Abstract
Neddylation is a posttranslational modification that plays important roles in regulating protein structure and function by covalently conjugating NEDD8, an ubiquitin-like small molecule, to the substrate. Here, we report that Parkinson's disease (PD)-related parkin and PINK1 are NEDD8 conjugated. Neddylation of parkin and PINK1 results in increased E3 ligase activity of parkin and selective stabilization of the 55 kDa PINK1 fragment. Expression of dAPP-BP1, a NEDD8 activation enzyme subunit, in Drosophila suppresses abnormalities induced by dPINK1 RNAi. PD neurotoxin MPP+ inhibits neddylation of both parkin and PINK1. NEDD8 immunoreactivity is associated with Lewy bodies in midbrain dopaminergic neurons of PD patients. Together, these results suggest that parkin and PINK1 are regulated by neddylation and that impaired NEDD8 modification of these proteins likely contributes to PD pathogenesis.
INTRODUCTION
Parkinson's disease (PD) is the most frequent neurodegenerative movement disorder affecting ∼1% of the population over age 65 (1). Most PD cases are sporadic. Mutations in several genes, however, are associated with the familial form of PD. These genes include SNCA (2), parkin (3), UCHL1 (4), PINK1 (5), DJ-1 (6), LRRK2 (7,8), ATP13A2 (9), GIGYF2 (10), Omi/HTRA2 (11), PLA2G6 (12) and FBXO7 (13). Recent linkage analysis and genome-wide association studies have identified additional genetic loci as PD risk factors, such as MAPT, BST1, GAK and HLA-DRB5 (1,14). Understanding the pathophysiological functions of these genes will help define PD etiology and design novel strategies for early diagnosis and treatment. Among these proteins, parkin is a RING/HECK hybrid ubiquitin E3 ligase with an ubiquitin-like domain at its N-terminus and a RING-IBR-RING domain at its C-terminus (3,15). It mediates ubiquitination and degradation of multiple proteins (16). In addition, parkin has an important role in protecting neurons against various insults and maintaining mitochondrial integrity (17–24). Recent studies suggest that parkin also functions as a tumor suppressor in multiple cancers (25,26). PINK1 is a putative kinase with an N-terminal mitochondrial targeting signal (5). In addition to the full-length protein, a 55 kDa PINK1 fragment with a truncated N-terminal segment is detected in the cytosol, suggesting the presence of cellular PINK1 processing and multi-compartmental functions for PINK1 (5,16). Several potential substrates of PINK1 have been identified despite the unclear biological consequence of PINK1-mediated protein phosphorylation (27–29). PINK1 functions in common pathways with parkin to maintain mitochondrial integrity, quality control and transport (17,21,24,30). Moreover, parkin, PINK1 and DJ-1 form an E3 ligase complex (the PPD complex) to promote the degradation of mis/unfolded proteins (16). Nevertheless, the cellular mechanism for the regulation of parkin and PINK1 is largely unknown.
NEDD8 is an ubiquitin-like small molecule that is covalently conjugated to proteins to regulate protein functions. Cullins are the first protein groups known to be NEDD8 conjugated. The NEDD8 conjugation of cullins activates the cullin-ring ligase complex. The conjugation process, also known as neddylation, is catalyzed by an enzymatic pathway similar to ubiquitination with distinct enzymes, such as APP-BP1/Uba3 heterodimer (E1), Ubc12 or UbeF2 (E2), and Dcn1 or Dcn1-like proteins (E3) (31–33). Several ubiquitin E3 ligases such as c-Cbl, mdm2 and mammalian IAPs can act as NEDD8 E3 ligases (34–36), suggesting the presence of diverse NEDD8 substrates besides cullin proteins and a potential crosstalk between ubiquitination and neddylation. An increasing number of non-cullin proteins is found to be neddylation regulated, including transcriptional factor p53 and BCA3 (36,37). Neddylation of the epidermal growth factor receptor (EGFR) and some ribosomal proteins modulates the stability of these proteins (35,38). In PD, specific immunoreactivity to NEDD8 is detected in Lewy bodies, suggesting that protein neddylation is involved in PD pathogenesis (39,40). Little is known, however, about the molecular role of neddylation in PD development.
In the present study, we identified that PD-associated parkin and PINK1 are NEDD8 modified. Neddylation results in increased parkin E3 ligase activity and stabilization of PINK1 55 kDa fragment. Expression of APP-BP1 in Drosophila suppresses dPINK1 RNAi-induced ommatidial degeneration, abnormal wing phenotype and male sterility.
