Abstract
Living cells are structured to create a range of microenvironments that support specific chemical reactions and processes. Understanding how cells function therefore requires detailed knowledge of both the subcellular architecture and the location of specific molecules within this framework. Here we review the development of two correlated cellular imaging techniques that fulfill this need. Cells are first imaged using cryogenic fluorescence microscopy to determine the location of molecules of interest that have been labeled with fluorescent tags. The same specimen is then imaged using soft X-ray tomography to generate a high-contrast, 3D reconstruction of the cells. Data from the two modalities are then combined to produce a composite, information-rich view of the cell. This correlated imaging approach can be applied across the spectrum of problems encountered in cell biology, from basic research to biotechnological and biomedical applications such as the optimization of biofuels and the development of new pharmaceuticals.
Keywords: fluorescence, localization, organelles, phenotype, reconstruction
INTRODUCTION
Living cells are home to a breathtaking range of interconnected molecular interactions and chemical reactions (1), most of which are absolutely essential for proper cell function. Obviously, many of these reactions can be replicated individually in vitro in the laboratory, but the conditions in a test tube differ enormously from those in the tightly packed interior of a cell. The effects of this molecular crowding are profound; molecules are significantly less mobile inside a cell, and macromolecular interactions take place much more readily (2, 3). For example, the equilibrium constant for the dimerization of a 40-kDa protein is 10 to 40 times greater inside a cell than in a dilute solution (4). For higher-order oligomerization the consequences of molecular crowding are even more apparent. The equilibrium constant for the tetramerization of the same 40-kDa protein may be as much as 1,000- to 100,000-fold greater inside a cell compared with in solution (4). As a result, understanding of the chemical processes that take place in living systems relies primarily on data from in vivo experiments. However, obtaining this type of information remains a major challenge. Many reactions that take place inside a cell occur between molecules that are present in extremely low concentrations. For example, if an enzyme is present at cellular concentration of 1 nM—a concentration frequently used in in vitro biochemistry experiments—this equates to only one molecule of the enzyme being present in a cell (unless the cell in question is particularly large) (4). Quantification of molecules at such low concentrations in the milieu of other molecules in a living cell presents an insurmountable technological challenge for most analytical techniques. Imaging—specifically fluorescence-based—has proven to be an efficient route to determining the location and estimating the concentration of low-copy-number molecules inside a cell (5–11). However, as mentioned above, this information alone is insufficient if we are to truly understand how living cells function (12).
Cells, particularly eukaryotic cells, are very complex structures. All higher-order cells are partitioned into functionally distinct spaces by the creation of membrane-bound, subcellular volumes termed organelles (13, 14). Organizing the interior of the cell in this way allows radically different chemical microenvironments to coexist inside a single cell as well as each environment to be tuned and optimized for a particular set of biochemical processes. The organelles therefore have the ability to play discrete roles within a cell and to contribute either independently or synergistically to the overall function of the cell. By virtue of organelles’ relatively small size, the reaction conditions inside an organelle can be altered radically by what might appear to be an insignificant change in molecular composition. For example, if a subcellular vesicle has a volume of 6 × 10−20 liters, the generation of one free proton within such a small volume will result in the vesicle having a pH of 6.0. If 50 additional free protons are generated, the same vesicle will be at pH 5.0 (4). Clearly, this variation can have a major impact on reaction kinetics and determine which particular enzymes are fully functional.
Cells not only carry out specific chemical reactions but also perform them in vast numbers, the scale of which is perhaps best illustrated by considering the number of reactions that take place in a human body. Humans are composed of approximately 1013 cells (and typically an additional 1014 microbial cells). Each cell performs thousands, or even millions, of chemical reactions per second (4). Furthermore, it is not enough that these reactions simply take place. They must also occur at the right points in time, in the correct subcellular locations, and in a specific order (15). Errors in timing or location can lead to abnormal cell behavior or cause premature cell death. This fact has significant ramifications for public health as cellular malfunction is the root cause of most diseases. Consequently, powerful economic and social benefits can be gained from using imaging to identify aberrant, disease-causing molecular interactions and then using this knowledge to guide, for example, the development of new pharmaceuticals and clinical protocols (16, 17).
In this review we discuss two correlated imaging techniques that have the capacity to inform on the location of reacting molecules and provide a high-definition visual description of subcellular architecture. These imaging techniques are carried out serially on the same sample of cells, and the data from the two modalities are merged to form a composite view that is significantly greater than the sum of its component parts. This allows fundamental questions in biology to be addressed, namely, which molecules are interacting, where in the cell, when, and with what consequences?
The first of these techniques is cryogenic lightmicroscopy. Instruments capable of imaging cells at low temperature have been in use for many years. Until recently, all these microscopes have used relatively low-numerical aperture air lenses (18). Such microscopes are only capable of producing only low-magnification images that contained significant aberrations owing to the refractive index mismatch between air and the specimen (18). Recently, a new generation of cryogenic microscopes has become available that uses liquid propane or other organic molecules as an immersion fluid. In addition to index-matching the fluid to the specimen, the use of organic liquids means this type of microscope can be equipped with high–numerical aperture optics (12, 18), and thus image at high magnification without significant aberrations. In addition to conventional bright-field imaging, cryo-light microscopes can be used to collect fluorescence images (18).
Fluorescence microscopy is ubiquitous in modern biology and is a key technology in both basic research and clinical diagnosis (12). In most cases fluorescence microscopes are used to determine the cellular location and relative concentration of molecules that have been labeled with fluorescent tags. These days labeling is generally done using genetically encoded molecules, such as the family of fluorescent proteins (FPs) derived from aquatic organisms (11, 19, 20).This is a relatively simple process, and in most cases a protein of interest can be labeled using commercially available kits containing the necessary DNA and other reagents. Fluorescence imaging is a very sensitive technique. Modern microscopes equipped with high-sensitivity cameras and laser light sources can detect the signal from labeled molecules that are present in very low abundance. Indeed, with care the fluorescence signal from single molecules can be imaged (9, 21, 22). Fluorescence imaging can therefore be used to gain insight into which potential reaction partners are present, their approximate concentrations, and their relative locations (19). However, the subcellular structural organization of the cell is not observed by this imaging method (12).This leads to many uncertainties. On the basis of fluorescence imaging, two species of molecule may appear to be in relatively close proximity inside the cell, but this does not necessarily mean they are physically capable of interacting or that they are in the correct chemical environment for a productive reaction to occur. The subcellular architecture controls these factors; for example, molecules may be spatially separated by a membrane that is not seen in fluorescence imaging. Therefore, in addition to molecular localization, the cellular architecture must be imaged in fine detail. This is the role of soft X-ray tomography (SXT), the second imaging technique we discuss in this review.
