Abstract
A significant proportion of disease-causing mutations affect precursor-mRNA splicing, inducing skipping of the exon from the mature transcript. Using F9 exon 5, CFTR exon 12 and SMN2 exon 7 models, we characterized natural mutations associated to exon skipping in Haemophilia B, cystic fibrosis and spinal muscular atrophy (SMA), respectively, and the therapeutic splicing rescue by using U1 small nuclear RNA (snRNA). In minigene expression systems, loading of U1 snRNA by complementarity to the normal or mutated donor splice sites (5′ss) corrected the exon skipping caused by mutations at the polypyrimidine tract of the acceptor splice site, at the consensus 5′ss or at exonic regulatory elements. To improve specificity and reduce potential off-target effects, we developed U1 snRNA variants targeting non-conserved intronic sequences downstream of the 5′ss. For each gene system, we identified an exon-specific U1 snRNA (ExSpeU1) able to rescue splicing impaired by the different types of mutations. Through splicing-competent cDNA constructs, we demonstrated that the ExSpeU1-mediated splicing correction of several F9 mutations results in complete restoration of secreted functional factor IX levels. Furthermore, two ExSpeU1s for SMA improved SMN exon 7 splicing in the chromosomal context of normal cells. We propose ExSpeU1s as a novel therapeutic strategy to correct, in several human disorders, different types of splicing mutations associated with defective exon definition.
INTRODUCTION
Splicing errors represent a significant amount of disease-causing mutations. Considering only changes at canonical splice sites, ∼15% of mutations were originally estimated to induce aberrant splicing (1) and similar frequencies are recorded in human disease databases [for example, 12% in cystic fibrosis (CF) and 10% in coagulation FIX (FIX) deficiency (Haemophilia B)]. However, the occurrence of splicing defects is significantly increased (∼50%) when genomic variants are systematically evaluated for their effect on precursor-mRNA (pre-mRNA) processing (2,3). This unexpected effect is largely due to mutations located in non-canonical regulatory elements and highlights the difficulty in correctly predicting the consequence of genomic variants on pre-mRNA splicing (4–8).
To correctly identify the exonic sequences from the larger non-coding introns, the splicing machinery recognizes several cis-acting elements on the nascent transcripts (9,10). The core elements required for splicing are moderately conserved and consist of the classical or canonical 5′ and 3′ splice sites (named also donor and acceptor sites) and include the polypyrimidine tract as well as the branch site near the 3′ss. The consensus motif of the 5′ss is made of nine-nucleotide sequence, CAG/GURAGU where R is a purine and, with the exception of the nearly obligate GU dinucleotide, the other positions are quite degenerated. Recognition of the exon requires also splicing regulatory elements that are classified, depending on their location and effect on splicing, as exonic/intronic splicing enhancers (ESE and ISE), exonic/intronic splicing silencers (ESS and ISS) (9,10) or composite exonic regulatory elements of splicing (CERES) (7). The exonic elements are composed by largely degenerated sequences, overlap with the coding capacity and interact with splicing factors with a positive (SR proteins) or negative (hnRNPs) effect on the exon recognition. In general, factors associated to the splice sites and to the exonic splicing regulatory elements promote a network of interactions across the exon. As a result, the final outcome of a splicing decision depends on the equilibrium between multiple positive and negative interactions over the exon in a process called exon definition (11).
Aberrant exon skipping is a common splicing defect caused by disruption of the network of interactions that define the exon. It results from mutations at the consensus donor or acceptor sites, at the polypyrimidine tract or in the exonic regulatory elements. In general, mutations at the consensus donor site reduce the complementarity with the U1 small nuclear RNA (snRNA) (see below), substitutions at the 3′ss polypyrimidine tract interfere with the complexes assembled on the 3′ss and exonic mutations affect exonic regulatory elements.
An early event in exon definition is the recognition of the donor site by the U1 snRNP (small nuclear RiboNuclearParticle). U1 snRNP is composed by a 164 bp long U1 snRNA associated to several protein factors. To initiate splicing, the 5′ end of the U1 snRNA interacts by complementarity with the moderately conserved sequence of the 5′ss. Interestingly, ∼40, 22 and 5% of normal 5′ss contains, respectively, two, three or four mismatches toward the U1 snRNA (12). U1 snRNAs with a modified 5′ tail that base pair exactly to the mutant donor sites have been used to correct 5′ss mutation (13–19). For example, we previously showed that a U1 snRNA complementary to the +5G/A mutant of the intron 7 donor splice site of coagulation F7 rescued splicing and protein biosynthesis and function in minigene systems (14,15). However, this correction approach appears to be limited to single mutations and, through the ability of modified U1 snRNAs to target other donor sites, it can potentially interfere with splicing of other pre-mRNAs.
Here, to study the effect of mutations on splicing and identify a correction strategy on an exon basis, we considered three genes relevant in human pathology, the coagulation F9, the Cystic Fibrosis Transmembrane Regulator (CFTR) and the SMN2 (Survival of Motoneuron 2), whose mutations are associated to coagulation factor IX (Haemophilia B), CF and spinal muscular atrophy (SMA), respectively. Expression analysis of several substitutions at the donor splice sites (in F9 exon 5 and CFTR exon 12), at the polypyrimidine tract (in F9 exon 5) and at exonic regulatory elements (in CFTR exon 12 and in SMN2 exon 7), indicated their ability to cause aberrant splicing and particularly exon skipping. Screening of U1 snRNAs binding at non-conserved intronic sequences downstream the exon identified for each case an unique exon-specific U1 snRNA (ExSpeU1) able to correct different types of mutations. In SMN, two ExSpeU1-corrected exon 7 skipping in the normal chromosomal context. Moreover, in a FIX splicing-competent expression system, an ExSpeU1 restored, for several mutations, protein secretion and procoagulant function to an extent that, if achieved in vivo, would be therapeutic.