RESULTS
Neddylation of parkin and PINK1
To study regulation of the parkin/PINK1/DJ-1 E3 ligase complex (16), we expressed FLAG-tagged PINK1 in SH-SY5Y cells, immunoprecipitated exogenous PINK1 and analyzed PINK1 interactome with mass spectrometry. The resulting PINK1 interactors include several essential components for NEDD8 conjugation: NEDD8, APP-BP1, UBC12 and COP9 signalosome proteins (Table 1). This result suggests that PINK1 or its interacting proteins are NEDD8 modified. To examine neddylation of parkin and PINK1 in the cell, we expressed parkin and PINK1 in HEK293 cells individually with NEDD8. Immunoprecipitation followed by immunoblotting for NEDD8 showed that immunoprecipitated parkin produced NEDD8-positive smear bands above the molecular weight of parkin (Fig. 1A). Likewise, PINK1 immunoprecipitation resulted in a NEDD8-positive smear band above the molecular weight of PINK1 (Fig. 1D). In contrast, in the absence of NEDD8, the NEDD8-positive band was not detected with either parkin or PINK1 immunoprecipitation (Fig. 1A and D). In addition, immunoprecipitation of α-synuclein in the presence of NEDD8 did not result in the NEDD8-positive band (Fig. 1A), demonstrating the specificity of NEDD8 conjugation of parkin and PINK1.
Table 1.
Interaction of PINK1 with neddylation machinery
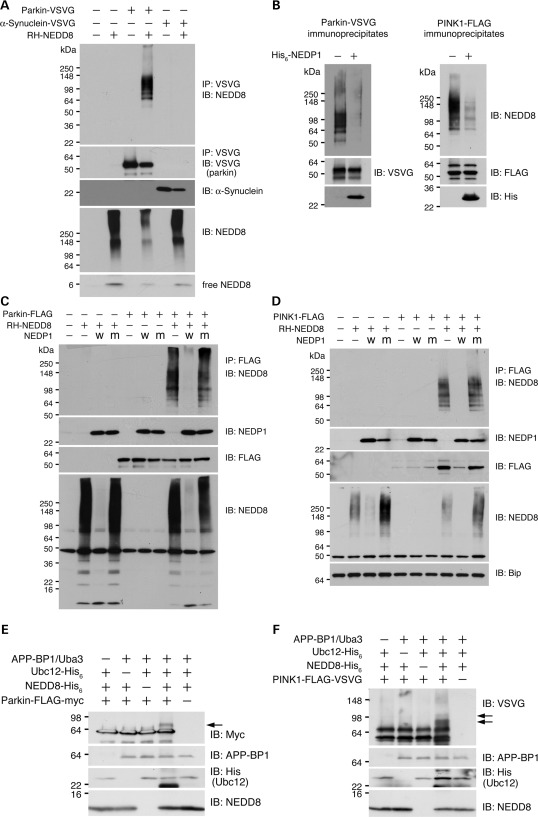
Figure 1.
Neddylation of parkin and PINK1. (A) Neddylation of parkin in transfected cells. Cells were transfected with control empty vector, parkin-VSVG, α-synuclein-VSVG with/without NEDD8 as shown on top of the panel. Parkin and α-synuclein were immunoprecipitated (IP) followed by immunoblotting (IB) with an anti-NEDD8 antibody (top panel). Immunoprecipitated parkin, α-synuclein and expression of NEDD8 were shown. For parkin-expressing cells, half of the protein amount was used for immunoprecipitation and immunoblotting compared with the other cells. (B) In vitro deneddylation by NEDP1. Immunoprecipitated parkin (left panel) and PINK1 (right panel) were treated with recombinant NEDP1 followed by detection of NEDD8 (top panel), parkin or PINK1 (middle panel), and NEDP1 (bottom panel). (C) In vivo deneddylation of parkin by NEDP1. Cells were cotransfected wild-type (W) or mutant (M) NEDP1 with parkin. Parkin was immunoprecipitated followed by detection of NEDD8 (top panel), NEDP1 (second panel to the top), parkin (third panel to the top) and total NEDD8 (bottom panel). (D) In vivo deneddylation of PINK1 by NEDP1. Cells were cotransfected wild-type (W) or mutant (M) NEDP1 with PINK1. PINK1 was immunoprecipitated followed by detection of NEDD8 (top panel), NEDP1 (second panel to the top), PINK1 (third panel to the top) and total NEDD8 (second panel to the bottom). Bip was detected as a loading control (bottom panel). Note that NEDD8 coexpression results in accumulation of the 55 kDa PINK1 fragment (lanes 8 and 10). (E and F) In vitro neddylation of parkin and PINK1. Purified recombinant parkin (E) and PINK1 (F) were mixed with different combination of neddylation components (indicated on the top of the figure). The reaction mixtures were detected for parkin (E, top panel) or PINK1 (F, top panel), APP-BP1 (second panel to the top), Ubc12 (third panel to the top) and NEDD8 (bottom panel). Arrows indicate NEDD8-modified proteins. Note that higher molecular weight parkin or PINK1 is detected only in reaction containing parkin/PINK1 and all neddylation components.
