-Based Inhibitors of ATX.
The first reference corresponds with the displayed structure and the following references refer to similar inhibitor structures.
RA, residual ATX activity.
PI, percentage inhibition.
One class of lipid-based inhibitors includes thiophosphates.44−46 An example of this class is thiophosphate 3 (IC50 = 0.6 μM, bis-pNPP) depicted in Table 2. The reported thiophosphates also act as agonists or antagonists for LPA1–3 receptors.44−46 A general danger of lipid-based ATX inhibitors is that they may act on downstream LPA/S1P receptors due to their structural similarity with LPA and S1P.
There is also a class of ATX inhibitors based on cyclic phosphatidic acid (cPA) that is a naturally occurring analog of LPA where the sn-2 hydroxy group forms a five-membered ring with the sn-3 phosphate.47,48 cPA analog 4 (Table 2, IC50 = 0.14 μM, bis-pNPP) has no significant agonist activity at LPA receptors. Recently, a sulfur analogue of cPA, 3-O-thia-cPA (5), has been reported as an ATX inhibitor (Table 2).49
Another class of lipid-based ATX inhibitors is based on α-bromomethylene phosphonates like BrP-LPA (6, Table 2).50,51 In addition, phosphonate 6 acts as a pan-LPA1–4 receptor antagonist. α-Bromobenzyl phosphonates are well-known protein tyrosine phosphatase inhibitors that target the active site covalently.52 Whether phosphonate 6 inhibits ATX in a covalent manner is unknown.
An inhibitor that is expected to bind ATX covalently is flouromethylphenyl phosphate 7 (Table 2).53 The binding mechanism of 7 with ATX is postulated to be the same as for ATX-ABP, as depicted in Scheme 2. Inhibitor 7 shows time-dependent inhibition of ATX.
Interestingly, phosphorylated FTY720 (FTY720-P, 8, Table 2) inhibits ATX with a Ki of 0.2 μM using pNP-TMP as an ATX assay substrate.54,55 FTY720-P is a synthetic analogue of S1P and acts as an S1P1 receptor antagonist (EC50 = 5 nM)55 that activates this receptor, causing its internalization and subsequent polyubiquitination, leading to proteasomal degradation of the S1P1 receptor. This results in the unresponsiveness of lymphocytes to S1P.56
Potencies of the previously described lipid-based inhibitors all have been determined in assays using unnatural ATX substrates (bis-pNPP, pNP-TMP, CPF4, and FS3) as reporter molecules. Therefore, it is difficult to compare potencies of distinct inhibitors and their effect on LPC hydrolysis. Using assays based on LPC hydrolysis gives a better indication whether ATX inhibitors could have an in vivo effect on LPA production by ATX. The most potent (IC50 = 5.6 nM, LPC) lipid-based inhibitor measured in an LPC hydrolysis assay is S32826 (9, Table 2).39,57,58 This phosphonate inhibitor is a result of screening 13 000 small molecules for their ability to inhibit ATX. Unfortunately, the poor in vivo stability and/or bioavailability of the compound did not permit further use in animal models.
A last lipid-like inhibitor class is based on a tyrosine building block, and an example of this class is inhibitor 10, which has a micromolar potency (Ki = 1.0 μM, LPC, Table 2).59−61
A frequently observed phenomenon for ATX inhibitors is their incapability to fully inhibit ATX. Examples are inhibitors 3, 4, and 9 depicted in Table 2 that have residual ATX activity (RA) for ATX.
7. Small Molecule Inhibitors of ATX
Since 2008, many small molecule inhibitors have been reported in both academic and patent literature (Table 3). The most potent inhibitor (IC50 = 1.7 nM, LPC) reported to date is PF-8380 (11).62,63 Interestingly, this compound reported by employees of Pfizer62 is based on an inhibitor described in a Merck KGaA patent application.63 Due to the high potency and favorable pharmacokinetic properties, PF-8380 is a suitable tool compound for in vivo evaluation of ATX inhibition.
Table 3. Activities of Small Molecule Inhibitors of ATX.
The first reference corresponds with the displayed structure, and the following references refer to similar inhibitor structures.