RESULTS
Effects of F9 exon 5 and CFTR exon 12 mutations on pre-mRNA splicing
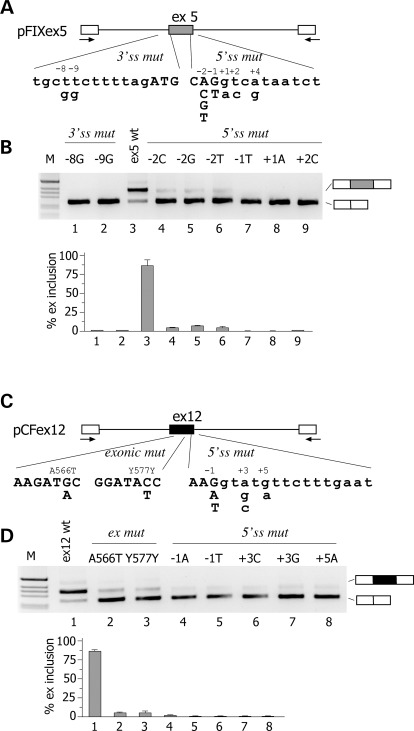
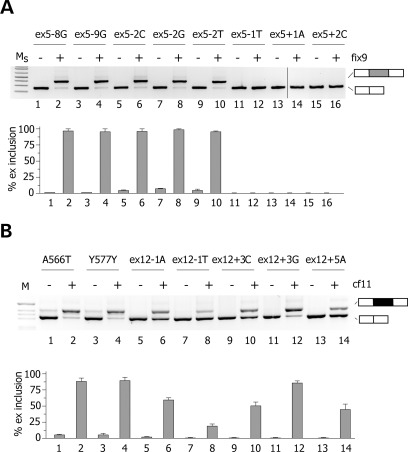
We evaluated nine different natural mutations associated with Haemophilia B deficiency (Fig. 1A) and seven mutations associated to CF (Fig. 1C). The list of the mutations analysed and their associated clinical phenotype are shown in Supplementary Material, Table S1. Mutations are numbered according to their position relative to the donor or acceptor site. The mutations in the F9 gene consists of two T to G transversions at the 3′ss polypyrimidine tract and seven substitutions at or near the 5′ss of exon 5. In the CFTR exon 12, we studied five donor-site substitutions and, as models for exonic mutations that affect splicing, two codon changes (A566T and Y577Y) located in the previously reported CERES element (7) (Supplementary Material, Table S1). To test their effect on splicing, the substitutions were inserted in appropriate minigenes followed by analysis of the splicing pattern in eukaryotic cells. For F9 exon 5, we prepared a novel minigene, whereas the CFTR exon 12 minigene has been previously described and extensively validated (7,8,20). Normal F9 ex 5 wt minigene showed that the exon was not completely included (∼85%) in the mature mRNA (Fig. 1B, lane 3). This pattern of splicing is entirely consistent with the in vivo splicing pattern of the F9 gene in human liver, where ∼90% of the exon is included (Supplementary Material, Fig. S1). The appreciable level of alternative splicing indicates that the exon 5 is poorly defined, very likely because of the presence of a weak 5′ss. In fact, it differs significantly from the consensus donor site at positions 3, 5 and 6, with a C, a U and an A, respectively (Supplementary Material, Fig. S2). Expression studies of the F9 ex 5 mutations revealed that the majority of them induced aberrant pre-mRNA processing (Fig. 1A). The −8 and −9 transversions of the polypyrimidine tract and the mutations at the consensus 5′ss at positions −2, −1, +1 and +2 induced complete exon skipping. On the other hand, all mutations in CFTR exon 12, either located at the donor sites (in position −1, +3 and +5) or in the exon (A566T and Y577Y), induced exon skipping (Fig. 1D). In is worth noting that the A566T and Y577Y exonic variants affect exonic splicing regulatory elements (7), which are important for correct exon definition. Thus, in two model systems, several natural mutations either at the splice sites or at exonic regulatory elements induce exon skipping.
Figure 1.
Effect of natural mutations on F9 exon 5 and CFTR exon 12 pre-mRNA splicing. (A and C) Schematic representation of the central region of the pFIX exon 5 and pCFex12 minigenes, respectively. Exonic sequences are boxed, introns are lines and the arrows represent primers for PCR amplification. The position of genomic variants relative to the splice sites is indicated. Exonic and intronic sequences are in upper and lower case, respectively. (B) Analysis of pFIX exon 5-spliced transcripts. HeLa cells were transfected with pFIXex5 wt or mutant minigenes and splicing pattern evaluated by RT–PCR. Amplified products were resolved on a 2% agarose gel. The identity of the bands is indicated on the right of the panel. M is the molecular 1 Kb marker. Lower panel shows the quantification of the percentage exon 5 inclusion (mean ± SD of three independent experiments done in duplicate). (D) Analysis of pCF exon 12 spliced transcripts. HeLa cells were transfected with pFIX ex12 wt or mutant minigenes and splicing pattern evaluated by RT–PCR. Amplified products were resolved on a 2% agarose gel. Lower panel shows the quantification of the percentage exon 12 inclusion (mean ± SD of three independent experiments done in duplicate).
U1 snRNAs complementary to the mutant 5′ss rescue some defective donor sites
We initially focused on mutations at donor sites, which reduce their complementarity to the 5′ tail of the U1 snRNA. To understand for each mutation the role of U1 snRNA in exon recognition, we prepared U1 snRNAs expression plasmids with compensatory changes that completely restore base-pairing with the wt or mutant 5′ss (Supplementary Material, Figs S2 and S3). These U1 snRNAs were cotransfected with the corresponding minigene variants followed by the analysis of the splicing pattern.
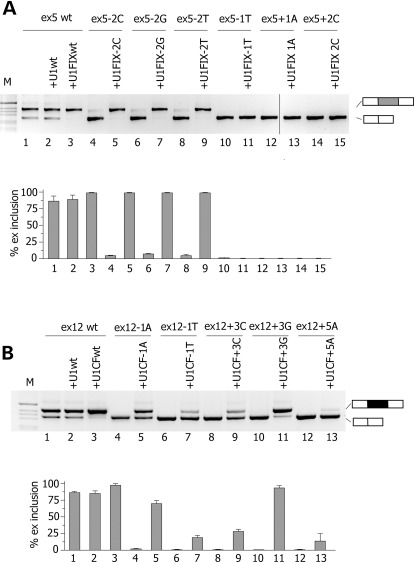
Cotransfection of U1FIXwt with the ex5 wt minigene improved the exon recognition inducing complete inclusion of the exon (Fig. 2A, lanes 1 and 3). Noticeably, the three synonymous changes at position −2 showed a remarkable increase in the percentage of exon inclusion after cotransfection of the corresponding complementary U1 snRNAs, with a negligible presence of aberrant transcripts (Fig. 2A, lanes 4–9). On the contrary, the splicing pattern of the other FIX mutations −1T, +1A and +2C was not affected by the corresponding complementary U1 snRNAs.
Figure 2.
Effect of increased complementarity of U1 snRNAs to normal and mutated donor splice site on pre-mRNA splicing. (A) Effect of complementary U1 snRNA to the F9 exon 5 5′ss. The upper panel shows the analysis of spliced transcripts. Minigenes were transfected in HeLa cells with the empty vector (lanes 1, 4, 6, 8, 10, 12, 14) or with modified U1s (lanes 3, 5, 7, 9, 11, 13, 15) and splicing pattern evaluated by RT–PCR. Amplified products were resolved on a 2% agarose gel. The identity of the U1 snRNAs is shown in Supplementary Material, Figure S2. Lower panel is the quantification of exon 5 splicing pattern after co-transfection with modified U1 snRNAs. Data are expressed as means ± SD of three independent experiments done in duplicate. (B) Effect of complementary U1 snRNAs to the CFTR exon 12 5′ss. Minigenes were transfected in HeLa cells with the empty vector (lanes 4, 6, 8) or with modified U1s (lanes 3, 5, 7, 9) and the splicing pattern evaluated by RT–PCR. Amplified products were resolved on a 2% agarose gel. The identity of the U1 snRNAs is shown in Supplementary Material, Figure S2. Lower panel is the quantification of exon 12 splicing pattern. Data are expressed as means ± SD of three independent experiments done in duplicate.
Minigene experiments with the CFTR exon 12 showed that all mutations were rescued by the corresponding complementarity U1 snRNAs. Whereas the −1T, +3C and +5A were slightly enhanced (Fig. 2B), the severe splicing defects caused by −1A and +3G mutations were efficiently corrected by the complementary U1 snRNAs, reaching ∼75 and ∼95% of exon inclusion. Cotransfection of U1 CFwt with the CFex12 wt also improved the splicing pattern (Fig. 2B, lane 3).