In the past, high-resolution visualization of cell structure was almost exclusively the domain of electron microscopy. However, the physical characteristics of electrons impose restrictions on the size of cell that can be imaged intact, typically a thickness limit of 400–500 nm (23). Many prokaryotes are sufficiently small, but virtually all higher-order cells, including unicellular eukaryotes such as yeasts, are substantially larger than this maximum size. Thus, prior to imaging the cell must be sectioned into thin strips or ablated using a technique such as ion milling (23, 24). This time-consuming task severely restricts the throughput of cells that can be imaged and makes collecting data from a large number of cells technically challenging to the point of being impractical. To overcome these limitations, several groups began to pursue the notion of imaging cells using X-rays as the illumination source (25–29). Soft X-rays within a region of the spectrum known as the water window can penetrate cells that are up to 15 µm thick (25). In addition, soft X-rays with these energies are attenuated strongly by carbon and nitrogen relative to their attenuation by water. Moreover, the absorption of X-rays by the specimen adheres to the Beer-Lambert law and is therefore linear, quantitative, and a function of thickness and chemical composition. In a SXT reconstruction of a cell, highly solvated regions appear relatively transparent to X-rays compared with regions that are densely packed with biomolecules (30). Consequently, SXT images of biological specimens contain excellent contrast (31), without the use of potentially damaging contrast-enhancing stains, for example, metals such as osmium or platinum. After reconstruction, a cell imaged by SXT can be segmented by bounding regions that have similar attenuation characteristics [this is quantified for each voxel in the reconstruction and is known as the linear absorption contrast (LAC)]. For example, regions in a cell that are mostly composed of biomolecules, such as a lipid droplet, have very high LAC values compared to organelles that are typically much lower in biomolecule density, such as vacuoles. Once segmented, further analysis can be carried out on the internal structure of the organelles, again based on the LAC values. For example, regions of different density inside the nucleus can be delineated and segmented.
As with all microscopes, soft X-ray microscopes produce only 2D projections of the specimen. Cells are sufficiently complex that their internal structures appear confusingly superimposed in projection, and interpretation therefore requires three dimensions. Fortunately, acquiring the third dimension is relatively straightforward and is simply a matter of collecting a series of projection images at angular increments around a rotation axis and then computing a 3D tomographic reconstruction of the specimen.
We begin by describing the process of imaging cells using SXT. We have chosen to describe the two techniques in this order for practical reasons; SXT is the dominant technique in terms of the scale and complexity of the required instrumentation (12). Cryo-light microscopes either can function as stand-alone instruments or can be incorporated into a soft X-ray microscope to make correlated imaging data from the same specimen as simple as possible (12, 18).
VISUALIZATION OF CELL STRUCTURE: SOFT X-RAY MICROSCOPY
The first X-ray microscope was developed in the latter half of the 1940s by Kirkpatrick & Baez (36). This microscope used grazing-incidence reflective optics to focus the X-rays onto the specimen. This was an exciting development for X-ray science and materials research (37, 38), but unfortunately this type of microscope proved unsuitable for biological imaging. Imaging cells is a recent development and required the development and incorporation of Fresnel zone optical systems into soft X-ray microscopes situated at third-generation synchrotron light sources. Almost as soon as these microscopes were commissioned, they began to produce clear projection images of biological specimens (30, 31, 39, 40). This progress sparked a resurgence in efforts to develop soft X-ray microscopes specifically for biological imaging.
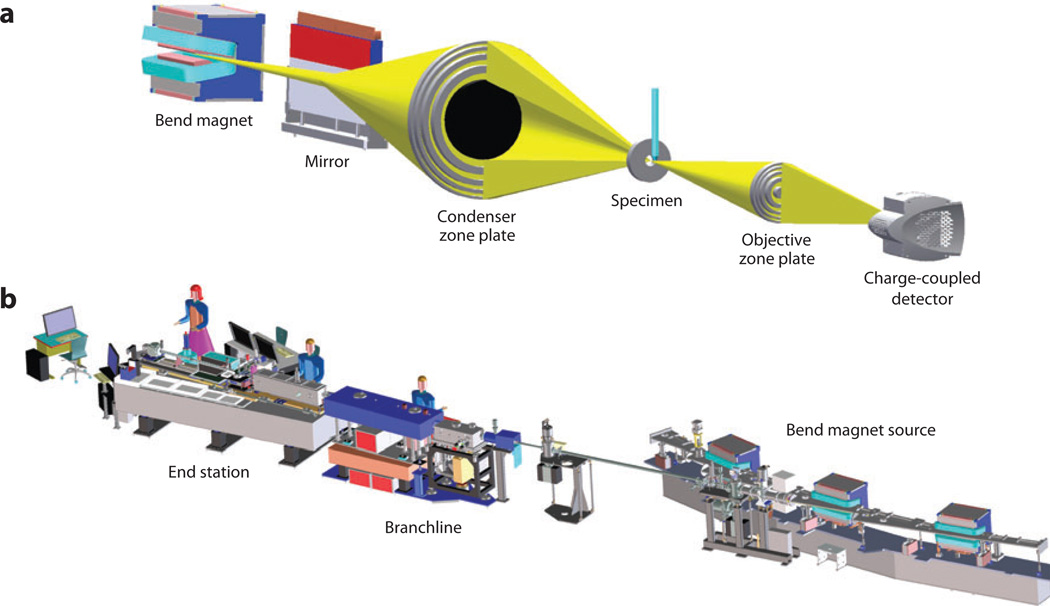
Conceptually, a soft X-ray microscope has an optical configuration similar to that of a simple, full-field light microscope. That is to say, a condenser lens focuses the illumination onto the specimen, and an objective lens refocuses unabsorbed light onto a detector (41). In practice, the physical nature of X-rays—in particular their low refractive index—means that conventional glass optics cannot be used in an X-ray microscope. Instead, soft X-ray microscopes are typically equipped with optical elements such as Fresnel zone plates. In this instrument configuration one zone plate acts as the microscope condenser and the other as the objective, as shown in Figure 1a. However, instead of a zone plate, a glass capillary waveguide can be used as the condenser. Both types of condensers have inherent advantages and disadvantages, but they produce images of similar resolution and quality. To avoid confusion and eliminate discussion that is not particularly germane to this review, we restrict our discussion to soft X-ray microscope designs that employ Fresnel zone plates as condensers; an example of this instrument is shown in Figure 1b. Fresnel zone plates contain radially symmetric rings known as Fresnel zones (25, 41). In a soft X-ray microscope these alternate between being opaque and transparent toward X-ray photons. In operation, a soft X-ray beam diffracts around the opaque zones (25). The zones can be spaced so that the light diffracted by each zone constructively interferes at the desired focus (for the condenser zone plate, the focal point typically would be at the specimen). Zones become narrower and more closely packed as one progresses outward from the center of the zone plate, until the outermost zone is reached (41). In the simplest case, the spacing in the outermost zone of the objective zone plate defines the maximum spatial resolution obtainable from a soft X-ray microscope optical system (41, 42). To date, soft X-ray imaging of cells has been carried out with objective zone plates with a spatial resolution of 25 to 50 nm (12, 43). However, lens designers and fabricators continue to make bold advances toward improving zone plate efficiency and spatial resolution. Zone plates have now been produced with a spatial resolution better than 15 nm (42). Installation of these new, spatially enhanced lenses in existing soft X-ray microscopes is a relatively trivial technical matter. Unfortunately, as the spatial resolution of the lens increases, the depth of field decreases (12). Consequently, these high-spatial-resolution zone plates have a depth of field shallower than the thickness of most eukaryotic cells. Overcoming this issue will require the development of techniques such as a combination of deconvolution with tomography (12).
