Abstract
Motor neuron degeneration in amyotrophic lateral sclerosis (ALS) has a familial cause in 10% of patients. Despite significant advances in the genetics of the disease, many families remain unexplained. We performed whole-genome sequencing in five family members from a pedigree with autosomal-dominant classical ALS. A family-based elimination approach was used to identify novel coding variants segregating with the disease. This list of variants was effectively shortened by genotyping these variants in 2 additional unaffected family members and 1500 unrelated population-specific controls. A novel rare coding variant in SPAG8 on chromosome 9p13.3 segregated with the disease and was not observed in controls. Mutations in SPAG8 were not encountered in 34 other unexplained ALS pedigrees, including 1 with linkage to chromosome 9p13.2–23.3. The shared haplotype containing the SPAG8 variant in this small pedigree was 22.7 Mb and overlapped with the core 9p21 linkage locus for ALS and frontotemporal dementia. Based on differences in coverage depth of known variable tandem repeat regions between affected and non-affected family members, the shared haplotype was found to contain an expanded hexanucleotide (GGGGCC)n repeat in C9orf72 in the affected members. Our results demonstrate that rare coding variants identified by whole-genome sequencing can tag a shared haplotype containing a non-coding pathogenic mutation and that changes in coverage depth can be used to reveal tandem repeat expansions. It also confirms (GGGGCC)n repeat expansions in C9orf72 as a cause of familial ALS.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder clinically characterized by muscular weakness due to degeneration of upper and lower motor neurons. Median survival is limited by respiratory failure to 3 years after disease onset (1). Extra-motor involvement such as frontal executive deficits or behavioral problems is seen in up to 50% of patients, but the clinical criteria for frontotemporal dementia (FTD) are met in only ∼5% of patients (2). Adequate treatments are lacking. Familial and sporadic cases are clinically indistinguishable. In most instances, accumulations of the protein TDP-43 are encountered at post-mortem examination of the brain and spinal cord (3). Although only 10% of ALS is familial, the discovery of disease genes accelerated research into the pathogenic mechanism of motor neuron degeneration (4). Three genes are relatively frequently encountered in familial ALS. Mutations in SOD1 are responsible for ∼20% of familial ALS (5) and mutations in TARDPB and FUS are each encountered in 3–5% (6–9). Mutations in ANG, VAPB, OPTN and VCP are less often causing ALS (10–13). Roughly 70% of ALS families remain unexplained after routine genetic testing. Using linkage analysis in larger ALS kindreds, loci at chromosome 18q (14) and 20p (15) have been identified. In addition, several families with ALS and FTD have been linked to chromosome 9q (16) and 9p (17–26). Next-generation sequencing techniques such as exome-sequencing or whole-genome sequencing will accelerate the discovery of disease genes.
In this study, we used whole-genome sequencing in a small kindred of familial ALS in which mutations in SOD1, TARDBP, FUS, ANG, ATXN2 and GRN were previously excluded (27–29). We sought for genetic variants that segregated with the disease in this family and were absent in controls.
RESULTS
Description of the pedigree
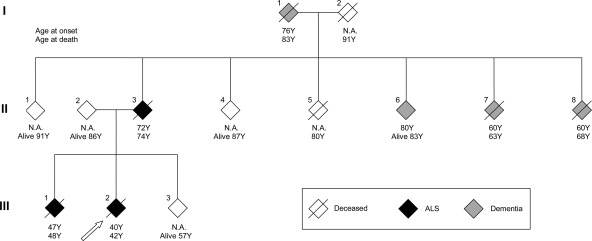
A small two-generation pedigree with classical ALS was studied (Fig. 1). The index patient (III:2) developed dysarthria and dysphagia at the age of 40. A progressive bulbar and facial weakness was noted, followed by weakness of the right hand 4 months later. No changes in cognition or personality were apparent, but a pseudobulbar affect with easy laughing was present. On clinical examination, a combination of upper and lower motor neuron signs were found in the bulbar region and in the limbs. Signs of lower motor neuron degeneration were present in three body regions on electrodiagnostic testing. The muscle weakness was steadily progressive and the patient died 22 months after disease onset. Around that time, ALS was diagnosed in one of the parents at the age of 72 (II:3). Subject II:3 suffered from progressive bulbar weakness with dysphagia and dysarthria. After 1 year, progressive muscle weakness and atrophy in the limbs with upper motor neuron signs were noted. Lower motor neuron signs were present in three body regions on electrodiagnostic testing. Death occurred at the age of 74. Three years later, at the age of 47, subject III:1 presented with difficulty walking caused by muscle weakness and atrophy in the right leg. At presentation, signs of lower and upper motor neuron loss in three body regions confirmed the diagnosis. Electrodiagnostic testing confirmed lower motor neuron involvement in three body regions. The disease progressed to the upper limbs and survival was limited to 16 months after disease onset. There was no history of ALS in generation I. I:1 and I:2 died at the age of 83 and 90, respectively. I:2 developed dementia presenting with memory complaints at the age of 76 years, but no motor problems were observed. Three siblings of generation two developed dementia. In 2 of them (II:7 and II:8), this was around the age of 60 years. Detailed clinical information and DNA was only available from subjects II:1–4 and subjects III:1–3.