These results indicate that the three synonymous −2 variants in F9 exon 5 and the donor 5′ss mutations in CFTR exon 12 do not completely disrupt the 5′ss function. They produce a defective donor site, whose recognition by complementary U1 snRNA variants recovers correct exon inclusion.
U1 snRNAs complementary to intronic sequences downstream of the 5′ss correct aberrant splicing
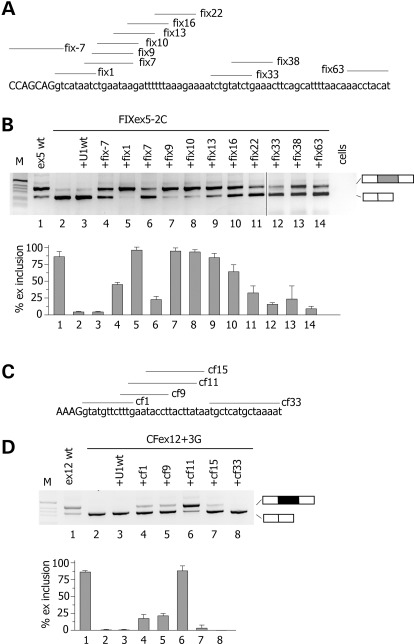
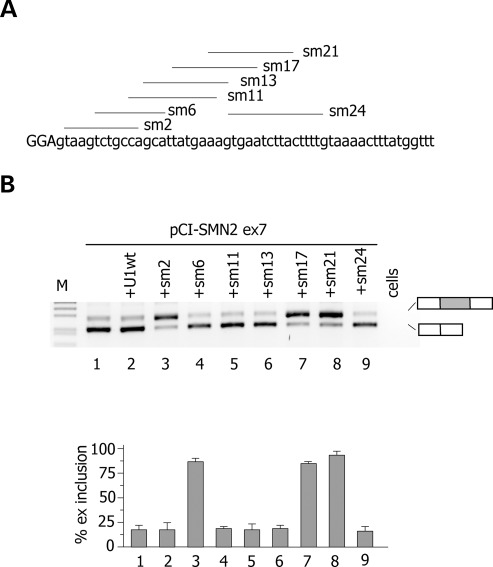
To promote exon definition, U1 snRNP does not necessarily have to perfectly bind at the 5′ss. Some atypical 5′ss are recognized by U1 snRNA shifted by one nucleotide (21) and U1 snRNA complementary to downstream intronic sequences were originally reported to promote exon inclusion in model gene system (22,23). Thus, we tested the effect of U1 snRNA-binding downstream mutant CFTR exon 12 and F9 exon 5. For this purpose, we prepared different U1 snRNAs, whose 5′ tails were modified to base pair at variable distance downstream of the 5′ss of F9 exon 5 and CFTR exon 12 (Fig. 3). This approach was also extended to the extensively investigated SMN2 exon 7, where a weak constitutive 5′ss and a synonymous exonic substitution are associated to exon skipping (24–26). In F9, we systematically screened U1 snRNAs from position −7 till position 63 relative to the donor site junction and the analysis was performed on the ex5 −2C mutant minigene. As shown in Figure 3A, all U1 snRNAs tested reduced exon 5 skipping, indicating their positive effect on exon definition. In particular fix 1, fix 9 and fix 10 U1 snRNAs showed the strongest effect with a nearly complete rescue of aberrant splicing. In CFTR exon 12, only the U1 snRNA variant cf11 induced a significant (70%) rescue of the splicing pattern in the +3G mutant (Fig. 3D). The other U1 snRNAs had a modest effect (cf1 and cf9) or no effect (cf15 and cf33) on splicing (Fig. 3D). In the SMN2 minigene, ∼20% of exon 7 is included in the final transcript, whereas cotransfection of sm2, sm17 and sm21 resulted in >80% of exon inclusion (Fig. 4B).
Figure 3.
Identification of ExSpeU1s in F9 exon 5 and CFTR exon 12. (A and C) Schematic representation of binding sites of the ExSpeU1s in F9 exon 5 and CFTR exon 12, respectively. Donor site and downstream intronic region are shown and exonic and intronic sequences are in upper and lower case, respectively. Lines correspond to the target-binding sequences of ExSpeU1s U1 snRNAs on the nascent pre-mRNA. (B) Analysis of F9 exon 5-spliced transcripts. The FIXex5-2C mutant minigene was transfected in HeLa cells alone (lane 1), with the empty vector (lane 2), or with plasmids encoding for the different ExSpeU1s. The pattern of splicing was evaluated by RT–PCR and amplified products were resolved on a 2% agarose gel. Lower panel is the quantification of exon 5 splicing pattern. (D) Analysis of CFTR exon 12-spliced transcripts. The mutant ex12 + 3G minigene was transfected in HeLa cells alone (lane 1), with the empty vector (lane 2), or with plasmids encoding for the ExSpeU1s. The pattern of splicing was evaluated by RT–PCR and amplified products were resolved on a 2% agarose gel. Lower panel is the quantification of exon 5 splicing pattern.
Figure 4.
Identification of ExSpeU1s in SMN2 exon 7. (A) Schematic representation of binding sites of the ExSpeU1s in SMN2 exon 7. Donor site and downstream intronic region are shown and exonic and intronic sequences are in upper and lower case, respectively. Lines correspond to the target-binding sequences of ExSpeU1s on the nascent pre-mRNA. (B) Analysis of SMN2 exon 7 spliced transcripts. The pCI-SMN2 minigene was transfected in HeLa cells alone (lane 1), with the U1wt (lane 2), or with plasmids encoding for ExSpeU1s. The pattern of splicing was evaluated by RT–PCR and amplified products were resolved on a 2% agarose gel. Lower panel is the quantification of SMN2 exon 7 splicing pattern. Data are the means ± SD of three experiments done in duplicate.
These results in three different gene systems indicate that U1 snRNAs binding downstream the 5′ss corrected exon skipping caused by defective 5′ss.
SMN-specific U1 snRNAs rescue exon 7 splicing in normal cells
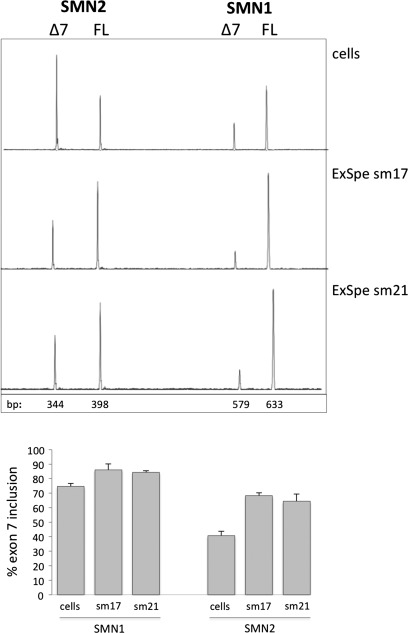
To study the effect of different ExSpeU1s in the chromosomal context, we focussed on SMN exon 7. Normal individuals have two SMN copies, SMN1 and SMN2, that mainly differ for a synonymous substitution in exon 7. This substitution induces SMN2 exon 7 skipping and because the majority of SMA patients lack SMN1, its splicing correction is at the basis of several therapeutic attempts (27,28). To evaluate the potential therapeutic effect of SMN-specific U1s in SMA, we transfected sm17 and sm21 in HEK293 cells and evaluated the endogenous pattern of splicing. Semiquantitative analysis of splicing isoforms showed that sm17 and sm21 induced a significant increase in the percentage of SMN exon 7 inclusion. In particular, transfection of the two ExSpeU1s increased the percentage of SMN2 exon 7 from ∼40 to ∼70% (Fig. 5).