Figure 1.
(a) The optical configuration of a bend magnet–based transmission soft X-ray microscope (XM-2) located at the Advanced Light Source, Berkeley, California. The specimen sits between the objective and the condenser optical elements. In this example, both are Fresnel zone plates (other microscopes use capillary waveguides in place of a zone plate as the condenser). (b) A computer-aided design (CAD) model. This figure illustrates the complexity and scale of this microscope—for example, the detector is located 22 m from the bend magnet. X-rays are transmitted from the bend magnet to the specimen via the branchline instrumentation. The end station–i.e., the part of the microscope where users mount specimens—is completely enclosed in an environmentally controlled room. In practice, the instrument has the feel of a relatively simple laboratory-based light microscope.
Sources of Soft X-Rays
Synchrotrons are currently the optimal source of illumination photons for a soft X-ray microscope (27, 43, 44). These light sources produce incredibly intense beams of soft X-rays that can be readily collimated and focused onto tiny specimens (44). The world’s first soft X-ray microscope to be designed specifically for biological experiments is located at the Advanced Light Source of Lawrence Berkeley National Laboratory, California. This synchrotron facility is the brightest source of soft X-rays in the world and therefore ideally suited to this type of instrument. In the wake of the success of this instrument, many other soft X-ray microscopes are now under construction or commissioned at synchrotron light sources around the world. However, absolute reliance on synchrotrons as sources of X-rays for these microscopes has inherent disadvantages: Relatively few of these facilities exist, and all are heavily oversubscribed. To address this potential shortfall, significant effort is being directed toward the development of small, laboratory-sized X-ray sources (commonly referred to as “tabletop” X-ray sources) (45–48). These instruments will soon make it feasible for individual research groups to have in-house SXT capabilities.
Specimen Preparation
Specimen preparation is an important factor in determining the final quality and fidelity of any biological imaging technique. Artifacts generated by specimen preparation protocols cannot be mitigated or eliminated post facto; they can only be accounted for. In SXT the specimen is flash cooled to cryogenic temperatures prior to imaging (12, 30, 43, 49, 52). Typically, the specimen is mounted in an appropriate holder and rapidly cooled to liquid-nitrogen temperature by moving the holder into a stream of cold helium gas or by rapidly plunging the specimen into a cryogen such as liquid propane (30). Both methods cool the specimen quickly enough to promote the formation of amorphous ice rather than crystalline ice. This means that the structure of the ice inside the cell remains similar to that of the liquid before freezing, and structural features are retained in their native state.
Specimen Mounting
Prior to imaging, specimens must be mounted in a suitable holder. Many possible holder configurations exist, all of which meet the criteria for mechanical stability during data collection. However, freedom of rotation is an important consideration. Cylindrical holders offer unfettered rotation around a central axis (12, 30); conversely, if the specimen sits on top of a flat holder, a limit is imposed on the maximum rotation that can be used, leading to a “missing wedge” of data that can be accessed (12). Consequently, cylindrical holders are preferred, and their use is therefore described below.
In practice, glass capillaries should be sized to match the cells being imaged (excess solution surrounding cells adds background noise to the image).Capillaries can be manufactured easily from thin-walled capillary tubes. Prior to loading with cells, thin glass microcapillaries are heated and then pulled using a micropipette puller to form an extended narrow tip. The tip region is typically 1 mm long with a diameter of 4 to 15 µm. This configuration combines mechanical stability with a long viewing length along which cells can be imaged (30). Multiple fields of view can be imaged along the length of the capillary simply by translating the holder in the microscope. Cells, suspended in a suitable medium, are loaded into the capillary using a regular pipette. Typically, capillaries are loaded with 1 µL of suspension containing between 10,000 and 100,000 cells. For most cell types, the capillary force is sufficient to draw the cells into the tapered region of the capillary to be imaged. In some instances (e.g., if the cells are particularly sticky or irregularly shaped), it may be necessary to use an adapted centrifuge to move the cells into the narrow bore of the capillary. In general, the total time needed to mount cells in a capillary holder is short enough that cells are rarely damaged in the process. An additional advantage to using glass capillaries for mounting cells is the ability to carry out fluorescence tomography or, at the very least, be able to view the cell without the mount impinging on the field of view.
Soft X-Ray Imaging in Three Dimensions
Imaging a typical cell by SXT requires the collection of projection images at angular increments over a 180° range (most commonly this means collection of 180 projection images at 1° rotational increments) (12, 30). Consequently, two challenges arise. First, repeated exposure of the specimen to a beam of X-rays could lead to a significant cumulative radiation dose and therefore open the prospect of radiation damage, resulting in the generation of artifacts in the images. However, it has long been accepted and repeatedly demonstrated that cooling the specimen to cryogenic temperatures reduces radiation damage. In practice, if a biological specimen is cooled to liquid nitrogen temperature, as many as 1,000 soft X-ray microscope images can be collected with no apparent signs of radiation damage (12, 30). The second issue is one of structures moving or changing inside the specimen during the time required to collect many exposures. With a high-spatial-resolution technique such as soft X-ray microscopy, even relatively small movements inside the cell during data collection have a profound impact on the fidelity of the calculated tomogram. Thus, the specimen must be fixed prior to imaging. The two possible methods for doing this are chemical fixatives and cryogenic immobilization (49).Chemical fixation is potentially quite damaging to cellular structures (23) and frequently causes the collapse of highly solvated structures such as vacuoles, leading to concerns about other possible damage to fine structures (23). Consequently, cryogenic immobilization is considered to be by far the better option; this method is virtually instantaneous and retains fine structural details inside the cell (23). Accordingly, cryo-immobilization is the principal method used in SXT to minimize artifacts owing to internal movements during data collection and/or radiation damage (12, 43, 50). Accordingly, the specimen is mounted on a cryo-rotation stage (cryo-stage) which both maintains the specimen at cryogenic temperature and allows it to be rotated around an axis (or axes).