To verify that the observed smear bands were NEDD8 modification of parkin and PINK1, we treated parkin and PINK1 immunoprecipitates with a recombinant NEDD8-specific deneddylation enzyme NEDP1 in vitro. NEDP1 treatment markedly reduced the NEDD8-immunoreactive high-molecular-weight smear bands of parkin and PINK1 (Fig. 1B). In addition, the expression of wild-type NEDP1, but not the isopeptidase-deficient NEDP1 mutant, abolished neddylation of parkin (Fig. 1C) and PINK1 (Fig. 1D). These results suggest that both parkin and PINK1 are specifically modified by NEDD8 conjugation.
Previous reports indicate that cullins are mononeddylated and appear as a single modified band. However, neddylated parkin and PINK1 appear as smear bands, indicating hyperneddylation. To exclude the possible contribution of polyubiquitination to the observed smear bands, we treated parkin and PINK1 immunoprecipitates with a recombinant ubiquitin-specific deubiquitinating enzyme Usp2 that non-specifically cleaves all ubiquitin conjugates to ubiquitin monomers (41,42). We observed little change to the NEDD8 smear bands associated with high-molecular-weight parkin or PINK1. The Usp2 activity was demonstrated by abolishing parkin polyubiquitination with Usp2-cc treatment. These results suggest that parkin and PINK1 are likely polyneddylated or multiple-mononeddylated.
To further confirm the neddylation of parkin and PINK1, we performed in vitro neddylation analysis using purified parkin and PINK1. NEDD8-modified parkin (Fig. 1E) and PINK1 (Fig. 1F) were detected only with the presence of the completed neddylation components. Together, the results support the notion that parkin and PINK1 are modified by NEDD8 conjugation.
Increased parkin E3 ligase activity by NEDD8 modification
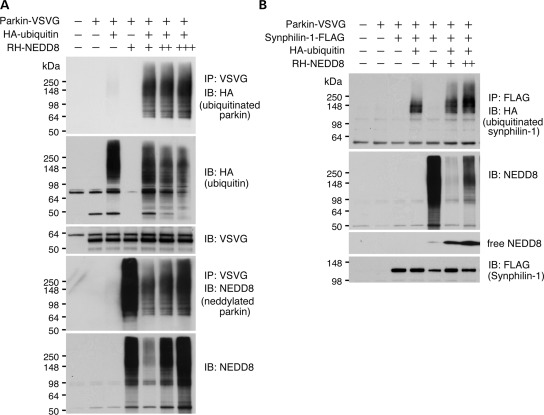
To determine the functional consequence of neddylation, we analyzed the E3 ligase activity of parkin in the presence of NEDD8. The results revealed that parkin ubiquitination was dose-dependently upregulated with increased neddylation (Fig. 2A). Likewise, ubiquitination of synphilin-1, another parkin substrate, was also upregulated with increased NEDD8 in a dose-dependent manner (Fig. 2B). This observation is likely specific to parkin substrates because the total ubquitination is inhibited with increased NEDD8. One explanation of this observation is that ubiquitination and neddylation compete for target proteins other than parkin in the cell. NEDD8 and ubiquitin are known to modify overlapping sets of lysine residues of certain substrates (35,36). The results suggest that NEDD8 modification potentiates parkin E3 ligase activity.
Figure 2.
Ectopic expression of NEDD8 increases parkin E3 ligase activity. (A) Cells expressing parkin, ubiquitin and NEDD8 in different combination shown on the top of the figure. Parkin ubiquitination (top panel), total ubiquitination (second panel to the top), parkin (third panel to the top), parkin neddylation (forth panel to the top) and total neddylation (bottom panel) were shown. Number of ‘+’ represents the quantity of NEDD8. Note that increased detection of parkin ubiquitination is seen with the increase in NEDD8. (B) Cells expressing synphilin-1, parkin, ubiquitin and NEDD8 in different combinations shown on the top of the figure. Synphilin-1 ubiquitination (top panel), total neddylation (middle panel) and expression of synphilin-1 were shown. Number of ‘+’ represents the quantity of NEDD8. Note that increased detection of synphilin-1 is seen with the increase in NEDD8.