Figure 5.
SMN exon 7 splicing correction mediated by sm17 and sm21 ExSpeU1s in normal cells. The upper panel shows a representative experiment in which the fluorescently labelled RT–PCR amplified fragments were separated on denaturing capillary electrophoresis. HEK393 cells were transfected with the indicated ExSpeU1s and amplified fragments were digested with DdeI to obtain SMN1 and SMN2 exon 7 inclusion (FL) and exclusion (Δ7) fragments. The size of the fragments is indicated. Lower panel shows the percentage of SMN1 and SMN2 exon 7 inclusion in ExSpeU1-treated and -untreated cells and data are expressed as means ± SD of three independent experiments done in duplicate. Transfection efficiency in the different experiments was 75–85%.
A unique exon-specific U1 snRNA can rescue splicing of defective splice site mutants and exonic variants
We subsequently tested the effect of the most active U1 snRNAs (fix9 for FIX and cf11 for CFTR), named exon-specific U1 snRNAs (ExSpeU1), on all splicing mutations. In F9 exon 5, fix9 ExSpeU1 completely rescued the defective donor sites of the three synonymous mutations in position −2 (Fig. 6A, lanes 5–10). At variance, the −1T, +1A and +2C 5′ss mutations, not corrected by loading the U1 snRNA directly on the donor site (Fig. 2A), were also not influenced by ExSpeU1 fix9 (Fig. 6A, lanes 11–16). Interestingly, cotransfection of ExSpeU1 fix9 induced nearly complete exon inclusion of the two −8G and −9G intronic variants of the polypyrimidine tract (Fig. 5A, lanes 1–4). In CFTR exon 12, ExSpeU1 cf11 rescued all 5′ss mutations with different efficiency. After ExSpeU1 cf11 cotransfection, the +3G mutation showed ∼85% of exon inclusion (Fig. 6B); the percentage of exon inclusion in −1A, +3C and +5A increased to ∼50%, whereas in −1T it improved to ∼20% (Fig. 6B). Intriguingly, we observed complete splicing correction by ExSpeU1 cf11 of the two exonic A566T and Y577Y mutations (Fig. 6B, lanes 1–4). Sequencing of ExSpeU1-induced products showed correct usage of the normal intron–exon junction and no activation of cryptic splice sites. To clarify if the enhancing effect is specific for the U1 snRNA particle, we inserted the tail of two active ExSpeU1s, fix10 and cf11, in the U7 snRNA. U7 is not involved in splicing and a modified version has been extensively used to express antisense RNA sequences (29–35). The resulting U7 fix10 and U7 cf11 were cotransfected with −8G, −2T mutants and +3G and Y577Y, respectively. In contrast to the ExSpeU1s, the corresponding U7s had no effect (Supplementary Material, Fig. S4), suggesting that U1 snRNP-specific proteins are required for splicing rescue and that ExSpeU1s are not just targeting intronic sequences with silencer function.
Figure 6.
Splicing correction mediated by fix9 and cf11 U1 ExSpeU1s on natural mutations. (A) HeLa cells were transfected with the indicated FIX exon 5 mutant minigenes alone (even lanes) or along with fix9 U1 ExSpeU1 (odd lanes). Splicing pattern was evaluated by RT–PCR and amplified products were resolved on a 2% agarose gel. Ms is the molecular 1 Kb plus marker. Lower panel shows the quantification of exon 5 splicing pattern. Percentage of exon inclusion is expressed as means ± SD of three experiments done in duplicate. (B) HeLa cells were transfected with the indicated CFTR exon 12 mutant minigenes alone (even lanes) or along with cf11 ExSpeU1 (odd lanes). Splicing pattern was evaluated by RT–PCR and amplified products were resolved on a 2% agarose gel. Lower panel shows the quantification of exon 12 splicing pattern. Percentage of exon inclusion is expressed as means ± SD of three experiments done in duplicate.
Altogether, these data indicate that recruitment of ExSpeU1 by complementarity on intronic sequences downstream of the exon corrects multiple splicing mutations located either at 5′ or 3′ splice sites or in exonic regulatory elements. This provides a novel therapeutic strategy exploiting a unique ExSpeU1 to correct a panel of splicing defects.
Rescue of splicing by the F9 exon5-specific U1 snRNA results in FIX biosynthesis and coagulant activity
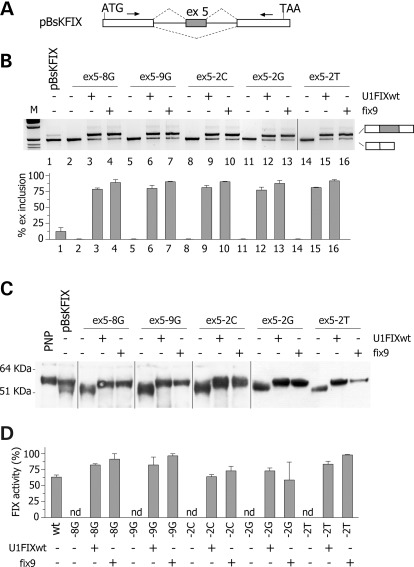
To establish whether the ExSpeU1-mediated correction of splicing of the different mutants result in a consistent rescue of protein biosynthesis and function, we took advantage of the FIX model. FIX is a serine protease secreted from cells that can be finely monitored by protein and functional assays. The splicing mutations corrected by the ExSpeU1 fix9 (the three −2 variants at the 5′ss and the two transversions at the polypyrimidine tract) were tested in a splicing-competent cDNA minigene context (pBsKFIX) (Fig. 7A). In this minigene, exon 5 was inserted in FIX cDNA transcript along with part of its intronic sequences in a manner that the correction of exon skipping produces a normal transcript with secretion of a functional protein (Fig. 7A). The pBsKFIX exon 5 wt and mutant minigenes were expressed in BHK cells with or without the modified U1 snRNAs complementary to the F9 donor site (U1FIXwt) or to the downstream intronic sequence (fix9).
Figure 7.
Rescue of splicing and FIX protein function by ExSpeU1. (A) Schematic representation of the pBsK-FIX minigene. Exonic sequences are boxed, introns are lines, the arrows represent primers for PCR amplification and the dotted lines indicate the two possible exon 5 alternative splicing events. The position of the ATG and TAA codons is indicated. The minigene is not drawn in scale. (B) Analysis of pBsK-FIX exon 5 spliced transcripts. pBsK-FIX exon 5 normal and mutant minigenes were transfected in BHK cells alone or with the indicated ExSpeU1s. The splicing pattern was evaluated by RT–PCR and amplified products were resolved on a 2% agarose gel. M is the molecular 1 Kb marker. Lower panel shows the quantification of the percentage exon 5 inclusion. Data are expressed as means ± SD of three independent experiments done in duplicate. (C) Western blotting of FIX in the BHK conditioned medium. PNP is pooled normal plasma. (D) FIX coagulant activity in the BHK conditioned medium. Data are expressed as means ± SD of three experiments done in duplicate.