Tomographic data collection is a relatively fast process. Typically, each projection image takes seconds (100-ms exposure followed by a delay while the detector reads out to a buffer). Consequently, it only takes a matter of minutes to collect the necessary 90 or 180 images. This appears to be well within the threshold of cumulative exposure required to cause observable radiation damage. Indeed, for most specimens, there is no discernible difference between an image collected at the start of a data series and that recollected after 180 (or even more) images have been collected from the same specimen. Of course, there is no suggestion that damage is not occurring; simply it is not apparently in 35- or 50-nm spatial resolution soft X-ray microscope images or reconstructions.
SXT is therefore highly analogous to the clinical practice of computed tomography (CT), in which X-ray projection images of the patient are taken at angular increments around an axis. This series of images is then used to compute a 3D representation of the patient, known as a tomogram, that can be used to diagnose disease or guide surgical procedures. In the same way that CT revolutionized aspects of clinical medicine, SXT has the potential to strongly impact our understanding of cell biology. A particular strength of the technique is its ability to visualize and quantify the phenotype of a cell or population of cells. For both basic research and more practical applications, the determination of changes in phenotype as a function of changes in specific genetic or environmental factors is fundamentally important (see Figure 2a,b). For example, a candidate drug molecule may have the desired effect on a particular reaction pathway in vitro; however, its usefulness as a potential pharmaceutical likely will be limited if the drug causes unwanted changes in the phenotype of the cell (32). This is only one simple example of how phenotypic information can be used. In recent work SXT has been used to determine the effects of genetic mutations in yeast on the structure and organization of the organelles (Figure 2b) (33), the phenotypic changes that occur during the infection of red blood cells with malarial parasites (Figure 3) (34), and the structure of mouse adenocarcinoma cells (Figure 4) (35).Data for the latter work were collected at the soft X-ray microscope (U41–TXM) located at the BESSY II synchrotron (Berlin, Germany). This microscope is fitted with a modified electron microscopy cryo-rotation stage and is therefore subject to limitations on the maximum specimen rotation (tilt). As a consequence, artifacts can be seen in the reconstruction due to missing data. In practice, the application and usefulness of these imaging modalities are limited only by our imagination (12).
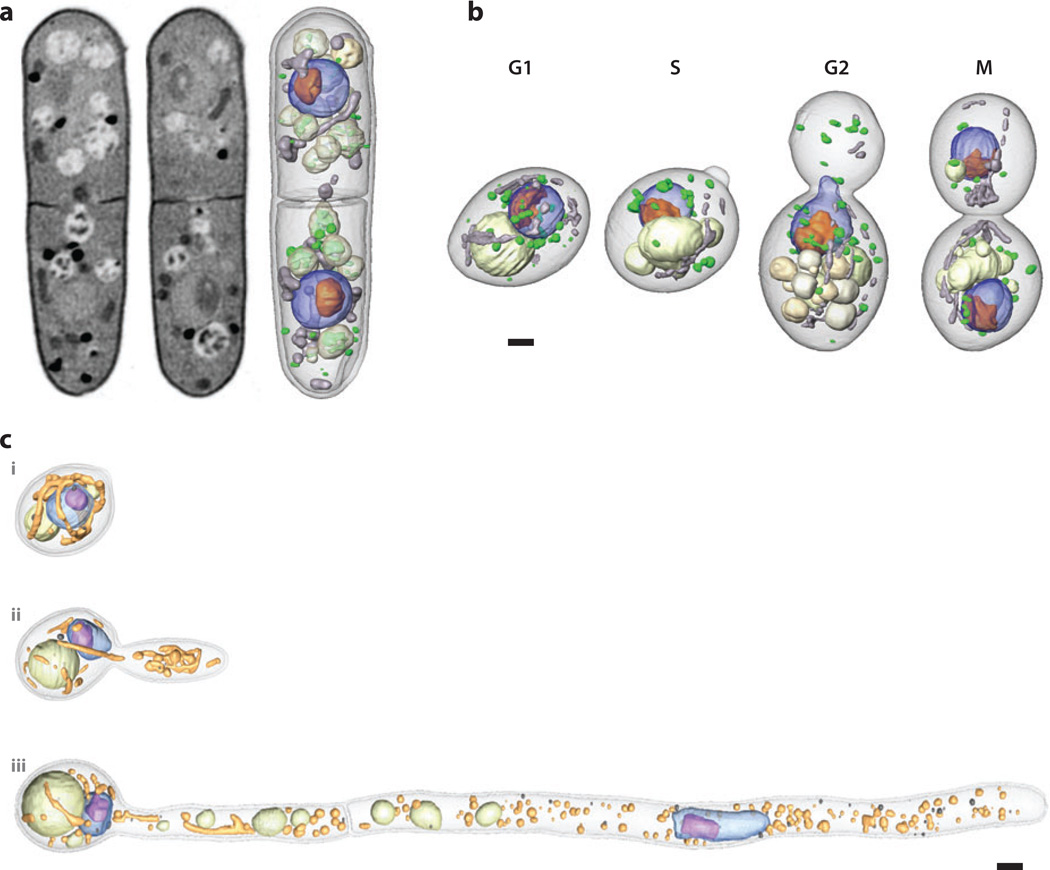
Figure 2.