Figure 1.
Pedigree of family with autosomal-dominant inheritance of ALS. The index patient is shown by the arrow. Age at disease onset and age at death are shown. Several other family members suffered from dementia, but no detailed clinical information or DNA was available from these family members. N.A. denotes not applicable.
Whole-genome sequencing
We performed whole-genome sequencing on the parents (II:2 not affected and II:3 affected) and three siblings (III:1 and III:2 affected, III:3 not affected) from this small kindred with classical rapidly progressive ALS. The inheritance pattern appeared to be autosomal dominant. Sequencing was performed at Complete Genomics (Mountain View, CA, USA), read mapping and variant detection was performed using the CGA tools®. Mapped bases after reference assembly and de novo assembly ranged from 188.4 to 338.2 Gb. The fraction of the genome sequenced in the five subjects was 89.8% on average (Supplementary Material, Table S1). The exome coverage was 96.6% on average and was consistently higher than the genome coverage for all five subjects. The average haploid coverage ranged from 44,1x to 74,2x (Supplementary Material, Table S1). At least 84.8% of the genome and 93.5% of the exome was sequenced with good coverage in all five subjects. The total single nucleotide variant (SNV) and small insertions/deletions (indels) count ranged from 3 045 998 to 3 280 751 and from 332 116 to 402 583, respectively, in the different subjects (Supplementary Material, Table S2).
First, the coding part of the genome was analyzed. The analysis flow is shown in Supplementary Material, Figure S1. In the coding region of each genome, 7947– 8666 missense variants, 24–56 nonsense variants and 302–496 frameshift variants were identified (Supplementary Material, Table S2). Using a family-based elimination approach, 414 missense mutations, 9 nonsense mutations and 8 frameshift mutations were retained that segregated with the disease in this family. After removing the variants also present in the most recent release of the 1000 Genomes Project or in dbSNP or in 9 previously sequenced genomes, 39 missense, 1 nonsense and 2 frameshift novel variants were selected as putative candidate causative mutations (Table 1).
Table 1.
Novel coding variants segregating with ALS in pedigree FALS3
| Gene | Gene name | Chrom | Variant | Confirmed | Present in 1500 population controls | Present in 2 family controls |
|---|---|---|---|---|---|---|
| NUP210L | nucleoporin 210kDa-like | 1 | p.V1196I | Yes | Yes | No |
| PMF1 | Polyamine-modulated factor 1 | 1 | p.R149W | Yes | Yes | No |
| KIF1B | Kinesin family member 1B | 1 | p.R18Q | Yes | No | Yes |
| KIF1B | Kinesin family member 1B | 1 | p.E1006G | Yes | Yes | Yes |
| RNF115 | Ring finger protein 115 | 1 | p.E239D | Yes | Yes | No |
| MAP3K6 | Mitogen-activated protein kinase 6 | 1 | p.R196W | Yes | No | Yes |
| C1orf59 | Chromosome 1 open reading frame 59 | 1 | p.V12I | Yes | Yes | Yes |
| SCMH1 | Sex comb on midleg homolog 1 | 1 | p.P266S | Yes | Yes | Yes |
| TDRD10 | Tudor domain containing 10 | 1 | p.R250C | Yes | Yes | No |
| AGXT | Alanine-glyoxylate aminotransferase | 2 | p.A280E | Yes | No | Yes |
| MYO7B | Myosin VIIB | 2 | p.T1124K | Yes | Yes | Yes |
| CCDC148 | Coiled-coil domain containing 148 | 2 | p.Y307F | Yes | Yes | Yes |
| ATG4B | ATG4 autophagy related 4 homolog B | 2 | p.R90Q | Yes | Yes | Yes |
| RNF123 | Ring finger protein 123 | 3 | p.A956V | Yes | No | No |
| DPPA2 | Developmental pluripotency associated 2 | 3 | p.A157S | Yes | Yes | Yes |
| SH3TC1 | SH3 domain and tetratricopeptide repeats 1 | 4 | p.L1088M | Yes | Yes | Yes |
| BRD2 | Bromodomain containing 2 | 6 | p.G565D | Yes | No | Yes |