Probably caused by the pBsKFIX minigene context, this construct showed exon 5 inclusion in ∼10% of the transcripts (Fig. 7, lane 1), while experiments with the pTB minigene (Fig. 1) and in vivo (Supplementary Material, Fig. S3) showed a ∼85% inclusion. In spite of this inefficient exon 5 recognition, the secreted FIX levels in the conditioned medium were appreciable by ELISA (13 ± 1 ng/ml), western blotting (Fig. 7C) and coagulation assays (Fig. 7D). In particular, western blotting revealed the presence of a band corresponding to the full-length FIX form (∼60 kDa), which migrated as plasma-derived FIX. In addition, a band corresponding to a smaller size FIX variant (∼51 kDa) was also clearly detectable. This arises from translation of the in frame FIX mRNA form lacking exon 5 (Fig. 7C), leading to a FIX molecule lacking 43 amino acids of the second epidermal growth factor like domain 2 (EGF2). Notably, the mRNA splicing and western blot patterns showed remarkably different relative amount of the two isoforms, with the aberrant form predominating at the mRNA (Fig. 7B) but not at the protein (Fig. 7C) level, a finding compatible with inefficient biosynthesis of the protein without the EGF2 domain.
Transfection of the five pBsKFIX mutant minigenes showed complete skipping of the exon (Fig. 7B, lanes 2, 5, 8, 11, 14), and cotransfection of either U1FIXwt or fix9 ExSpeU1 induced a complete rescue of the splicing pattern. In all five mutants, the percentage of exon inclusion increased to level above 75% (Fig. 7B). The extent of splicing rescue mediated by these U1 snRNAs was remarkable, taking into account the inefficient exon 5 recognition in this context. Results from investigations at the protein level in the conditioned medium were consistent with those from splicing assays. The expression of the five mutants resulted in secretion of deleted FIX proteins (Fig. 7C), which did not display any appreciable coagulant activity (Fig. 7D). At variance, co-expression of variants with either U1FIXwt or fix9 ExSpeU1 induced the exclusive synthesis of the full-length FIX (Fig. 7C). This was paralleled by a complete rescue of FIX activity (Fig. 7D), which for most variants was higher than that measured for the pBsKFIX wt construct.
DISCUSSION
In this study, we provide a novel strategy to correct different types of natural splicing mutations using exon specific U1 snRNAs (ExSpeU1). ExSpeU1s bind by complementarity to intronic sequences downstream of the exon and rescue different types of splicing defects associated to exon skipping. ExSpeU1s are active on several 5′ss mutations in CFTR exon 12 and F9 exon 5, on two transversions at the polypyrimidine tract in F9 exon 5, on two exonic substitutions in CFTR exon 12 and on defective SMN2 exon 7 splicing (also due to an exonic variant). In this latter case, ExSpeU1 rescued SMN2 exon 7 splicing directly in the chromosomal context of normal cells, providing a new therapeutic strategy for SMA. On the other hand, in the model of Haemophilia B, a unique ExSpeU1 induced complete splicing correction of five different F9 mutations, thus resulting in a complete rescue of protein biosynthesis and coagulation activity.
A limited number of studies have explored the role and potential therapeutic effect of U1 snRNAs on splicing correction of donor site mutations (13–19). In all cases, the modified tails of U1 snRNA have few nucleotide changes in comparison to the WT sequences and base pair exactly to the mutant donor sites. Thus, their therapeutic effect is restricted to each mutation and the potential complementarity of these modified U1 snRNAs to other normal 5′ss might affect splicing of other exons, in particular those alternatively spliced. The ExSpeU1s we have developed here, which bind to non-conserved intronic sequences, have two major advantages, (i) they do not interact directly with normal 5′ss and (ii) correct different types of splicing defects associated to exon skipping. The mutations rescued by ExSpeU1s affect three different splicing regulatory elements important for exon recognition: the 5′ss, the polypyrimidine tract of the 3′ss and the exonic splicing regulatory sequences. To our knowledge, this is the first time that an U1 snRNA variant is shown to correct exon skipping due to polypyrimidine tract and exonic mutants. SMN2 is a paradigmatic example of exon skipping caused by a synonymous variant in an exonic regulatory element and its correction is at the basis of several therapeutic attempts in SMA (27,28).
Compared with classical gene replacement therapies, ExSpeU1-mediated rescue has a number of additional advantages and stimulating perspectives. The direct splicing correction maintains the regulation of the gene expression in the correct cell-specific chromosomal context under the control of endogenous transcription and pre-mRNA processing regulatory elements. The short length of the ExSpeU1s cassette (∼500 bp) can also be useful in gene therapy of splicing mutations in large genes such as CFTR and F8, whose full-length transcript can represent a limiting step for their insertion in viral vectors such as AAV. On the other hand, to achieve the optimal therapeutic rescue, the expression level could be modulated in vivo by varying the number of ExSpeU1 cassette in the viral vector. In the case of dominant-negative mutations where the replacement therapy is not feasible, the splicing correction mediated by ExSpeU1 will act directly reducing the amount of the mutated toxic protein. Moreover, binding of the ExSpeU1s to intronic sequences, which are not conserved, will significantly reduce the possibility of off-target events. Although binding of ExSpeU1s to unrelated sequences not directly involved in splicing might have an effect on other pre-mRNA processing steps (36), our result indicate that it will be possible to design ExSpeU1s with improved specificity by inserting subtle changes in sequence target and/or length. For example, ExSpeU1s sm17 and sm21 in SMN2 (Fig. 3B) or fix9, fix10 and fix13 in F9 exon 5 (Fig. 3B) bind at nearby or overlapping intronic sequences and are functionally active. Future studies in cellular and animal models will be required to address this point.
According to the classical exon definition model, a network of interactions across the exon put in contact the splicing complexes assembled on the splice sites (11,19,37). This includes the recognition of the 5′ss by the U1 snRNP, binding of U2AFs to the polypyrimidine tract and identification of exonic splicing regulatory elements by SR proteins. The mutations we have analysed here affect differently these regulatory elements and probably block splicing at the first step when the exon has to be recognized co-transcriptionally. At this stage, recruitment of U1 snRNA at the 5′ss or at downstream sequences by means of ExSpeU1s can rescue the different defects. This occurs not only for mutations at the consensus 5′ss that directly interfere with U1 snRNA binding but also for mutations located at distance from the donor site in the polypyrimidine tract or in exonic regulatory elements. In these cases, ExSpeU1s probably compensate the missing interactions and facilitate the formation of the correct network of splicing factors over the exon.