Another very potent ATX inhibitor is the boronic acid HA155 (12, IC50 = 5.7 nM, LPC, Table 3).36,38 This molecule resulted from screening ∼40 000 small molecules followed by medicinal chemistry efforts on the resulting screening hit. In the original screening hit, a carboxylic acid moiety was replaced by a boronic acid moiety, designed to target the threonine oxygen nucleophile in the ATX active site. HA130,36,38 a positional boronic acid isomer of HA155, together with PF-8380 are the only two inhibitors to date that have been demonstrated to lower LPA levels in vivo (rat or mice).
Recently, other boronic acid-based inhibitors have been reported64 as a result of a structure-based study resulting in E-HA219 (13, Table 3), a potent inhibitor of ATX (IC50 = 5.3 nM, LPC).
A common feature of the three most potent inhibitors reported so far (11–13, Table 3) is that they share a long linear and flexible structure, which is also reflected in the structure of LPC and LPA. Probably this structural feature helps the inhibitors to accommodate to the lipid binding site, since they bind to the ATX active site. Most of the other reported small molecules inhibitors lack this structural feature.
Several Merck KGaA patent applications claim ATX inhibitors (15–18) with in vitro potencies between 0.1 and 10 μM (LPC).65−71 The structural diversity of these inhibitors is large, as can be judged from Table 3.
Screening a phosphodiesterase targeted inhibitor library revealed that vinpocetine (19, Table 3), a PDE type 1 inhibitor (IC50 = 8–50 μM), inhibits ATX with an IC50 value of 122 μM using LPC as a substrate.72 In addition, screening a second library consisting of kinase inhibitors known to target the ATP binding site in kinases resulted in the discovery of damnacanthal (20, Table 3), a p56lck tyrosine kinase inhibitor (IC50 = 17–620 nM), as ATX inhibitor (IC50 = 139 μM, LPC).72
Several ATX screening programs used libraries from the National Cancer Institute (NCI). This led for example to the discovery of arsenic acid NSC-48300 (21, Table 3) as ATX inhibitor, reported by two independent groups.38,73,74 It is a competitive inhibitor that could suggest that the arsenic acid moiety in 21 acts as a nonhydrolyzable phosphate mimic. In addition, NSC-9616 (22, Table 3) has been identified as ATX inhibitor from screening an NCI library.75
The first virtual screen for ATX inhibitors led to the discovery of H2L-7905958 (23, Table 3).76 This inhibitor has a Ki value of 1.9 μM (FS3) for ATX.77 At the time of this virtual screen the ATX structure was not resolved. Therefore, the authors generated a homology model of ATX based on a Xanthomonas axonopodis pV. citri (Xac) ENPP structure and used this homology model for virtual screening. Recently, a follow-up study has been reported describing new analogues of 23.77
Another ATX inhibitor is bithionol (24, Table 3),73,74 a known metal ion chelating molecule like l-histidine and EDTA.78 The latter two inhibit ATX by scavenging metal ions in solution, which are required for ATX activity. However, inhibitor 24 appears to act directly on ATX and not via metal chelation, since the Ki value of 24 for ATX is 2 orders of magnitude lower than the used metal ion concentration.
8. ATX Structure and Inhibitor Design
In 2011 the crystal structure of ATX had been resolved independently by two groups.26,79 Aside from the unliganded ATX structure, structures of ATX with different species of LPA26 and the inhibitor HA155 liganded ATX structure79 have been reported. Binding of HA155 to the ATX active site is predominantly driven by hydrophobic interactions and by a boronic acid moiety binding to the threonine oxygen nucleophile in the ATX active site (Figure 2B).79 The latter binding was predicted because the boronic acid moiety in HA155 was designed and introduced to target the threonine oxygen nucleophile in the ATX active site. In addition, the ATX–HA155 structure showed that one of the boronic acid hydroxyl moieties is simultaneously tethered by the two zinc ions in the ATX active site (Figure 2B). Therefore, the boronic acid moiety not only targets the threonine oxygen nucleophile but also the two zinc ions that are essential for the catalytic activity of ATX. Another feature of HA155 binding to the ATX active site is that its 4-fluorobenzyl moiety binds to the hydrophobic lipid binding pocket of ATX (Figure 2A), preventing the alkyl chain of the lipid from binding to this pocket.26,79
Figure 2.