| TULP1 | Tubby like protein 1 | 6 | p.A496T | Yes | No | Yes |
| CUTA | CutA divalent cation tolerance homolog | 6 | p.C22X | Yes | – | Yes |
| PSORS1C2 | Psoriasis susceptibility 1 candidate 2 | 6 | 280delC | Yes | – | Yes |
| CCDC132 | Coiled-coil domain containing 132 | 7 | p.V770I | Yes | No | Yes |
| SPAG8 | Sperm-associated antigen 8 | 9 | p.I121V | Yes | No | No |
| IFNA7 | Interferon, alpha 7 | 9 | p.V129L | Yes | Yes | No |
| NDOR1 | NADPH dependent diflavin oxidoreductase 1 | 9 | p.R30G | Yes | – | Yes |
| GPR133 | G protein-coupled receptor 133 | 12 | p.S265Y | Yes | Yes | Yes |
| MTMR15 | Myotubularin related protein 15 | 15 | p.E240K | Yes | Yes | Yes |
| ICT1 | Immature colon carcinoma transcript 1 | 17 | p.E171D | Yes | No | Yes |
| DNAH17 | Dynein, axonemal, heavy chain 17 | 17 | p.Q4387E | Yes | No | Yes |
| IFT20 | Intraflagellar transport 20 homolog | 17 | p.R105Q | Yes | Yes | Yes |
| DDX52 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 | 17 | p.D5G | Yes | Yes | Yes |
| EVI2A | Ecotropic viral integration site 2A | 17 | p.G187S | Yes | Yes | Yes |
| LRRC37B | Leucine rich repeat containing 37B | 17 | p.E510Q | Yes | – | Yes |
| KRTAP4–8 | Keratin associated protein 4–8 | 17 | p.C30X | Yes | Yes | No |
| ZNF229 | Zinc finger protein 229 | 19 | p.G513R | Yes | Yes | No |
| CCDC8 | Coiled-coil domain containing 8 | 19 | p.N318K | Yes | Yes | No |
| ZNF534 | Zinc finger protein 534 | 19 | 1510delC | Yes | Yes | Yes |
| PLIN4 | Perilipin 4 | 19 | p.V917I | Yes | Yes | No |
| SLCO4A1 | Solute carrier organic anion transporter family, member 4A1 | 20 | p.V263I | Yes | Yes | No |
| ARFGAP1 | ADP-ribosylation factor GTPase activating protein 1 | 20 | p.K278E | Yes | Yes | No |
| COL9A3 | Collagen, type IX, alpha 3 | 20 | p.P476R | Yes | Yes | No |
| TSPYL2 | Testis-specific Y-encoded-like protein 2 | X | p.R353C | Yes | Yes | Yes |
| FAM47B | Family with sequence similarity 47, member B | X | p.G14V | Yes | Yes | Yes |
Validation of novel coding variants segregating with disease
Next, a Sequenom MassAssay was performed for all candidate mutations. To check the quality of the results obtained from the whole-genome sequencing, the assay was run on the five original DNA samples. This assay failed in validating four variants due to the primer design. These variants were checked by Sanger sequencing or Taqman assay. All variants identified in the original whole-genome sequencing study were confirmed (Table 1).
In order to limit the list of candidate mutations, controls were analyzed. For this purpose, we used three groups of population-specific controls (900 healthy controls and 600 breast cancer controls with self-declared Flemish ethnicity for 3 generations, and 2 additional non-affected family members, being subject II:1 and II:4). We hypothesized that the novel variants (segregating with disease and not previously reported) that are also encountered in population controls or in two elderly healthy family members are unlikely to be the causative mutation. More likely, such variants represent very rare or population-specific variants. Using this strategy, all but two variants could be excluded. Both sets of controls provided complementary information, 27 variants could be excluded by genotyping the population controls and 28 by genotyping two additional unaffected family members (Table 1). The A956V variant in RNF123 (ring finger protein 123) and the I121V variation in SPAG8 (sperm-associated antigen 8) were not encountered in any of the controls. Of interest, SPAG8 is located on chromosome 9p13.3, just outside the narrow locus shared by all ALS-FTD families linked to 9p21 (17–26), but inside the larger region linked to ALS-FTD in several families. We therefore further studied the SPAG8 gene in ALS.