Interestingly, the two ExSpeU1s for SMN2 (sm17 and sm21) are partially complementary to a previously reported intronic splicing silencer, which binds to the negative splicing factor hnRNPA1 (38). Targeting this ISS with antisense oligonucleotides resulted in inclusion of SMN2 exon 7 in model minigenes and in cellular systems (39) and was recently shown to improve the phenotype in a SMA mouse model (27). Thus, it is possible that ExSpeU1s facilitates recruitment of the splicing machinery on the exon interfering with intronic splicing regulatory sequences located downstream the donor site. However, this mechanism seems unlikely for F9 exon 5 and CFTR exon 12, as modified U7 particles binding to the target intronic sequences had no effect on splicing (Supplementary Material, Fig. S4). It is possible that ExSpeU1 acts at different levels depending on the architecture and relative strength of splicing regulatory elements in each exon. More in general, ExSpeU1s-mediated induction of exon inclusion could be used to regulate the synthesis of specific alternatively spliced isoforms for therapeutic purposes. For example, therapeutic induction of specific alternative splicing isoforms affect tumour progression (40), angiogenesis (41,42) or aging (43), and modified U1 snRNAs have been recently used to promote splicing to inhibit HIV replication (44).
Even if the disease-causing splicing mutations here investigated have a comparable effect on mature transcript, they behave differently regarding their sensitivity to ExSpeU1-mediated correction. The −1T +1T and +2C mutations in F9 exon 5 did not respond to the ExSpeU1 fix9, whereas the 5′ss mutations in CFTR exon 12 showed a variable response to cf11 ExSpeU1: in this case, the rescue efficiency was optimal for the +3G, intermediate for the −1A, +3C and +5A and low for the −1T (Fig. 5). As we have previously suggested (19), the different rescue efficiency to ExSpeU1 indicates that donor site mutations are mechanistically different. Interestingly, mutations that respond to ExSpeU1 (Fig. 2) were also sensitive to U1 snRNA complementary to the mutated 5′ss sequence (Fig. 5), suggesting a common effect of U1 snRNAs regardless of its loading position. Non-responsive mutations could affect the splicing progression after the exon definition step and their rescue would require the complementation with additional splicing factors. Even if mutations in positions +1 and +2 are not expected to respond to ExSpeU1 due to the obligate presence of the GT dinucleotide at the 5′ss, the mechanism that regulates the rescue efficiency of the other mutations is less obvious. Some positions are known to interact with other splicing factors in later splicing steps (like the −1 and +5 positions to U5 snRNA and U6 snRNA, respectively) (45,46). More studies are necessary to fully address the mechanism and identify the factors involved in determining the ExSpeU1 responsiveness of this type of splicing defects at the donor site.
In conclusion, a single ExSpeU1 appropriately loaded by complementarity to intronic sequences downstream of the exon rescues correct splicing impaired by different mutations and recover the corresponding protein biosynthesis and function in model gene systems. The functional levels obtained in the model of Haemophilia B and in SMN2 would produce in vivo a therapeutic correction of the bleeding defect and of the SMA phenotype, respectively. ExSpeU1s constitute a promising novel therapeutic strategy to correct splicing defects associated to defective exon definition in several human disorders.
MATERIALS AND METHODS
Hybrid minigene constructs
pCF exon 12 minigene has been previously described (7) and exonic and donor splice site mutations were introduced substituting the AccI-BamHI cassette by polymerase chain reaction (PCR)-mediated site-directed mutagenesis. To obtain the pFIX exon 5 minigene, the FIX exon 5 fragment consisting of the last 314 bp of intron 4, exon 5 (129 bp) and the first 278 bp of intron 5 was amplified from normal genomic DNA using FIXex5dir and FIXex5rev and cloned into the pTB NdeI-minigene (7). Polypyrimidine tract and donor splice site mutations were introduced in pFIXex5 between the unique PstI and XbaI sites of pFIX exon 5 by PCR-mediated site-directed mutagenesis. pCI-SMN2 exon 7 was obtained from Dr Adrian Krainer (CSHL, NY, USA). Modified and exon-specific U1 snRNAs were created by replacing the sequence between the sites BclI and BglII with oligonucleotides as previously reported (47). Sequences of oligonucleotides are provided in Supplementary Material, Table S2. Hybrid minigenes were verified by the sequence analysis. Modified U7 snRNAs (U7 fix10 and U7 cf11) were created by PCR amplification of U7SmOPT vector using cfsh11U7 and FIXsh10U7 and Sp6 primers. PCR products were digested with HindIII and StuI and ligated into HindIII/StuI sites of the U7SmOPT vector and the resulting clones verified by the sequence analysis.
The splicing-competent FIX cDNA expression cassette was synthesized by GeneScript Inc. (Piscataway, NJ, USA) and its sequence is available upon request. The cassette, inserted in the pCDNA 3.1+ backbone to make the pBsK-FIX, consists of a simian virus 40 (SV40) promoter followed by (i) the human FIX cDNA sequence from exons 1 to 4 and the 5′ portion (544 bp) of intron 4, (ii) a unique NdeI restriction site, (iii) the last 314 bp of intron 4, exon 5 and the 5′ portion (278 bp) of intron 5, (iv) a NdeI restriction site, (v) the 3′ end (908 bp) of intron 5 followed by the cDNA sequence from exons 6 to 8, and (vi) the SV40 polyadenylation site. The FIX cDNA fragments were amplified with high-fidelity DNA polymerase from the pCMV5-FIX vector. The NdeI–NdeI cassettes containing the last 314 bp of intron 4, the exon 5 and the 5′ portion (278 bp) of intron 5 were subcloned into pBsK-FIX to make the mutant variants for investigation of rescue at the protein level.
Analysis of hybrid minigene expression and SMN splicing
HeLa (human cervical carcinoma), HEK393 and BHK cell lines were grown in Dulbecco's modified Eagle's medium with Glutamax I (Gibco) (DMEM with glutamine, sodium pyruvate, pyridoxine and 4.5 g/l glucose) supplemented with 10% fetal calf serum (Euro Clone) and Antibiotic Antimycotic (Sigma) according to the manufacturer's instructions. HeLa cells grown on six well plates were transfected with Effectene reagents (Qiagen) according to the manufacturer's protocol. 0.5 µg of hybrid minigenes were transfected either alone or with 0.5 µg of wt/mutant U1 snRNA-encoding plasmids. Total RNA extraction was performed after 24 h of incubation using TRIreagent (Invitrogen) and reverse transcription reaction was carried out as described (47). alpha2,3 and Bra2 oligonucleotides were used for amplification of pCF exon 12 and pFIX exon 5; T7-F2 and E8-75 + 5′R oligonucleotides for pCI-SMN2. The conditions used for the PCRs were 94°C for 5 min for the initial denaturation, 94°C for 45 s, 56°C for 45 s, 72°C for 45 s for 35 cycles and 72°C for 10 min for the final extension. PCR products were resolved on 2% agarose gel electrophoresis. Quantification of exon inclusion was performed using the ImageJ software.
For SMN2 exon 7 analysis, HEK393 cells grown in six wells were transfected with 2 μg of each ExSpeU1s expression plasmid with the calcium-phosphate method and RT–PCR performed with E8-467-R and fluorescently labelled FAM-E6-F primers. Reactions were incubated at 95°C for 3 min followed by 35 cycles at 95°C for 30 s, 55°C for 30 s and 72°C for 36 s. Amplified fragments were digested with DdeI and separated on denaturing capillary electrophoresis (ABI-3100). Quantification of intensity of SMN1 and SMN2 exon 7 inclusion and exclusion bands was performed with Peakscanner™ software.