(a) Soft X-ray tomographic reconstruction of the yeast Schizosaccharomyces pombe. Two orthoslices are shown from different regions in the tomographic reconstruction of a cell (left, center) illustrating the range of contrast (i.e., differences in linear absorption coefficient values that are a consequence of variation in biomolecular composition). Light-colored regions have relatively low attenuation and are therefore highly solvated. Dark regions are highly attenuating and therefore composed mostly of biomolecules (e.g., lipid droplets, membranes). The image on the right shows the same reconstruction after segmentation. The nucleus is shown in blue, the nucleoli in orange, mitochondria in gray, vacuoles in light gray, and lipid droplets in green. Panel a reprinted from Reference 12. (b) Segmented soft X-ray tomographic reconstructions of the yeast Saccharomyces cerevisiae at key stages of the cell cycle. The nucleus is shown in blue, the nucleoli in orange, mitochondria in gray, vacuoles in light gray, and lipid droplets in green. Panel b reprinted from Reference 60. (c) Soft X-ray microscopes typically have a field of view that is on the order of 15 to 20 µm. However, imaging cells that are longer than this is straightforward, especially if the cells are mounted in capillary tubes. Projection data sets are collected, and then the specimen is translated along the rotation axis until an adjacent region of the cell comes into view. This process is repeated until the entire cell has been imaged. Once the data have been reconstructed, the individual data sets are computationally stitched together to form a single volumetric reconstruction. In the example shown, cells of the different phenotypes displayed by the pathogenic yeast Candida albicans were imaged using soft X-ray tomography. The longest of these cells—those with the hyphal phenotype—were approximately 50 µm long and their images were therefore composite reconstructions from five individual data sets. The nucleus is shown in blue, the nucleoli in purple, mitochondria in yellow, vacuoles in pale green, and lipid droplets in dark gray. Scale bars = 1 µm. Panel c reprinted from Reference 32.
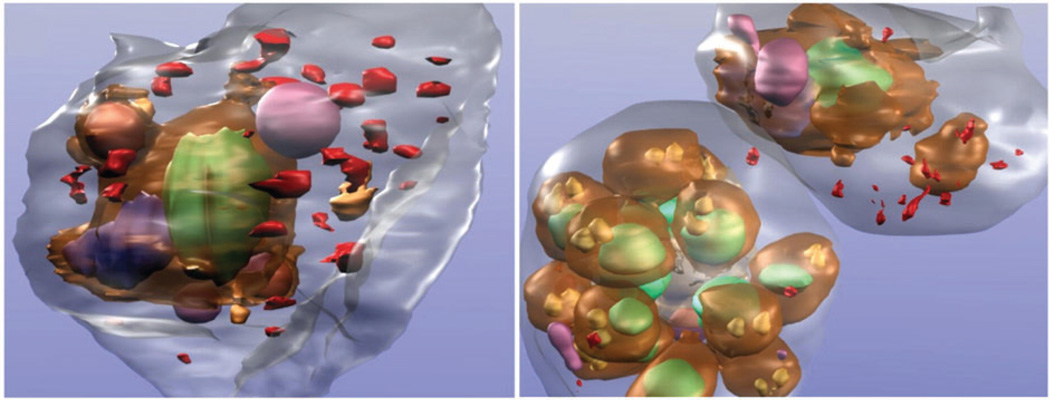
Figure 3.
Protein localization using immunolabeling methods. Plasmodium falciparum-infected erythrocytes expressing green fluorescent protein (GFP) chimeras of a Maurer’s cleft resident protein, the membrane-associated histidine-rich protein 1 (MAHRP1), were permeabilized and labeled with mouse anti-GFP antiserum followed by 6-nm gold-labeled protein A. The size of the gold particles was increased by silver enhancement. Dark deposits are observed in the red blood cell cytoplasm consistent with labeling of the Maurer’s clefts. Parasite structures: surface of parasite (brown), nuclei (green), digestive vacuole (blue), and rhoptry organelles (gold). Other structures seen in RBC cytoplasm: hemoglobin-containing vesicles (magenta) and membranous structures (red). Figure reprinted from Reference 34.
Figure 4.
An X-ray tomographic reconstruction of mouse adenocarcinoma cells showing many subcellular organelles. Abbreviations: ER, endoplasmic reticulum; L, lysosomes; M, mitochondria; NM, nuclear membrane; NP, nuclear pore; PM, plasma membrane; V, vesicle. Scale bar = 0.39 µm. Figure reprinted from Reference 35.
As discussed below, cryo-immobilization has one major additional advantage in terms of correlated imaging: Fluorescent labels are significantly more resistant to photobleaching when imaged at low temperature (9, 51).
LOCALIZATION OF MOLECULES
Immunolabeling
Initial efforts to localize molecules in a SXT reconstruction focused on adaptation of immunolabeling methods that are standard practice in electron microscopy/tomography. In these methods, the molecule of interest is immunolabeled with an electron-dense tag (40) or with a molecule that contains an absorption edge located within the water window (e.g., vanadium or titanium) (53). The basic principle behind this method is simple: The label attenuates soft X-rays significantly more strongly than any biological features, thus allowing the label to be identified unambiguously in a soft X-ray microscope image. An example of this process is shown in Figure 3 (34). Unfortunately, immunolabeling methods have significant disadvantages. Most seriously, the specimen must be chemically fixed and incubated with membrane-permeabilizing agents, such as detergents or organic solvents, to allow ingress of large antibodies into the interior of the cell (23). Clearly, disruption of the integrity of cellular membranes likely results in proteins and other molecules leaking out of organelles, or even the cell itself, and also causes damage to cellular substructures (23). Moreover, once inside the cell, the antibody may have limited access to the target epitope-containing molecule. In addition, the metal marker, which is at the opposite end of the antibody, is a significant distance from the epitope. Therefore, immunolabeling is a less-than-optimal solution to the problem (12). A much better option is to use genetically encoded fluorescent labels to localize molecules of interest, and to correlate this data directly with SXT images. Fortunately, the methods and instruments needed to perform this technique have now been developed. In particular, Le Gros et al. (18) created the world’s first cryogenic light microscope capable of operation at high numerical aperture. Integration of this light-based modality with a soft X-ray microscope (XM-2, a soft X-ray microscope for biological imaging, located at the Advanced Light Source, Berkeley, California) allows a specimen to be imaged with two different techniques (43). Correlation of the data from these two imaging methods–cryo-light and SXT—is straightforward; both techniques are carried out on the same instrument after a simple translation of the specimen holder.
Fluorescence Labeling
Localization of molecules by correlated fluorescence and X-ray imaging has many distinct advantages. The literature contains a wealth of readily available fluorescence data. In addition, most biological laboratories have a fluorescent microscope and routinely utilize the power of genetically encoded fluorescent tags, such as green FP and its many derivatives, in their day-to-day research efforts (6, 11, 20). Consequently, a wealth of knowledge exists on molecular localization based on this technique. However, information on the environment surrounding the labeled molecule is lacking, and it is often difficult, if not impossible, to delineate the precise cellular location of a molecule on the basis of fluorescence data alone. Fundamentally important questions frequently remain unanswered, such as “Is the molecule inside or outside the organelle?” This can best be determined by localizing the molecule in the context of a high-spatial-resolution reconstruction. A secondary advantage to the ability to carry out correlated light and SXT imaging is that the fluorescence data can resolve ambiguities over the assignment of a segmented region in a reconstruction. Such ambiguities occur, for example, when functionally different organelles are similar in size and have similar LAC values, or in cases in which the segmented objects approach the limits of the technique in terms of size and/or difference in LAC from neighboring cellular contents.