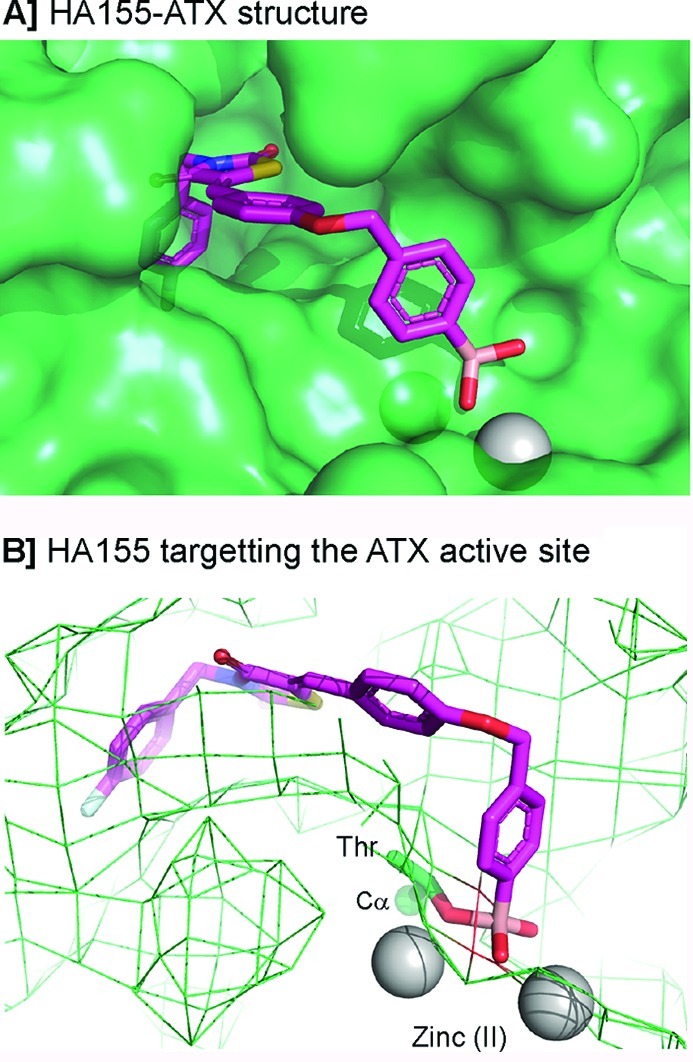
The inhibitor liganded ATX structure (PDB ID 2XRG). (A) Binding of HA155 with ATX. (B) Boronic acid in HA155 targeting the threonine (Thr) oxygen nucleophile and two zinc ions in the ATX active site.
The ATX structure liganded with HA155 triggered an ATX structure-based inhibitor design.64 This study led to E-HA219 (IC50 = 5.3 nM, LPC, Table 3) a hydantoine analogue of HA155 (IC50 = 5.7 nM, LPC, Table 3). Remarkably, E-HA219 is an E-isomer while HA155 is a Z-isomer. To understand how E-HA219 binds to ATX, molecular docking experiments were performed that suggested a binding pose for E-HA219 different from that of the original binding pose of HA155 or from the Z-isomer of HA219 for ATX. This study predicted that the 4-fluorobenzyl moiety in HA155 and E-HA219 binds differently to the hydrophobic pocket in ATX. This finding may be used to design new inhibitors that fully exploit the hydrophobic pocket in ATX, opening further options for inhibitor design.
9. Concluding Remarks
ATX is an attractive therapeutic target for LPA-related diseases. In the past decade, many ATX inhibitors have been discovered and developed, ranging from metal chelators to lipid and lipid-based inhibitors to small molecule inhibitors. Over the last 3 years many patents on ATX inhibitors have appeared from pharmaceutical industry and academia emphasizing the interest on ATX as drug target. Finally, the recently reported crystal structures of ATX will aid medicinal chemistry efforts to further develop ATX inhibitors into therapeutic agents.