We sequenced the gene in a cohort of unexplained familial ALS patients seen at the neuromuscular reference center in Leuven, in whom mutations in SOD1, TARDBP, FUS, ANG, ATXN2 and GRN were previously excluded (27–29). The 8 exons and intron–exon boundaries of SPAG8 were sequenced in 43 ALS patients belonging to 33 unrelated pedigrees. Three additional missense changes (D37A, S110G and R349S) were found in two different pedigrees, but none of them appeared to be pathogenic. The D37A variant was found in ALS patients belonging to a pedigree with slowly progressive ALS, and the S110G and the R349S variant were observed in a family with dementia and ALS. The R349S variant was also observed in 5/1073 controls. The other two variants were absent in controls, but did not segregate with disease within the respective pedigrees. In addition, no mutations in SPAG8 were found in an ALS patient from a family with proven linkage to the 9p13.2–21.3 locus (19). Therefore, mutations in SPAG8 are unlikely to be pathogenic.
Fine-mapping of the shared haplotype around the SPAG8 gene
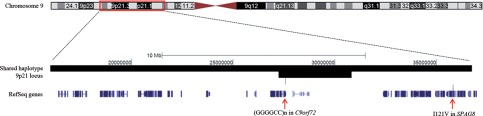
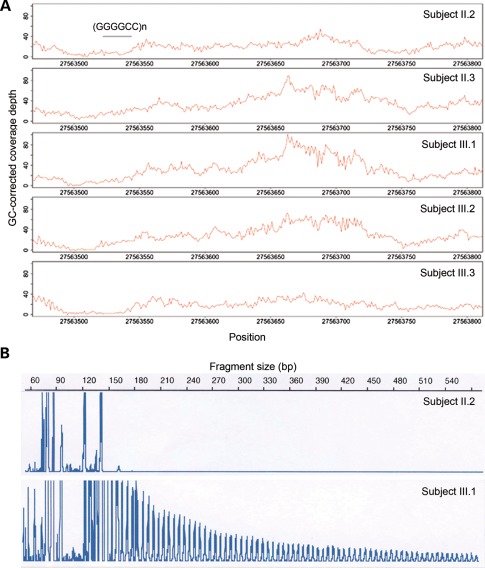
Since SPAG8 is located near a locus shared by all ALS-FTD families with linkage to 9p21, we hypothesized that this variant was tagging a causal non-coding mutation in this family. We determined the shared haplotype around the I121V variant by analyzing the known SNPs on chromosome 9p. The shared haplotype containing the variant was 22.7 Mb in size (14 041 725–36 734 729) and overlapped with the core 9p21 locus shared by all ALS-FTD families (Fig. 2). Within the overlapping region of the segregating haplotype and the core 9p linkage region, which consisted of 3.6 Mb (27 228 617–30 809 382), we analyzed all the novel non-coding variants. Nineteen novel variants segregating with disease were identified, of which none was located in conserved regions. It is therefore unlikely that any of these variants is causing the disease phenotype. We then assessed structural variants predicted by the CGA tools®. None of the structural variants identified was segregating with disease. Finally, since simple tandem repeats can also be involved in causing neurodegenerative diseases, we assessed whether variable tandem repeat expansions were present in affected family members. Since tandem repeats are often missed by paired-end short-read sequencing technologies due to mapping errors, leading to changes in coverage depth in and around tandem repeats (30), we assessed coverage depth within the core 9p linkage region. Theoretically, two different types of mapping errors can occur when assessing tandem repeat extensions: (i) if both ends of the paired-end read can effectively be mapped, the coverage depth in and around the repeat will be increased, or (ii) if one end of the paired-end read fails to map in the tandem repeat, there will be low coverage within the repeat region but increased coverage around the repeat site due to correct mapping of the other paired-end outside of the repeat region. We therefore investigated both scenarios in all intra-genic repeat regions within the core 9p linkage region. For the first scenario, seven tandem repeats with increased coverage at the repeat site segregating with disease in our pedigree were identified (Supplementary Material, Table S3). For the second scenario, only one tandem repeat with increased coverage depth around the repeat site was found. In particular, a peak of increased coverage depth in the 300 bps following the repeat was observed in affected family members (II:3, III:1 and III:2) but not in non-affected members (II:2 and III:3) (Fig. 3A). The repeat identified turned out to be the very recently described hexanucleotide (GGGGCC)n repeat in intron 1 of C9orf72 (31–33) as a novel cause of familial ALS and FTD. The repeat expansion in C9orf72 could not be observed in the aligned sequences. However, when amplifying the repeat region using polymerase chain reaction (PCR) affected family members were homozygous for a short repeat (repeat size of 5), whereas non-affected family members were heterozygous for two different repeat lengths (repeat size of 2 and 5). An unamplifiable repeat expansion can explain this observation. Using repeat-primed PCR, the expanded hexanucleotide repeat was revealed and was shown to segregate with disease (Fig. 3B). Although this technique does not allow accurate measurements of the repeat size, it was estimated to be ∼80 in this kindred, which is within the range described in ALS-FTD patients (range 30 to >100, up to 23 in controls) (31,32).