CRYO-FLUORESCENCE MICROSCOPY
The development of high–numerical aperture cryogenic light microscopy is a significant development in its own right and is not just a useful imaging adjunct to SXT. It has been long understood that cryo-cooling significantly increases the useful working life of a fluorescent label, before it gets photobleached (9). Consequently, cryogenic immersion microscopy meets a long-standing need in biological and biomedical imaging by providing a means of imaging fluorescent probes that photobleach quickly at room temperature. Although low-temperature microscopes have long been used to image frozen cells, all these instruments operated in air, and therefore they could use only lenses with relatively low numerical apertures. The development of a cryogenic immersion lens allows frozen specimens to be imaged at high resolution by using index-matched cryogens. This is an important factor because it opens up the door to the use of super-resolution fluorescent imaging techniques on specimens containing labels with greatly enhanced fluorescence properties.
Over the years, significant progress has been made toward enhancing genetically encodable fluorescent labels, in particular in terms of using families of FPs derived from jellyfish and corals (6, 11, 20, 21, 54–57). Most of this work has been aimed at increasing the range of available colors, reducing oligomerization of the molecules to produce physically smaller tags, and so on. This work has been fruitful; genetically encoded tags have now evolved into highly versatile research tools. However, there has been little focus on, or success in, extending the fluorescent lifetime of these molecules (54). This is a significant hurdle because the highest possible spatial resolution determination of a fluorescence signal, and therefore the precision with which relative protein locations can be determined, is a trade-off between image collection and photon damage (58). By cryogenically cooling the specimen it becomes possible to collect not only correlated light microscopy data but high-quality data on the location of specific molecules inside the cell (18). Consequently, the addition of high–numerical aperture cryogenic light microscopy to a soft X-ray microscope is the combination of two powerful, unique techniques (59) (Figure 5).
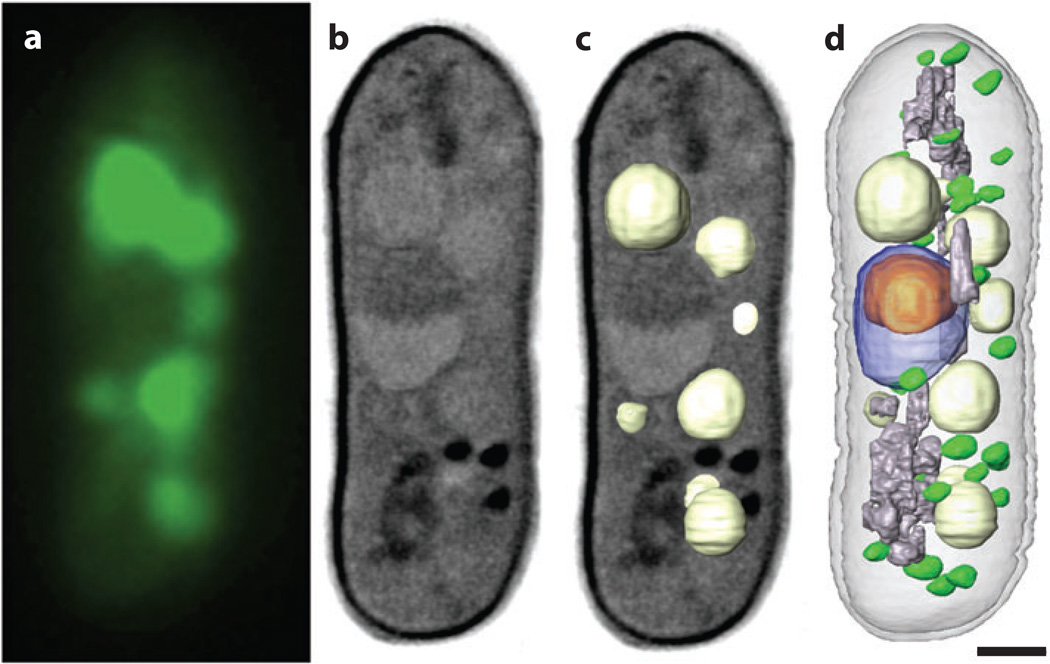
Figure 5.
Correlated soft X-ray tomography and cryo-light imaging (wide-field fluorescence). (a) The vacuoles fluorescently labeled and imaged by cryo-light microscopy. (b,c) Slices through the volumetric reconstruction calculated from soft X-ray tomography data, with the vacuoles shown as segmented volumes in panel c. The segmented vacuoles correlate closely with the locations determined from cryo-light microscopy. (d) The same cell after the major organelles have been segmented. The nucleus is shown in blue, the nucleoli in orange, mitochondria in gray, vacuoles in light gray, and lipid droplets in green. Scale bar = 1 µm.
SUMMARY AND FUTURE PROSPECTS
Soft X-ray tomography is a unique imaging technique capable of providing unprecedented insights into the structure of cells and tissue specimens that are held in a near-native state (43). Work to date using this technique only hints at the spectrum of cell types amenable to this kind of imaging, and the types of questions that can be answered. In the future, SXT will be employed on an ever-expanding range of specimen types, including cells whose dimensions and complexities push the limits of the technique. This is particularly true for complex high-order eukaryotic cells.
The spatial resolution that can be achieved with SXT depends on the optical system installed in the microscope. In the past these nanofabricated objects were both expensive and difficult to manufacture. However, the revolution in nanofabrication technology has made production of Fresnel lenses efficient and comparatively low cost. Moreover, it is now possible to manufacture such optics with increasingly finer structural features. This is enormously important in terms of the spatial resolution that can be obtained using SXT, because the distance between the outer zones in a Fresnel optic determines the maximum spatial resolution that can be achieved in a soft X-ray microscope. State-of-the-art soft X-ray Fresnel lenses are capable of resolving features in test objects with nearly 10-nm spatial resolution (42). Rapid progress is being made on the development of methods that combine tomography with through-focus deconvolution techniques. This work is in the early stages of evolution but builds on a firm theoretical and experimental foundation generated by many researchers in electron and light microscopy. Once completed, this combination of methods will allow ultra-high-resolution imaging to be carried out on thick specimens in a near-native state. Consequently, use of high-resolution zone plates with shallow depth of field will undoubtedly soon become a common feature in SXT.