Acknowledgments
We thank Anastassis Perrakis for critically reading the manuscript. This work was supported by grants from The Netherlands Organization for Scientific Research (NWO), the Dutch Cancer Society (KWF), and The Netherlands Proteomics Centre supported by The Netherlands Genomics Initiative.
Glossary
Nonstandard Abbreviations
- ABTS
2,2′-azinobis(3-ethylbenzothiazoline-6-sulfonic acid)
- ApnA
diadenosine polyphosphates
- ATP
adenosine-5′-triphosphate
- ATX
autotaxin
- ATX-ABP
ATX activity-based probe
- bis-pNPP
bis(p-nitrophenyl) phosphate
- cPA
cyclic phosphatidic acid
- EDTA
ethylenediaminetetraacetic acid
- ENPP
ectonucleotide pyrophosphatase and phosphodiesterase
- FRET
Förster fluorescence resonance energy transfer
- HRP
horseradish peroxidase
- HTS
high-throughput screening
- HVA
homovanillic acid
- LC–MS/MS
liquid chromatography–tandem mass spectrometry
- LPA
lysophosphatidic acid
- LPC
lysophosphatidylcholine
- LPP
lipid phosphate phosphatases
- lysoPLD
lysophospholipase D
- NAD+
nicotinamide adenine dinucleotide
- NCI
National Cancer Institute
- PC
proprotein convertases
- pNP-TMP
thymidine 5′-monophosphate p-nitrophenyl ester
- S1P
sphingosine 1-phosphate
- SMB
somatomedin B
- SPC
sphingosylphosphorylcholine
- TLC
thin layer chromatography
- UDP
uridine diphosphate
- Xac
X. axonopodis pV. citri
Biographies

Harald Albers obtained his M.S. degree in Chemical Engineering at Eindhoven University of Technology (TUe), The Netherlands, in 2007 with honors. During his training in organic chemistry in the laboratory of Prof. E. W. Meijer (TUe), he developed C3-symmetrical discotics functionalized with MRI labels, which have the ability to self-assemble into a supramolecular contrast agent for MRI. In 2007, he started his graduate studies at The Netherlands Cancer Institute (NKI), which resulted in the discovery and further development of boronic acid-based inhibitors of autotaxin, a lipid phosphodiesterase that holds promise as a drug target for anticancer therapy.

Huib Ovaa received his Ph.D. degree for his work in organic synthesis in the laboratory of the late Prof. J. H. van Boom (Leiden University, The Netherlands) studying olefin metathesis reactions on sugar derivatives, in 2001. During subsequent postdoctoral work with H. L. Ploegh (Harvard Medical School, Boston, MA), chemical synthesis was used to study proteasome action and activity of deubiquitylating enzymes. This was followed by a position as an instructor of pathology at the same institute. Since 2004, he has headed a chemical biology group at The Netherlands Cancer Institute (NKI), developing inhibitors of autotaxin and studying targeted protein degradation and antigen presentation.