Figure 2.
Shared haplotype containing I121V variant in SPAG8 map of chromosome 9, showing the location of the core 9p21 locus containing the MOBKL2B, IFNK and C9orf72 genes and the shared haplotype containing the I121V variant in SPAG8.
Figure 3.
Expanded (GGGGCC)n repeat in C9orf72 in affected family members. (A) GC-corrected coverage depth reveals region with increased coverage depth in affected family members only in region following (GGGGCC)n repeat in C9orf72. (B) PCR products of repeat-primed PCR demonstrate repeat expansion in affected family members by the presence of a tail of stutter amplification. An example of a non-affected and affected family member is shown.
DISCUSSION
Next-generation sequencing technologies are expected to accelerate the discovery of novel disease genes (34). In patients with familial ALS or ALS-FTD, ∼50% of pedigrees remain unexplained. Very few studies using next-generation sequencing have been performed in neurodegenerative disorders. Exome sequencing has revealed mutations in VCP in familial ALS (13), mutations in VPS35 in Parkinson's disease (35,36), mutations in DNMT1 in a rare form of dementia with hearing loss and neuropathy (37) and mutations in DNAJC5 in adult-onset neuronal ceroid lipofuscinosis (38). No whole-genome studies in neurodegenerative disorders have been reported.
Because of the challenge of filtering and interpreting the large number of identified variants, the few whole-genome sequencing studies performed have focused on missense mutations, nonsense mutations or small frameshift variants (insertions or deletions), similar to exome sequencing studies (39,40). Due to the mapping of short reads, other genetic variants, such as structural variants of 50–500bp or repeat expansions, are difficult to extract from the aligned sequences. In our small kindred of autosomal-dominant ALS, whole-genome sequencing data of just five individuals (three affected and two unaffected) were used to find coding variants that segregated with disease. The list of candidates was dramatically reduced by the use of family controls and population-specific controls. Although we could not demonstrate that the most promising rare variant represents a pathogenic mutation, the availability of whole-genome sequencing data allowed us to delineate a shared haplotype of 22.7 Mb tagged by this rare coding variant. This haplotype completely overlapped with the core 9p linkage region and was shown to contain the very recently described hexanucleotide (GGGGCC)n repeat expansion in intron 1 of C9orf72 (31–33). This repeat expansion was not apparent from the aligned sequences, but was identified when assessing changes in coverage depth of known variable tandem repeat regions segregating with disease. The differences in coverage depth between affected and non-affected family members are possibly due half-mapped reads. In contrast to the studies that only very recently identified an expanded (GGGGCC)n repeat in C9orf72 using established linkage regions in large families with ALS-FTD, our whole-genome sequencing was able to pinpoint the same region using only a small kindred.
C9orf72 encodes an uncharacterized nuclear protein. How (GGGGCC)n in C9orf72 cause disease is unknown. The repeat expansion leads to loss of an alternatively spliced transcript, possibly to reduced protein expression and to the formation of nuclear RNA foci (31,32). Further studies are required to elucidate the disease mechanism of this novel cause of ALS-FTD with TDP-43 pathology.
Our study illustrates caveats in the interpretation of rare variants identified by current strategies for whole-genome sequencing and how rare variants can be useful in tagging a nearby non-coding true disease-causing mutation and how changes in coverage depth can reveal repeat expansions. In addition, the pathogenic nature of (GGGGCC)n repeat expansion in C9orf72 is further supported by our results.
MATERIALS AND METHODS
Subjects
Between 1994 and 2010, DNA samples from patients with familial ALS and controls seen at the neuromuscular clinic in Leuven were collected (28). Patients met the revised El Escorial (41) and Awaji criteria (42) of probable ALS. Blood samples were obtained after informed consent, and this study was approved by the local ethical committee of the KU Leuven. After excluding mutations in SOD1, TARDBP, FUS, ANG, ATXN2 and GRN (27–29), the cause of ALS remained unexplained in 46 patients belonging to 34 families. Three affected family members from pedigree FALS3 were used for whole-genome sequencing. Index patients from the other unexplained families and from an ALS pedigree with proven linkage to 9p (19) were used for validating the results obtained. Population-specific controls consisted of 900 healthy controls and 600 breast cancer controls.
Whole-genome sequencing
Sample DNA was extracted from whole blood using standard methods and sequenced at Complete Genomics (Mountain View, CA, USA) using unchained combinatorial probe anchor ligation chemistry on self-assembling DNA nanoballs (DNBs) (43). Raw reads were aligned to the reference genome (National Center for Biotechnology Information (NCBI) build 36). Mapped reads were assembled using their CGA Tool. Variants were called and scored using a local de novo assembly approach. Sequencing alignment and variant calling were also performed at Complete Genomics.