In the future, the most exciting technological developments will be in the continued development of methods for correlated cryo-fluorescence and X-ray imaging. This is truly innovative in terms of both instrumentation and potential impact on biomedical research. Using cryo-light microscopy in tandem with soft X-ray tomography allows the position of fluorescently tagged molecules to be viewed in the context of the framework produced by a high-resolution reconstruction of the cell. This prospect has already generated enormous community interest. In closing, we feel confident in saying that soft X-ray tomography has finally come of age and has emerged as a mainstream technique with wide applicability in areas as diverse as drug discovery, biomedical research, and basic cell biology.
ACKNOWLEDGMENTS
We thank Dr. Elizabeth A. Smith for reading this manuscript and providing valuable feedback. The authors gratefully acknowledge funding from the NIH National Center for Research Resources grant RR019664 and NIH NIDA grant DA030320, and the Department of Energy Office of Biological and Environmental Research under contract number DE-AC02-05CH11231.
Glossary
- Cryogen(ic)
typically temperatures below −160°C
- Soft X-rays
X-rays with energies less than 1 keV
- Tomography
a method for calculating a 3D volumetric reconstruction of the specimen from 2D microscope images
- Water window
soft X-rays between the K edges of oxygen (2.34 nm) and carbon (4.4 nm)
- Linear absorption coefficient (LAC)
calculated for each voxel in a soft X-ray tomographic reconstruction. Areas of similar LAC value are segmented (isolated) from the other cell contents
- Projection image
a single, 2D image from a tilt/rotation series
- Reconstruction
the process of computationally recombining aligned projection images to form a virtual, 3D volume
- Zone plate
a Fresnel optic used to focus X-rays in a soft X-ray microscope
- Synchrotron
a circular particle accelerator that generates brilliant beams of light and X-rays
- Amorphous ice
amorphous solid form of water produced by cooling liquid water quickly enough to prevent the formation of a crystal lattice
- CT
computed tomography
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Sci.; 2008. p. 1392. [Google Scholar]
- 2.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 3.Ellis RJ. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- 4.Comm. Defin. Adv. Concept. Basis Biol. Sci. 21st Century, Natl. Res. Counc. The Role of Theory in Advancing 21st-Century Biology: Catalyzing Transformative Research. Washington, DC: Natl. Acad. Press; 2008. [Google Scholar]
- 5.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 6.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 7.Gustafsson MGL. Extended resolution fluorescence microscopy. Curr. Opin. Struct. Biol. 1999;9:627–634. doi: 10.1016/s0959-440x(99)00016-0. [DOI] [PubMed] [Google Scholar]
- 8.Martínez-Rocha AL, Roncero MIG, López-Ramirez A, Mariné M, Guarro J, et al. Rho1 has distinct functions in morphogenesis, cell wall biosynthesis and virulence of Fusarium oxysporum. Cell. Microbiol. 2008;10:1339–1351. doi: 10.1111/j.1462-5822.2008.01130.x. [DOI] [PubMed] [Google Scholar]
- 9.Moerner WE, Orrit M. Illuminating single molecules in condensed matter. Science. 1999;283:1670–1676. doi: 10.1126/science.283.5408.1670. [DOI] [PubMed] [Google Scholar]
- 10.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J. Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 11.Tsien RY. Building and breeding molecules to spy on cells and tumors. FEBS Lett. 2005;579:927–932. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 12.McDermott G, Le Gros MA, Knoechel CG, Uchida M, Larabell CA. Soft X-ray tomography and cryogenic light microscopy: the cool combination in cellular imaging. Trends Cell Biol. 2009;19:587–595. doi: 10.1016/j.tcb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fagarasanu A, Fagarasanu M, Rachubinski RA. Maintaining peroxisome populations: a story of division and inheritance. Annu. Rev. Cell Dev. Biol. 2007;23:321–344. doi: 10.1146/annurev.cellbio.23.090506.123456. [DOI] [PubMed] [Google Scholar]
- 14.Warren G, Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- 15.Aloy P, Russell RB. Structural systems biology: modelling protein interactions. Nat. Rev. Mol. Cell Biol. 2006;7:188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- 16.Bullen A. Microscopic imaging techniques for drug discovery. Nat. Rev. Drug Discov. 2008;7:54–67. doi: 10.1038/nrd2446. [DOI] [PubMed] [Google Scholar]
- 17.Lang P, Yeow K, Nichols A, Scheer A. Cellular imaging in drug discovery. Nat. Rev. Drug Discov. 2006;5:343–356. doi: 10.1038/nrd2008. [DOI] [PubMed] [Google Scholar]
- 18.Le Gros MA, McDermott G, Uchida M, Knoechel CG, Larabell CA. High-aperture cryogenic light microscopy. J. Microsc. 2009;235:1–8. doi: 10.1111/j.1365-2818.2009.03184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 20.Tsien RY. Imagining imaging’s future. Nat. Rev. Mol. Cell Biol. 2003;4(Suppl.):SS16–SS21. [PubMed] [Google Scholar]
- 21.Dickson RM, Cubitt AB, Tsien RY, Moerner WE. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 22.Lounis B, Moerner WE. Single photons on demand from a single molecule at room temperature. Nature. 2000;407:491–493. doi: 10.1038/35035032. [DOI] [PubMed] [Google Scholar]
- 23.Leis A, Rockel B, Andrees L, Baumeister W. Visualizing cells at the nanoscale. Trends Biochem. Sci. 2009;34:60–70. doi: 10.1016/j.tibs.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Baumeister W. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr. Opin. Struct. Biol. 2002;12:679–684. doi: 10.1016/s0959-440x(02)00378-0. [DOI] [PubMed] [Google Scholar]
- 25.Attwood DT. Soft X-Rays and Extreme Ultraviolet Radiation: Principles and Applications. New York: Cambridge Univ. Press; 1999. p. 470. [Google Scholar]
- 26.Sayre D, Kirz J, Feder R, Kim DM, Spiller E. Transmission microscopy of unmodified biological materials: comparative radiation dosages with electrons and ultrasoft x-ray photons. Ultramicroscopy. 1977;2:337–349. doi: 10.1016/s0304-3991(76)91997-5. [DOI] [PubMed] [Google Scholar]