References
- Stracke M. L.; Krutzsch H. C.; Unsworth E. J.; Arestad A.; Cioce V.; Schiffmann E.; Liotta L. A. J. Biol. Chem. 1992, 267, 2524. [PubMed] [Google Scholar]
- Stefan C.; Jansen S.; Bollen M. Purinergic Signalling 2006, 2, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumura A.; Majima E.; Kariya Y.; Tominaga K.; Kogure K.; Yasuda K.; Fukuzawa K. J. Biol. Chem. 2002, 277, 39436. [DOI] [PubMed] [Google Scholar]
- Umezu-Goto M.; Kishi Y.; Taira A.; Hama K.; Dohmae N.; Takio K.; Yamori T.; Mills G. B.; Inoue K.; Aoki J.; Arai H. J. Cell Biol. 2002, 158, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K.; Herr D.; Mutoh T.; Chun J. Curr. Opin. Pharmacol. 2009, 9, 15. [DOI] [PubMed] [Google Scholar]
- Moolenaar W. H.; van Meeteren L. A.; Giepmans B. N. BioEssays 2004, 26, 870. [DOI] [PubMed] [Google Scholar]
- Sciorra V. A.; Morris A. J. Biochim. Biophys. Acta 2002, 1582, 45. [DOI] [PubMed] [Google Scholar]
- Brindley D.; Pilquil C. J. Lipid Res. 2009, 50, S225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M.; Okudaira S.; Kishi Y.; Ohkawa R.; Iseki S.; Ota M.; Noji S.; Yatomi Y.; Aoki J.; Arai H. J. Biol. Chem. 2006, 281, 25822. [DOI] [PubMed] [Google Scholar]
- van Meeteren L. A.; Ruurs P.; Stortelers C.; Bouwman P.; van Rooijen M. A.; Pradere J. P.; Pettit T. R.; Wakelam M. J.; Saulnier-Blache J. S.; Mummery C. L.; Moolenaar W. H.; Jonkers J. Mol. Cell. Biol. 2006, 26, 5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills G. B.; Moolenaar W. H. Nat. Rev. Cancer 2003, 3, 582. [DOI] [PubMed] [Google Scholar]
- Taghavi P.; Verhoeven E.; Jacobs J. J.; Lambooij J. P.; Stortelers C.; Tanger E.; Moolenaar W. H.; van Lohuizen M. Oncogene 2008, 27, 6806. [DOI] [PubMed] [Google Scholar]
- Nam S. W.; Clair T.; Campo C. K.; Lee H. Y.; Liotta L. A.; Stracke M. L. Oncogene 2000, 19, 241. [DOI] [PubMed] [Google Scholar]
- Liu S.; Umezu-Goto M.; Murph M.; Lu Y.; Liu W.; Zhang F.; Yu S.; Stephens L. C.; Cui X.; Murrow G.; Coombes K.; Muller W.; Hung M. C.; Perou C. M.; Lee A. V.; Fang X.; Mills G. B. Cancer Cell 2009, 15, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucharaba A.; Serre C. M.; Gres S.; Saulnier-Blache J. S.; Bordet J. C.; Guglielmi J.; Clezardin P.; Peyruchaud O. J. Clin. Invest. 2004, 114, 1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.; Wang D.; Iyer S.; Ghaleb A. M.; Shim H.; Yang V. W.; Chun J.; Yun C. C. Gastroenterology 2009, 136, 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda H.; Newton R.; Klein R.; Morita Y.; Gunn M.; Rosen S. Nat. Immunol. 2008, 9, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradere J. P.; Klein J.; Gres S.; Guigne C.; Neau E.; Valet P.; Calise D.; Chun J.; Bascands J. L.; Saulnier-Blache J. S.; Schanstra J. P. J. Am. Soc. Nephrol. 2007, 18, 3110. [DOI] [PubMed] [Google Scholar]