Bio-informatic analysis
Variants annotation
All variants from each genome were annotated independently using Annovar Tool. Coding variants were selected for further investigation. Coding variants include missense variants, nonsense variants and frameshift variants (insertions or deletions).
Family-based elimination approach
The analysis was performed under the assumption that the causative mutation should be shared by the three affected subjects (II:3, III:1 and III:2) but not present in the two unaffected subjects (II:2 and III:3). This approach was applied to all the coding variants.
Novel variants selection
To eliminate common germline polymorphisms from consideration, variants reported in dbSNP130 or in 1000 genome project 2010 July release were removed. Variants also identified in nine in-house Complete Genomics whole-genome sequenced germline DNA samples from gynecological cancer patients of Flemish origin were considered population-specific variants. They were removed as well from consideration. Only the variants not previously identified were considered putative candidate causative mutations.
Detection of the shared haplotype around the I121V variant in SPAG8
To determine this haplotype, we analyzed SNPs on chromosome 9. We first filtered out SNPs which were identical in both parents or for which neither of the parents were heterozygous. Using this list of unique heterozygous SNPs in the parents, the parental origin for each of these SNPs in the siblings can be determined. An analysis of consecutive SNPs in both parents and the three siblings was used to make an estimation of the haplotype blocks.
Detection of short tandem repeats in shared haplotype
Short tandem repeats are regions in the genome of minimum six consecutive bases built up by at least two identical blocks of 2–6bp. To locate all short tandem repeats in the haplotype block, we used a tool called ‘grepseq’ (http://code.google.com/p/grepseq/). This tool scans fasta-files in search for positions in the genome that match a given pattern (e.g. ‘short tandem repeat'). The coverage depth was analyzed in and around short tandem repeats of at least two consecutive repeats of 4–6bp. The coverage depth is a number provided by Complete Genomics that indicates the amount of times a given base has been sequenced by different reads. After correcting the coverage depth for the GC bias using a script from Complete Genomics (UCSC GC-track), we searched for short tandem repeats with unexpected coverage depth.
Genotyping
Variants identified by whole-genome sequencing were confirmed using a Sequenome MassAssay system (Sequenom, San Diego, CA, USA).
Variants that failed genotyping using Sequenom assays were typed in family and unrelated controls using bidirection dideoxy sequencing of amplified products or Taqman genotyping assays-by-design run on a 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions.
The hexanucleotide repeat region in intron 1 of C9orf72 was typed by PCR and repeat-primed PCR as previously described (31,32). The length of fragments generated by repeat-primed PCR was determined after running on an ABI3130xl sequencer, using GeneMapper software version 4.0 (Applied Biosystems).
SUPPLEMENTARY MATERIAL
FUNDING
S.H. and M.M. hold a PhD-fellowship and P.V.D. holds a clinical investigatorship of FWO-Vlaanderen. A.G. is supported by the Charcot Foundation and Onderzoeksfonds K.U.Leuven/Research Fund K.U.Leuven (OT/11/087). W.R. is supported through the E. von Behring Chair for Neuromuscular and Neurodegenerative Disorders. This work is supported by the VIB Tech Watch Fund, the Health Seventh Framework Programme (FP7/2007-2013, grant agreement no. 259867), by the Interuniversity Attraction Poles (IUAP) programme P6/43 of the Belgian Federal Science Policy Office and by the university of Leuven (GOA 11/014 and Methusalem).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients and their family members who participated in this study.
Conflict of Interest statement. None declared.