Transfection of BHK splicing-competent pBsK-FIX expression vector and reverse transcription was done as described (14). PCR reaction was carried out using pBsK-FIXdir and pBsK-FIXrev oligonucleotides. PCR products were resolved by 2% agarose gel electrophoresis. Liver RNA (First Choice Human Total RNA Survey Panel, Ambion, Inc.) was retrotranscribed in standard conditions and amplified with FIX140 and FIX279 primers.
FIX activity and protein assays
FIX coagulant activity was assessed by the aPTT coagulation assay (48). FIX antigen levels in the conditioned medium were evaluated by ELISA (Factor IX antigen, FIX; Affinity Biologicals, Ancaster, Canada). For western blotting analysis, 26 μl of the conditioned medium were incubated 5 min at 95°C and run on 4–12% SDS–PAGE (NuPAGE Bis–Tris gel, Invitrogen®, Carlsbad, CA, USA). Proteins were transferred onto a 0.2 μm nitrocellulose membrane (Whatman®, Dassel, Germany), which was blocked over night with PBS buffer supplemented with 0.1% Tween-20 (PBS-T) and 5% low fat dry milk (Bio-Rad, Hercules, CA, USA). Membranes were then incubated for 3 h at room temperature with an anti-Human F.IX peroxidase conjugated (GAFIX-APHRP; Affinity Biologicals). The Supersignal® West Femto reagent (Thermo Scientific, Rockford, IL, USA) was exploited for detection. Plasma derived FIX or rFIX-wt were used to optimize the assay.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by grants from the Telethon foundation (GGP09183), the Italian Cystic Fibrosis Foundation no (FFC#9/2009) adopted by Dedikato, CIV and Delegazione FFC del Lago di Garda, the Ministero dell'Università e della Ricerca (MIUR)-Progetti di Ricerca di Interesse Nazionale (PRIN), the University of Ferrara and Fondazione CARIFE. Funding to pay the Open Access publication charges for this article was provided by Comitato Telethon Fondazione Onlus.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Andres Muro and Marco Baralle for critical reading of the manuscript, Adrian Krainer for the pCI-SMN2 exon 7 minigene and D. Schümperli for the U7SmOPT plasmid.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Krawczak M., Reiss J., Cooper D.N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 2.Teraoka S.N., Telatar M., Becker-Catania S., Liang T., Onengut S., Tolun A., Chessa L., Sanal O., Bernatowska E., Gatti R.A., et al. Splicing defects in the ataxia-telangiectasia gene, ATM: underlying mutations and consequences. Am. J. Hum. Genet. 1999;64:1617–1631. doi: 10.1086/302418. doi:10.1086/302418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ars E., Serra E., Garcia J., Kruyer H., Gaona A., Lazaro C., Estivill X. Mutations affecting mRNA splicing are the most common molecular defects in patients with neurofibromatosis type 1. Hum. Mol. Genet. 2000;9:237–247. doi: 10.1093/hmg/9.2.237. doi:10.1093/hmg/9.2.237. [DOI] [PubMed] [Google Scholar]
- 4.Pagani F., Baralle F.E. Genomic variants in exons and introns: identifying the splicing spoilers. Nat. Rev. Genet. 2004;5:389–396. doi: 10.1038/nrg1327. doi:10.1038/nrg1327. [DOI] [PubMed] [Google Scholar]
- 5.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. doi:10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 6.Wang G.S., Cooper T.A. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat. Rev. Genet. 2007;8:749–761. doi: 10.1038/nrg2164. doi:10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 7.Pagani F., Stuani C., Tzetis M., Kanavakis E., Efthymiadou A., Doudounakis S., Casals T., Baralle F.E. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. doi:10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 8.Pagani F., Raponi M., Baralle F.E. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl Acad. Sci. USA. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. doi:10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M., Manley J.L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nilsen T.W., Graveley B.R. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. doi:10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berget S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 12.Carmel I., Tal S., Vig I., Ast G. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004;10:828–840. doi: 10.1261/rna.5196404. doi:10.1261/rna.5196404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baralle M., Baralle D., De Conti L., Mattocks C., Whittaker J., Knezevich A., Ffrench-Constant C., Baralle F.E. Identification of a mutation that perturbs NF1 agene splicing using genomic DNA samples and a minigene assay. J. Med. Genet. 2003;40:220–222. doi: 10.1136/jmg.40.3.220. doi:10.1136/jmg.40.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinotti M., Balestra D., Rizzotto L., Maestri I., Pagani F., Bernardi F. Rescue of coagulation factor VII function by the U1+5A snRNA. Blood. 2009;113:6461–6464. doi: 10.1182/blood-2009-03-207613. doi:10.1182/blood-2009-03-207613. [DOI] [PubMed] [Google Scholar]
- 15.Pinotti M., Rizzotto L., Balestra D., Lewandowska M.A., Cavallari N., Marchetti G., Bernardi F., Pagani F. U1-snRNA-mediated rescue of mRNA processing in severe factor VII deficiency. Blood. 2008;111:2681–2684. doi: 10.1182/blood-2007-10-117440. doi:10.1182/blood-2007-10-117440. [DOI] [PubMed] [Google Scholar]
- 16.Schmid F., Glaus E., Barthelmes D., Fliegauf M., Gaspar H., Nurnberg G., Nurnberg P., Omran H., Berger W., Neidhardt J. U1 snRNA-mediated gene therapeutic correction of splice defects caused by an exceptionally mild BBS mutation. Hum. Mutat. 2011;7:815–824. doi: 10.1002/humu.21509. [DOI] [PubMed] [Google Scholar]
- 17.Tanner G., Glaus E., Barthelmes D., Ader M., Fleischhauer J., Pagani F., Berger W., Neidhardt J. Therapeutic strategy to rescue mutation-induced exon skipping in rhodopsin by adaptation of U1 snRNA. Hum. Mutat. 2009;30:255–263. doi: 10.1002/humu.20861. doi:10.1002/humu.20861. [DOI] [PubMed] [Google Scholar]
- 18.Glaus E., Schmid F., Da Costa R., Berger W., Neidhardt J. Gene therapeutic approach using mutation-adapted U1 snRNA to correct a RPGR splice defect in patient-derived cells. Mol. Ther. 2011;19:936–941. doi: 10.1038/mt.2011.7. doi:10.1038/mt.2011.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Susani L., Pangrazio A., Sobacchi C., Taranta A., Mortier G., Savarirayan R., Villa A., Orchard P., Vezzoni P., Albertini A., et al. TCIRG1-dependent recessive osteopetrosis: mutation analysis, functional identification of the splicing defects, and in vitro rescue by U1 snRNA. Hum. Mutat. 2004;24:225–235. doi: 10.1002/humu.20076. doi:10.1002/humu.20076. [DOI] [PubMed] [Google Scholar]