- Tager A.; LaCamera P.; Shea B.; Campanella G.; Selman M.; Zhao Z.; Polosukhin V.; Wain J.; Karimi-Shah B.; Kim N.; Hart W.; Pardo A.; Blackwell T.; Xu Y.; Chun J.; Luster A. Nat. Med. 2008, 14, 45. [DOI] [PubMed] [Google Scholar]
- Pamuklar Z.; Federico L.; Liu S.; Umezu-Goto M.; Dong A.; Panchatcharam M.; Fulerson Z.; Berdyshev E.; Natarajan V.; Fang X.; van Meeteren L. A.; Moolenaar W. H.; Mills G. B.; Morris A. J.; Smyth S. S. J. Biol. Chem. 2009, 284, 7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer A.; Martens J.; Kulik W.; Rueff F.; Kuiper E.; van Buuren H.; van Erpecum K.; Kondrackiene J.; Prieto J.; Rust C.; Geenes V.; Williamson C.; Moolenaar W. H.; Beuers U.; Oude E. Gastroenterology 2010, 139, 1008. [DOI] [PubMed] [Google Scholar]
- Giganti A.; Rodriguez M.; Fould B.; Moulharat N.; Coge F.; Chomarat P.; Galizzi J.; Valet P.; Saulnier-Blache J.; Boutin J.; Ferry G. J. Biol. Chem. 2008, 283, 7776. [DOI] [PubMed] [Google Scholar]
- Murata J.; Lee Y.; Clair T.; Krutzsch H.; Arestad A.; Sobel M.; Liotta L.; Stracke M. J. Biol. Chem. 1994, 269, 30479. [PubMed] [Google Scholar]
- Jansen S.; Stefan C.; Creemers J.; Waelkens E.; van Eynde A.; Stalmans W.; Bollen M. J. Cell Sci. 2005, 118, 3081. [DOI] [PubMed] [Google Scholar]
- Gijsbers R.; Aoki J.; Arai H.; Bollen M. FEBS Lett. 2003, 538, 60. [DOI] [PubMed] [Google Scholar]
- Nishimasu H.; Okudaira S.; Hama K.; Mihara E.; Dohmae N.; Inoue A.; Ishitani R.; Takagi J.; Aoki J.; Nureki O. Nat. Struct. Mol. Biol. 2011, 18, 205. [DOI] [PubMed] [Google Scholar]
- Clair T.; Aoki J.; Koh E.; Bandle R.; Nam S.; Ptaszynska M.; Mills G.; Schiffmann E.; Liotta L.; Stracke M. Cancer Res. 2003, 63, 5446. [PubMed] [Google Scholar]
- Postma F. R.; Jalink K.; Hengeveld T.; Moolenaar W. H. EMBO J. 1996, 15, 2388. [PMC free article] [PubMed] [Google Scholar]
- Hla T.; Lee M.; Ancellin N.; Paik J.; Kluk M. Science 2001, 294, 1875. [DOI] [PubMed] [Google Scholar]
- Ishii I.; Fukushima N.; Ye X.; Chun J. Annu. Rev. Biochem. 2004, 73, 321. [DOI] [PubMed] [Google Scholar]
- Spiegel S.; Milstien S. Nat. Rev. Mol. Cell Biol. 2003, 4, 397. [DOI] [PubMed] [Google Scholar]
- Tokumura A.; Miyake M.; Yoshimoto O.; Shimizu M.; Fukuzawa K. Lipids 1998, 33, 1009. [DOI] [PubMed] [Google Scholar]
- van Meeteren L. A.; Ruurs P.; Christodoulou E.; Goding J. W.; Takakusa H.; Kikuchi K.; Perrakis A.; Nagano T.; Moolenaar W. H. J. Biol. Chem. 2005, 280, 21155. [DOI] [PubMed] [Google Scholar]
- Scherer M.; Schmitz G.; Liebisch G. Clin. Chem. 2009, 55, 1218. [DOI] [PubMed] [Google Scholar]
- Murph M.; Tanaka T.; Pang J.; Felix E.; Liu S.; Trost R.; Godwin A.; Newman R.; Mills G. Methods Enzymol. 2007, 433, 1. [DOI] [PubMed] [Google Scholar]
- Albers H. M.; Dong A.; van Meeteren L. A.; Egan D. A.; Sunkara M.; van Tilburg E. W.; Schuurman K.; van Tellingen O.; Morris A. J.; Smyth S. S.; Moolenaar W. H.; Ovaa H. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S.; Horiuti Y. J. Biochem. 1978, 83, 677. [DOI] [PubMed] [Google Scholar]