References
- 1.Rowland L.P., Shneider N.A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 2.Phukan J., Pender N.P., Hardiman O. Cognitive impairment in amyotrophic lateral sclerosis. Lancet. Neurol. 2007;6:994–1003. doi: 10.1016/S1474-4422(07)70265-X. [DOI] [PubMed] [Google Scholar]
- 3.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 4.Dion P.A., Daoud H., Rouleau G.A. Genetics of motor neuron disorders: new insights into pathogenic mechanisms. Nat. Rev. Genet. 2009;10:769–782. doi: 10.1038/nrg2680. [DOI] [PubMed] [Google Scholar]
- 5.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J.P., Deng H.X., et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 6.Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Vande Velde C., Bouchard J.P., Lacomblez L., Pochigaeva K., Salachas F., et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 7.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwiatkowski T.J, Jr, Bosco D.A., Leclerc A.L., Tamrazian E., Vanderburg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 9.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenway M.J., Andersen P.M., Russ C., Ennis S., Cashman S., Donaghy C., Patterson V., Swingler R., Kieran D., Prehn J., et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat. Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura A.L., Mitne-Neto M., Silva H.C., Richieri-Costa A., Middleton S., Cascio D., Kok F., Oliveira J.R., Gillingwater T., Webb J., et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004;75:822–831. doi: 10.1086/425287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maruyama H., Morino H., Ito H., Izumi Y., Kato H., Watanabe Y., Kinoshita Y., Kamada M., Nodera H., Suzuki H., et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 13.Johnson J.O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V.M., Trojanowski J.Q., Gibbs J.R., Brunetti M., Gronka S., Wuu J., et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hand C.K., Khoris J., Salachas F., Gros-Louis F., Lopes A.A., Mayeux-Portas V., Brewer C.G., Brown R.H, Jr., Meininger V., Camu W., et al. A novel locus for familial amyotrophic lateral sclerosis, on chromosome 18q. Am. J. Hum. Genet. 2002;70:251–256. doi: 10.1086/337945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sapp P.C., Hosler B.A., McKenna-Yasek D., Chin W., Gann A., Genise H., Gorenstein J., Huang M., Sailer W., Scheffler M., et al. Identification of two novel loci for dominantly inherited familial amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2003;73:397–403. doi: 10.1086/377158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosler B.A., Siddique T., Sapp P.C., Sailor W., Huang M.C., Hossain A., Daube J.R., Nance M., Fan C., Kaplan J., et al. Linkage of familial amyotrophic lateral sclerosis with frontotemporal dementia to chromosome 9q21-q22. JAMA. 2000;284:1664–1669. doi: 10.1001/jama.284.13.1664. [DOI] [PubMed] [Google Scholar]
- 17.Morita M., Al-Chalabi A., Andersen P.M., Hosler B., Sapp P., Englund E., Mitchell J.E., Habgood J.J., de Belleroche J., Xi J., et al. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66:839–844. doi: 10.1212/01.wnl.0000200048.53766.b4. [DOI] [PubMed] [Google Scholar]
- 18.Valdmanis P.N., Dupre N., Bouchard J.P., Camu W., Salachas F., Meininger V., Strong M., Rouleau G.A. Three families with amyotrophic lateral sclerosis and frontotemporal dementia with evidence of linkage to chromosome 9p. Arch. Neurol. 2007;64:240–245. doi: 10.1001/archneur.64.2.240. [DOI] [PubMed] [Google Scholar]
- 19.Vance C., Al-Chalabi A., Ruddy D., Smith B.N., Hu X., Sreedharan J., Siddique T., Schelhaas H.J., Kusters B., Troost D., et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2–21.3. Brain. 2006;129:868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- 20.Boxer A.L., Mackenzie I.R., Boeve B.F., Baker M., Seeley W.W., Crook R., Feldman H., Hsiung G.Y., Rutherford N., Laluz V., et al. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. J. Neurol. Neurosurg. Psychiatry. 2011;82:196–203. doi: 10.1136/jnnp.2009.204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson J.P., Williams N.M., Majounie E., Waite A., Stott J., Newsway V., Murray A., Hernandez D., Guerreiro R., Singleton A.B., et al. Familial frontotemporal dementia with amyotrophic lateral sclerosis and a shared haplotype on chromosome 9p. J. Neurol. 2011;258:647–655. doi: 10.1007/s00415-010-5815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rollinson S., Mead S., Snowden J., Richardson A., Rohrer J., Halliwell N., Usher S., Neary D., Mann D., Hardy J., et al. Frontotemporal lobar degeneration genome wide association study replication confirms a risk locus shared with amyotrophic lateral sclerosis. Neurobiol. Aging. 2011;32:758 e751–758 e757. doi: 10.1016/j.neurobiolaging.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 23.Le Ber I., Camuzat A., Berger E., Hannequin D., Laquerriere A., Golfier V., Seilhean D., Viennet G., Couratier P., Verpillat P., et al. Chromosome 9p-linked families with frontotemporal dementia associated with motor neuron disease. Neurology. 2009;72:1669–1676. doi: 10.1212/WNL.0b013e3181a55f1c. [DOI] [PubMed] [Google Scholar]