- 20.Raponi M., Baralle F.E., Pagani F. Reduced splicing efficiency induced by synonymous substitutions may generate a substrate for natural selection of new splicing isoforms: the case of CFTR exon 12. Nucleic Acids Res. 2007;35:606–613. doi: 10.1093/nar/gkl1087. doi:10.1093/nar/gkl1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roca X., Krainer A.R. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat. Struct. Mol. Biol. 2009;16:176–182. doi: 10.1038/nsmb.1546. doi:10.1038/nsmb.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen J.B., Snow J.E., Spencer S.D., Levinson A.D. Suppression of mammalian 5′ splice-site defects by U1 small nuclear RNAs from a distance. Proc. Natl Acad. Sci. USA. 1994;91:10470–10474. doi: 10.1073/pnas.91.22.10470. doi:10.1073/pnas.91.22.10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang D.Y., Cohen J.B. U1 small nuclear RNA-promoted exon selection requires a minimal distance between the position of U1 binding and the 3′ splice site across the exon. Mol. Cell Biol. 1997;17:7099–7107. doi: 10.1128/mcb.17.12.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. doi:10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. doi:10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 26.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. doi:10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 27.Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. doi:10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorson C.L., Rindt H., Shababi M. Spinal muscular atrophy: mechanisms and therapeutic strategies. Hum. Mol. Genet. 2010;19:R111–R118. doi: 10.1093/hmg/ddq147. doi:10.1093/hmg/ddq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer K., Marquis J., Trub J., Nlend Nlend R., Verp S., Ruepp M.D., Imboden H., Barde I., Trono D., Schumperli D. Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum. Mol. Genet. 2009;18:546–555. doi: 10.1093/hmg/ddn382. doi:10.1093/hmg/ddn382. [DOI] [PubMed] [Google Scholar]
- 30.Gorman L., Suter D., Emerick V., Schumperli D., Kole R. Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc. Natl Acad. Sci. USA. 1998;95:4929–4934. doi: 10.1073/pnas.95.9.4929. doi:10.1073/pnas.95.9.4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madocsai C., Lim S.R., Geib T., Lam B.J., Hertel K.J. Correction of SMN2 Pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol. Ther. 2005;12:1013–1022. doi: 10.1016/j.ymthe.2005.08.022. doi:10.1016/j.ymthe.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 32.Liu S., Asparuhova M., Brondani V., Ziekau I., Klimkait T., Schumperli D. Inhibition of HIV-1 multiplication by antisense U7 snRNAs and siRNAs targeting cyclophilin A. Nucleic Acids Res. 2004;32:3752–3759. doi: 10.1093/nar/gkh715. doi:10.1093/nar/gkh715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asparuhova M.B., Marti G., Liu S., Serhan F., Trono D., Schumperli D. Inhibition of HIV-1 multiplication by a modified U7 snRNA inducing Tat and Rev exon skipping. J. Gene Med. 2007;9:323–334. doi: 10.1002/jgm.1027. doi:10.1002/jgm.1027. [DOI] [PubMed] [Google Scholar]
- 34.Francois V., Klein A.F., Beley C., Jollet A., Lemercier C., Garcia L., Furling D. Selective silencing of mutated mRNAs in DM1 by using modified hU7-snRNAs. Nat. Struct. Mol. Biol. 2011;18:85–87. doi: 10.1038/nsmb.1958. doi:10.1038/nsmb.1958. [DOI] [PubMed] [Google Scholar]
- 35.Uchikawa H., Fujii K., Kohno Y., Katsumata N., Nagao K., Yamada M., Miyashita T. U7 snRNA-mediated correction of aberrant splicing caused by activation of cryptic splice sites. J. Hum. Genet. 2007;52:891–897. doi: 10.1007/s10038-007-0192-8. doi:10.1007/s10038-007-0192-8. [DOI] [PubMed] [Google Scholar]
- 36.Beckley S.A., Liu P., Stover M.L., Gunderson S.I., Lichtler A.C., Rowe D.W. Reduction of target gene expression by a modified U1 snRNA. Mol. Cell Biol. 2001;21:2815–2825. doi: 10.1128/MCB.21.8.2815-2825.2001. doi:10.1128/MCB.21.8.2815-2825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faustino N.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. doi:10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 38.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008;82:834–848. doi: 10.1016/j.ajhg.2008.01.014. doi:10.1016/j.ajhg.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 2007;5:e73. doi: 10.1371/journal.pbio.0050073. doi:10.1371/journal.pbio.0050073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghigna C., De Toledo M., Bonomi S., Valacca C., Gallo S., Apicella M., Eperon I., Tazi J., Biamonti G. Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed: therapeutic potential of bifunctional oligonucleotides and indole derivatives. RNA Biol. 2010;7:495–503. doi: 10.4161/rna.7.4.12744. doi:10.4161/rna.7.4.12744. [DOI] [PubMed] [Google Scholar]
- 41.Nowak D.G., Amin E.M., Rennel E.S., Hoareau-Aveilla C., Gammons M., Damodoran G., Hagiwara M., Harper S.J., Woolard J., Ladomery M.R., et al. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: a novel therapeutic strategy for angiogenesis. J. Biol. Chem. 2009;285:5532–5540. doi: 10.1074/jbc.M109.074930. doi:10.1074/jbc.M109.074930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merdzhanova G., Gout S., Keramidas M., Edmond V., Coll J.L., Brambilla C., Brambilla E., Gazzeri S., Eymin B. The transcription factor E2F1 and the SR protein SC35 control the ratio of pro-angiogenic versus antiangiogenic isoforms of vascular endothelial growth factor-A to inhibit neovascularization in vivo. Oncogene. 2010;29:5392–5403. doi: 10.1038/onc.2010.281. doi:10.1038/onc.2010.281. [DOI] [PubMed] [Google Scholar]
- 43.Fong L.G., Vickers T.A., Farber E.A., Choi C., Yun U.J., Hu Y., Yang S.H., Coffinier C., Lee R., Yin L., et al. Activating the synthesis of progerin, the mutant prelamin A in Hutchinson-Gilford progeria syndrome, with antisense oligonucleotides. Hum. Mol. Genet. 2009;18:2462–2471. doi: 10.1093/hmg/ddp184. doi:10.1093/hmg/ddp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandal D., Feng Z., Stoltzfus C.M. Excessive RNA splicing and inhibition of HIV-1 replication induced by modified U1 small nuclear RNAs. J. Virol. 2010;84:12790–12800. doi: 10.1128/JVI.01257-10. doi:10.1128/JVI.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawa H., Shimura Y. Association of U6 snRNA with the 5′-splice site region of pre-mRNA in the spliceosome. Genes Dev. 1992;6:244–254. doi: 10.1101/gad.6.2.244. doi:10.1101/gad.6.2.244. [DOI] [PubMed] [Google Scholar]
- 46.Sontheimer E.J., Steitz J.A. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science. 1993;262:1989–1996. doi: 10.1126/science.8266094. doi:10.1126/science.8266094. [DOI] [PubMed] [Google Scholar]
- 47.Pagani F., Buratti E., Stuani C., Bendix R., Dork T., Baralle F.E. A new type of mutation causes a splicing defect in ATM. Nat. Genet. 2002;30:426–429. doi: 10.1038/ng858. doi:10.1038/ng858. [DOI] [PubMed] [Google Scholar]
- 48.Bernardi F., Dolce A., Pinotti M., Shapiro A.D., Santagostino E., Peyvandi F., Batorova A., Lapecorella M., Schved J.F., Ingerslev J., et al. Major differences in bleeding symptoms between factor VII deficiency and hemophilia B. J. Thromb. Haemost. 2009;7:774–779. doi: 10.1111/j.1538-7836.2009.03329.x. doi:10.1111/j.1538-7836.2009.03329.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.