- Albers H. M.; van Meeteren L. A.; Egan D. A.; van Tilburg E. W.; Moolenaar W. H.; Ovaa H. J. Med. Chem. 2010, 53, 4958. [DOI] [PubMed] [Google Scholar]
- Ferry G.; Moulharat N.; Pradere J.; Desos P.; Try A.; Genton A.; Giganti A.; Beucher-Gaudin M.; Lonchampt M.; Bertrand M.; Saulnier-Blache J.; Tucker G.; Cordi A.; Boutin J. J. Pharmacol. Exp. Ther. 2008, 327, 809. [DOI] [PubMed] [Google Scholar]
- Takakusa H.; Kikuchi K.; Urano Y.; Sakamoto S.; Yamaguchi K.; Nagano T. J. Am. Chem. Soc. 2002, 124, 1653. [DOI] [PubMed] [Google Scholar]
- Ferguson C.; Bigman C.; Richardson R.; van Meeteren L. A.; Moolenaar W. H.; Prestwich G. Org. Lett. 2006, 8, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli S.; Houben A.; Albers H. M.; van Tilburg E. W.; de Ru A.; Aoki J.; van Veelen P.; Moolenaar W. H.; Ovaa H. ChemBioChem 2010, 11, 2311. [DOI] [PubMed] [Google Scholar]
- Clair T.; Koh E.; Ptaszynska M.; Bandle R.; Liotta L.; Schiffmann E.; Stracke M. Lipids Health Dis. 2005, 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgam G.; Tsukahara R.; Makarova N.; Walker M.; Fujiwara Y.; Pigg K.; Baker D.; Sardar V.; Parrill A.; Tigyi G.; Miller D. Bioorg. Med. Chem. Lett. 2006, 16, 633. [DOI] [PubMed] [Google Scholar]
- Durgam G.; Virag T.; Walker M.; Tsukahara R.; Yasuda S.; Liliom K.; van Meeteren L. A.; Moolenaar W. H.; Wilke N.; Siess W.; Tigyi G.; Miller D. J. Med. Chem. 2005, 48, 4919. [DOI] [PubMed] [Google Scholar]
- Gududuru V.; Zeng K.; Tsukahara R.; Makarova N.; Fujiwara Y.; Pigg K.; Baker D.; Tigyi G.; Miller D. Bioorg. Med. Chem. Lett. 2006, 16, 451. [DOI] [PubMed] [Google Scholar]
- Baker D.; Fujiwara Y.; Pigg K.; Tsukahara R.; Kobayashi S.; Murofushi H.; Uchiyama A.; Murakami-Murofushi K.; Koh E.; Bandle R.; Byun H.; Bittman R.; Fan D.; Murph M.; Mills G.; Tigyi G. J. Biol. Chem. 2006, 281, 22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupte R.; Siddam A.; Lu Y.; Li W.; Fujiwara Y.; Panupinthu N.; Pham T.; Baker D.; Parrill A.; Gotoh M.; Murakami-Murofushi K.; Kobayashi S.; Mills G.; Tigyi G.; Miller D. Bioorg. Med. Chem. Lett. 2010, 20, 7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R.; Kato M.; Suzuki T.; Nakazaki A.; Nozaki E.; Gotoh M.; Murakami-Murofushi K.; Kobayashi S. Bioorg. Med. Chem. Lett. 2011, 21, 4180. [DOI] [PubMed] [Google Scholar]
- Prestwich G., Tigyi G., Jiang G., Zhang G., Gajewiak J., Zhang H., Xu X. Patent WO2008157361A1, 2008; 65 pp.
- Jiang G.; Xu Y.; Fujiwara Y.; Tsukahara T.; Tsukahara R.; Gajewiak J.; Tigyi G.; Prestwich G. ChemMedChem 2007, 2, 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Zhou B.; Liang F.; Wang W.; Huang Z.; Zhang Z. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrill-Baker A., Baker D., Montedonico L. Patent WO2010040080A1, 2010; 38 pp.