Stabilization of the 55 kDa fragment of PINK1 by neddylation
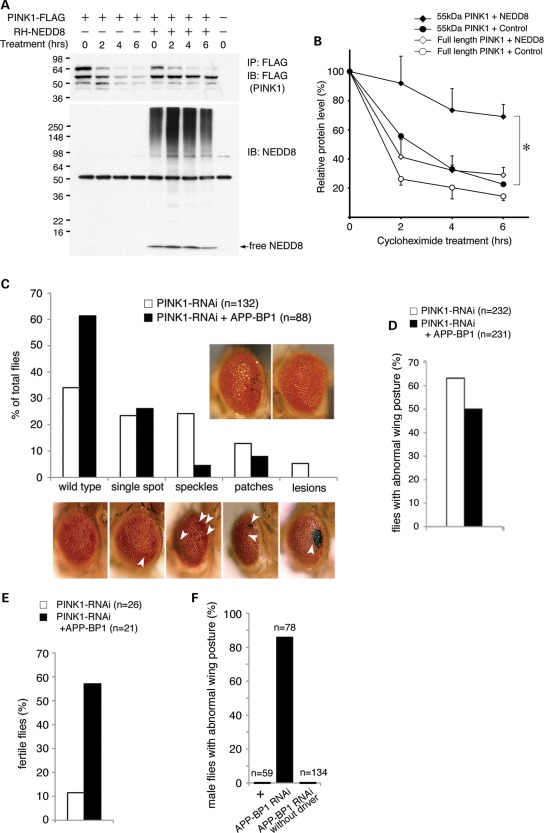
Ectopic expression of NEDD8 regulates the stability of EGFR and ribosomal protein L11 (35,38). We next examined the effect of PINK1 neddylation. With the expression of NEDD8, the steady-state level of a 55 kDa PINK1 fragment was increased (Fig. 1D). Cycloheximide-mediated chase analyses showed that the levels of both full-length PINK1 and its 55 kDa fragment were decreased rapidly with a half-life of less than 2 h (Fig. 3A and B). With the expression of NEDD8, the half-life of the full-length PINK1 remained as 2 h. Nevertheless, half-life of the PINK1 55 kDa fragment was increased to more than 6 h (Fig. 3A and B). The results indicate that PINK1 neddylation selectively increases the stability of the PINK1 55 kDa fragment.
Figure 3.
Enhanced neddylation stabilizes the 55 kDa PINK1 cleavage product in cells and rescues dPINK1-RNAi-induced abnormalities in Drosophila. (A and B) Stabilization of the 55 kDa PINK1 cleavage product by NEDD8. (A) Representative immunoblotting results for PINK1 and NEDD8 upon cycloheximide treatment. Duration of treatment is shown on the top of the figure. (B) Quantification of PINK1 and its cleavage product after cycloheximide treatment. The results are from two independent experiments. Values represent mean ± standard error. 55 kDa PINK1: the 55 kDa PINK1 fragment; Control: an empty vector. *P=0.0314 at 6 h treatment time point between 55 kDa PINK1 + NEDD8 and 55 kDa PINK1 + Control by an unpaired, two-tailed t-test. (C) dAPP-BP1 overexpression rescues dPINK1-RNAi-induced ommatidial degeneration. Quantification of flies in different eye phenotypic categories. The observed eye phenotypes are categorized into different phenotypic groups ranging in levels of severity shown at the bottom: including wild-type < single spot (least severe) < speckles < patches < lesion (most severe). Arrowheads indicate affected ommatidia of the eye. Representative external eye phenotypes of dPINK1-RNAi flies without (left) and with dAPP-BP1 overexpression (right) are inserted into the upper right of the graph. Note that the Chi-square test indicates significantly different eye phenotype distribution (P< 0.0001) between two groups of flies. (D) dAPP-BP1 overexpression rescues dPINK1-RNAi-induced abnormal wing posture. Note that the Chi-square test indicates significantly different wing phenotype distribution (P= 0.0058) between two groups of flies. (E) dAPP-BP1 overexpression rescues tubulin-GAL4 driven, dPINK1-RNAi-induced male sterility. Note that the Chi-square test indicates significant improved male fertility (P=0.0009) with coexpression of dAPP-BP1. (F) APP-BP1 knockdown results in the droopy wing phenotype. ‘+’ and ‘APP-BP1 RNAi’ indicate how24B-GAL4>dcr-2/+ flies and how24B-GAL4>dcr-2, dAPP-BP1 RNAi flies, respectively. ‘APP-BP1 RNAi without driver’ indicates UAS-dAPP-BP1 RNAi flies expressing no GAL4 driver.