- 24.Shatunov A., Mok K., Newhouse S., Weale M.E., Smith B., Vance C., Johnson L., Veldink J.H., van Es M.A., van den Berg L.H., et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet. Neurol. 2010;9:986–994. doi: 10.1016/S1474-4422(10)70197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gijselinck I., Engelborghs S., Maes G., Cuijt I., Peeters K., Mattheijssens M., Joris G., Cras P., Martin J.J., De Deyn P.P., et al. Identification of 2 Loci at chromosomes 9 and 14 in a multiplex family with frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Arch. Neurol. 2010;67:606–616. doi: 10.1001/archneurol.2010.82. [DOI] [PubMed] [Google Scholar]
- 26.Luty A.A., Kwok J.B., Thompson E.M., Blumbergs P., Brooks W.S., Loy C.T., Dobson-Stone C., Panegyres P.K., Hecker J., Nicholson G.A., et al. Pedigree with frontotemporal lobar degeneration—motor neuron disease and Tar DNA binding protein-43 positive neuropathology: genetic linkage to chromosome 9. BMC Neurol. 2008;8:32. doi: 10.1186/1471-2377-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemmens R., Race V., Hersmus N., Matthijs G., Van Den Bosch L., Van Damme P., Dubois B., Boonen S., Goris A., Robberecht W. TDP-43 M311V mutation in familial amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry. 2009;80:354–355. doi: 10.1136/jnnp.2008.157677. [DOI] [PubMed] [Google Scholar]
- 28.Van Damme P., Goris A., Race V., Hersmus N., Dubois B., Bosch L.V., Matthijs G., Robberecht W. The occurrence of mutations in FUS in a Belgian cohort of patients with familial ALS. Eur. J. Neurol. 2010;17:754–756. doi: 10.1111/j.1468-1331.2009.02859.x. [DOI] [PubMed] [Google Scholar]
- 29.Van Damme P., Veldink J.H., van Blitterswijk M., Corveleyn A., van Vught P.W., Thijs V., Dubois B., Matthijs G., van den Berg L.H., Robberecht W. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 2011;76:2066–2072. doi: 10.1212/WNL.0b013e31821f445b. [DOI] [PubMed] [Google Scholar]
- 30.Reumers J., De Rijk P., Zhao H., Liekens A., Smeets D., Cleary J., Van Loo P., Van Den Bossche M., Catthoor K., Sabbe B., et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. Nat. Biotechnol. 2011;30:61–68. doi: 10.1038/nbt.2053. [DOI] [PubMed] [Google Scholar]
- 31.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renton A.E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gijselinck I., Van Langenhove T., van der Zee J., Sleegers K., Philtjens S., Kleinberger G., Janssens J., Bettens K., Van Cauwenberghe C., Pereson S., et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet. Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 34.Tsuji S. Genetics of neurodegenerative diseases: insights from high-throughput resequencing. Hum. Mol. Genet. 2010;19:R65–R70. doi: 10.1093/hmg/ddq162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimprich A., Benet-Pages A., Struhal W., Graf E., Eck S.H., Offman M.N., Haubenberger D., Spielberger S., Schulte E.C., Lichtner P., et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am. J. Hum. Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vilarino-Guell C., Wider C., Ross O.A., Dachsel J.C., Kachergus J.M., Lincoln S.J., Soto-Ortolaza A.I., Cobb S.A., Wilhoite G.J., Bacon J.A., et al. VPS35 mutations in Parkinson disease. Am. J. Hum. Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein C.J., Botuyan M.V., Wu Y., Ward C.J., Nicholson G.A., Hammans S., Hojo K., Yamanishi H., Karpf A.R., Wallace D.C., et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat. Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noskova L., Stranecky V., Hartmannova H., Pristoupilova A., Baresova V., Ivanek R., Hulkova H., Jahnova H., van der Zee J., Staropoli J.F., et al. Mutations in DNAJC5, encoding cysteine-string protein alpha, cause autosomal-dominant adult-onset neuronal ceroid lipofuscinosis. Am. J. Hum. Genet. 2011;89:241–252. doi: 10.1016/j.ajhg.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rios J., Stein E., Shendure J., Hobbs H.H., Cohen J.C. Identification by whole-genome resequencing of gene defect responsible for severe hypercholesterolemia. Hum. Mol. Genet. 2010;19:4313–4318. doi: 10.1093/hmg/ddq352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lupski J.R., Reid J.G., Gonzaga-Jauregui C., Rio Deiros D., Chen D.C., Nazareth L., Bainbridge M., Dinh H., Jing C., Wheeler D.A., et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N. Engl. J. Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooks B.R., Miller R.G., Swash M., Munsat T.L. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral. Scler. Other Motor Neuron Disord. 2000;1:293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 42.Schrooten M., Smetcoren C., Robberecht W., Van Damme P. Benefit of the Awaji diagnostic algorithm for amyotrophic lateral sclerosis: a prospective study. Ann. Neurol. 2011;70:79–83. doi: 10.1002/ana.22380. [DOI] [PubMed] [Google Scholar]
- 43.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G., et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.