- 27.Kirz J, Jacobsen C, Howells M. Soft X-ray microscopes and their biological applications. Q. Rev. Biophys. 1995;28:33–130. doi: 10.1017/s0033583500003139. [DOI] [PubMed] [Google Scholar]
- 28.Schmahl G. Personal view: the development of X-ray microscopy with synchrotron radiation during the last two decades. Synchrotron Radiat. News. 2007;20:27–28. [Google Scholar]
- 29.Schmahl G, Rudolph D, Niemann B, Guttmann P, Thieme J, Schneider G. X-ray microscopy. Naturwissenschaften. 1996;83:61–70. [PubMed] [Google Scholar]
- 30.Le Gros MA, McDermott G, Larabell CA. X-ray tomography of whole cells. Curr. Opin. Struct. Biol. 2005;15:593–600. doi: 10.1016/j.sbi.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Larabell C, Le Gros M. Whole cell cryo X-ray tomography and protein localization at 50 micron resolution. Biophys. J. 2004;86:A185. [Google Scholar]
- 32.Uchida M, McDermott G, Wetzler M, Le Gros MA, Myllys M, et al. Soft X-ray tomography of phenotypic switching and the cellular response to antifungal peptoids in Candida albicans. Proc. Natl. Acad. Sci. USA. 2009;106:19375–19380. doi: 10.1073/pnas.0906145106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchida M, Sun Y, McDermott G, Knoechel C, Le Gros MA, et al. Quantitative analysis of yeast internal architecture using soft X-ray tomography. Yeast. 2011;28:227–236. doi: 10.1002/yea.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanssen E, Knoechel C, Klonis N, Abu-Bakar N, Deed S, et al. Cryo transmission X-ray imaging of the malaria parasite, P. falciparum. J. Struct. Biol. 2011;173:161–168. doi: 10.1016/j.jsb.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider G, Guttmann P, Heim S, Rehbein S, Mueller F, et al. Three-dimensional cellular ultra-structure resolved by X-ray microscopy. Nat. Methods. 2010;7:985–987. doi: 10.1038/nmeth.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirkpatrick P, Baez AV. Formation of optical images by x-rays. J. Opt. Soc. Am. 1948;38:766–774. doi: 10.1364/josa.38.000766. [DOI] [PubMed] [Google Scholar]
- 37.Matsuyama S, Mimura H, Yumoto H, Sano Y, Yamamura K, et al. Development of scanning x-ray fluorescence microscope with spatial resolution of 30 nm using Kirkpatrick-Baez mirror optics. Rev. Sci. Instrum. 2006;77:103102. [Google Scholar]
- 38.Ortega R, Bohic S, Tucoulou R, Somogyi A, Devès G. Microchemical element imaging of yeast and human cells using synchrotron X-ray microprobe with Kirkpatrick-Baez optics. Anal. Chem. 2004;76:309–314. doi: 10.1021/ac035037r. [DOI] [PubMed] [Google Scholar]
- 39.Schneider G. Cryo X-ray microscopy with high spatial resolution in amplitude and phase contrast. Ultramicroscopy. 1998;75:85–104. doi: 10.1016/s0304-3991(98)00054-0. [DOI] [PubMed] [Google Scholar]
- 40.Meyer-Ilse W, Hamamoto D, Nair A, Lelievre SA, Denbeaux G, et al. High resolution protein localization using soft X-ray microscopy. J. Microsc. 2001;201:395–403. doi: 10.1046/j.1365-2818.2001.00845.x. [DOI] [PubMed] [Google Scholar]
- 41.Andersen EH, Olynick DL, Hartneck BD, Veklerov G, Denbeaux G, et al. Nanofabrication and diffractive optics for high-resolution X-ray applications. J. Vac. Sci. Technol. 2000;18:2970–2975. [Google Scholar]
- 42.Chao WL, Harteneck BD, Liddle JA, Anderson EH, Attwood DT. Soft X-ray microscopy at a spatial resolution better than 15 nm. Nature. 2005;435:1210–1213. doi: 10.1038/nature03719. [DOI] [PubMed] [Google Scholar]
- 43.Larabell CA, Nugent KA. Imaging cellular architecture with X-rays. Curr. Opin. Struct. Biol. 2010;20:623–631. doi: 10.1016/j.sbi.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakdinawat A, Fischer P, Anderson E, Attwood D. Single-element objective lens for soft x-ray differential interference contrast microscopy. Opt. Lett. 2006;16:1559–1564. doi: 10.1364/ol.31.001564. [DOI] [PubMed] [Google Scholar]
- 45.Tuohimaa T, Otendal M, Hertz HM. Phase-contrast x-ray imaging with a liquid-metal-jet-anode microfocus source. Appl. Phys. Lett. 2007;91:074104. [Google Scholar]
- 46.Tuohimaa T, Ewald J, Schlie M, Fernandez-Varea JM, Hertz HM, Vogt U. A microfocus x-ray source based on a nonmetal liquid-jet anode. Appl. Phys. Lett. 2008;92:233509. [Google Scholar]
- 47.de Groot J, Johansson GA, Hemberg O, Hertz HM. Improved liquid-jet laser-plasma source for X-ray microscopy. J. Phys. IV. 2003;104:121–122. [Google Scholar]
- 48.Stollberg H, Pokorny M, Hertz HM. A vacuum-compatible wet-specimen chamber for compact X-ray microscopy. J. Microsc. 2007;226:71–73. doi: 10.1111/j.1365-2818.2007.01754.x. [DOI] [PubMed] [Google Scholar]
- 49.Weiss D, Schneider G, Niemann B, Guttmann P, Rudolph D, Schmahl G. Computed tomography of cryogenic biological specimens based on X-ray microscopic images. Ultramicroscopy. 2000;84:185–197. doi: 10.1016/s0304-3991(00)00034-6. [DOI] [PubMed] [Google Scholar]
- 50.Jensen GJ, Briegel A. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr. Opin. Struct. Biol. 2007;17:260–267. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss D, Schneider G, Vogt S, Guttmann P, Niemann B, et al. Tomographic imaging of biological specimens with the cryo transmission X-ray microscope. Nucl. Instrum. Methods Phys. Res. A. 2001;467:1308–1311. [Google Scholar]
- 53.Ashcroft JM, Gu W, Zhang T, Hughes SM, Hartman KB, et al. TiO2 nanoparticles as a soft X-ray molecular probe. Chem. Commun. 2008;2008:2471–2473. doi: 10.1039/b801392f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, et al. Improving the photo-stability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaner NC, Steinbach PA, Tsien RY. Aguide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 56.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 58.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McDermott G, Le Gros MA, Knoechel CG, Uchida M, Larabell CA. Soft X-ray tomography and cryogenic light microscopy: the cool combination in cellular imaging. Trends Cell Biol. 2009;19:587–595. doi: 10.1016/j.tcb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uchida M, Sun Y, McDermott G, Knoechel C, Le Gros MA, et al. Quantitative analysis of yeast internal architecture using soft X-ray tomography. Yeast. 2011;28:227–236. doi: 10.1002/yea.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]