- van Meeteren L. A.; Brinkmann V.; Saulnier-Blache J.; Lynch K.; Moolenaar W. H. Cancer Lett. 2008, 266, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine W.; Kiss G.; Liu J.; E S.; Gotoh M.; Murakami-Murofushi K.; Pham T.; Baker D.; Parrill A.; Lu X.; Sun C.; Bittman R.; Pyne N.; Tigyi G. Cell. Signalling 2010, 22, 1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oo M.; Thangada S.; Wu M.; Liu C.; Macdonald T.; Lynch K.; Lin C.; Hla T. J. Biol. Chem. 2007, 282, 9082. [DOI] [PubMed] [Google Scholar]
- Gupte R.; Patil R.; Liu J.; Wang Y.; Lee S.; Fujiwara Y.; Fells J.; Bolen A.; Emmons-Thompson K.; Yates C. R.; Siddam A.; Panupinthu N.; Pham T.; Baker D.; Parrill A.; Mills G.; Tigyi G.; Miller D. ChemMedChem 2011, 6, 922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G.; Madan D.; Prestwich G. Bioorg. Med. Chem. Lett. 2011, 21, 5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East J.; Kennedy A.; Tomsig J.; De Leon A.; Lynch K.; MacDonald T. Bioorg. Med. Chem. Lett. 2010, 20, 7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P.; Tomsig J.; McCalmont W.; Lee S.; Becker C.; Lynch K.; Macdonald T. Bioorg. Med. Chem. Lett. 2007, 17, 1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui P.; McCalmont W.; Tomsig J.; Lynch K.; Macdonald T. Bioorg. Med. Chem. 2008, 16, 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierse J.; Thorarensen A.; Beltey K.; Bradshaw-Pierce E.; Cortes-Burgos L.; Hall T.; Johnston A.; Murphy M.; Nemirovskiy O.; Ogawa S.; Pegg L.; Pelc M.; Prinsen M.; Schnute M.; Wendling J.; Wene S.; Weinberg R.; Wittwer A.; Zweifel B.; Masferrer J. J. Pharmacol. Exp. Ther. 2010, 334, 310. [DOI] [PubMed] [Google Scholar]
- Schiemann K., Schultz M., Blaukat A., Kober I. Patent WO2009046841A2, 2009; 152 pp.
- Albers H. M.; Hendrickx L. J.; van Tol R. J.; Hausmann J.; Perrakis A.; Ovaa H. J. Med. Chem. 2011, 54, 4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehle W., Kober I., Schiemann K., Schultz M., Wienke D. Patent WO2010060532A1, 2010; 142 pp.
- Staehle W., Kober I., Schiemann K., Schultz M., Wienke D. Patent DE102008059578A1, 2010; 66 pp.
- Schultz M., Schiemann K., Staehle W. Patent WO2011006569A1, 2011; 218 pp.
- Schultz M., Schiemann K., Staehle W. Patent DE102009033392A1, 2011; 109 pp.
- Schiemann K., Schultz M., Staehle W. Patent WO2010115491A2, 2010; 179 pp.
- Schiemann K., Schultz M., Staehle W., Kober I., Wienke D., Krier M. Patent WO2010063352A1, 2010; 177 pp.
- Schultz M., Schiemann K., Botton G., Blaukat A., Kober I. Patent WO2009046804A1, 2009; 163 pp.
- Moulharat N.; Fould B.; Giganti A.; Boutin J.; Ferry G. Chem.-Biol. Interact. 2008, 172, 115. [DOI] [PubMed] [Google Scholar]
- Saunders L. P.; Ouellette A.; Bandle R.; Chang W. C.; Zhou H.; Misra R. N.; De La Cruz E. M.; Braddock D. T. Mol. Cancer Ther. 2008, 7, 3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock D. Patent WO2009151644A2, 2009; 82 pp.
- North E. J.; Howard A.; Wanjala I.; Pham T.; Baker D.; Parrill A. J. Med. Chem. 2010, 53, 3095. [DOI] [PubMed] [Google Scholar]
- Parrill A.; Echols U.; Nguyen T.; Pham T.; Hoeglund A.; Baker D. Bioorg. Med. Chem. 2008, 16, 1784. [DOI] [PubMed] [Google Scholar]
- Hoeglund A.; Bostic H.; Howard A.; Wanjala I.; Best M.; Baker D.; Parrill A. J. Med. Chem. 2010, 53, 1056. [DOI] [PubMed] [Google Scholar]
- Fogg A. G.; Gray A.; Burns D. T. Anal. Chim. Acta 1970, 51, 265. [DOI] [PubMed] [Google Scholar]
- Hausmann J.; Kamtekar S.; Christodoulou E.; Day J.; Wu T.; Fulkerson Z.; Albers H. M.; van Meeteren L. A.; Houben A.; van Zeijl L.; Jansen S.; Andries M.; Hall T.; Pegg L.; Benson T.; Kasiem M.; Harlos K.; Kooi C.; Smyth S.; Ovaa H.; Bollen M.; Morris A.; Moolenaar W. H.; Perrakis A. Nat. Struct. Mol. Biol. 2011, 18, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]