Expression of neddylation E1 component dAPP-BP1 in Drosophila suppresses abnormalities induced by dPINK1 reduction
The NEDD8 system is conserved across species including Drosophila. Previously, we reported that inhibition of Drosophila PINK1 (dPINK1) via RNAi in Drosophila eyes, using GMR-GAL4 driver induces ommatidial degeneration resulting in black speckles, patches or lesions in eyes (Fig. 3C, bottom) (43). We next examined whether the dPINK1-RNAi-induced eye phenotypes are affected by neddylation. Expression of Drosophila APP-BP1 (dAPP-BP1), the NEDD8 activation enzyme E1 regulatory subunit, shows little effect on external eyes of wild-type flies. Nevertheless, expression of dAPP-BP1 in dPINK1-RNAi flies significantly suppressed eye phenotypes, including speckles, patches or lesions (Chi-square test, P< 0.0001 between dPINK1-RNAi+dAPP-BP1 and dPINK1-RNAi flies) (Fig. 3C). Stabilization of dPINK1 expected from dAPP-BP1 overexpression may compensate the dPINK1 reduction.
We further examined the effect of dAPP-BP1 on another dPINK1-RNAi fly line showing abnormal wing phenotypes (44). At 7 days of age, 63% of the dPINK1-RNAi flies with muscle-specific knockdown using Mhc-GAL4 (n= 232) displayed a drooped wing posture (Fig. 3D). With dAPP-BP1 overexpression, only 50% of dPINK1-RNAi flies (n= 231) showed the drooped wing phenotypes (Fig. 3D). Therefore, overexpression of dAPP-BP1 significantly inhibits the abnormal wing phenotype induced by dPINK1 RNAi (P=0.0058, Chi-square test). It is well documented that flies with either PINK1 null alleles or dPINK1 knockdown show reduced male fertility (17,22,45). Overexpression of dAPP-BP1 under tubulin-GAL4 driver greatly improves the male fertility of dPINK1-RNAi flies from 12 to 57% (Chi-square test, P= 0.0009, Fig. 3E).
Consistent with the results, knockdown of dAPP-BP1 using a different mesoderm-specific driver, how24B-GAL4>dcr-2, results in droopy wing phenotype in 86% of males (n=78, Fig. 3F). No wing phenotype was observed in control male flies with genotypes of how24B-GAL4>dcr-2 (0%, n=59) or dAPP-BP1-RNAi (0%, n=134). The results provide additional evidence to support regulation of PINK1 by neddylation.
Together, results from two independent dPINK1-RNAi fly lines suggest that Drosophila PINK1 function is likely regulated by protein neddylation.
Detection of parkin neddylation in human brain
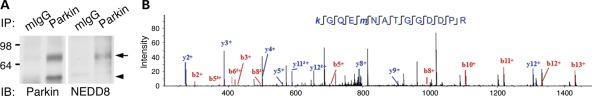
Next, we analyzed the neddylation of endogenous parkin in human brain tissues. Endogenous parkin was immunoprecipitated from the brain lysates of PD patients (Fig. 4A). Bands that were immunopositive both for parkin and NEDD8 were excised and subjected to analysis with mass spectrometry. The results identified a 13-amino acid-length parkin fragment with lysine 76 residue glycine–glycine modified (Fig. 4B). We were unable to analyze neddylation of PINK1 because anti-PINK1 antibody for immunoprecipitation is not available. The result provides supporting evidence of neddylation of endogenous parkin in human brain.
Figure 4.
Neddylation of endogenous parkin in human brain. (A) Parkin is immunopurified from human brain lysates. The purified parkin was verified by immunoblotting with parkin (IB: Parkin) and NEDD8 (IB: NEDD8) specific antibodies. On the top of the figure, mIgG indicates immobilized control mouse IgG, while Parkin indicates immobilized anti-parkin antibody. Arrowhead and arrow indicate unmodified and modified parkin, respectively. (B) The sequence and tandem mass spectrum of a neddylated peptide derived from parkin. K76 in small letter (k) is Gly–Gly modified.
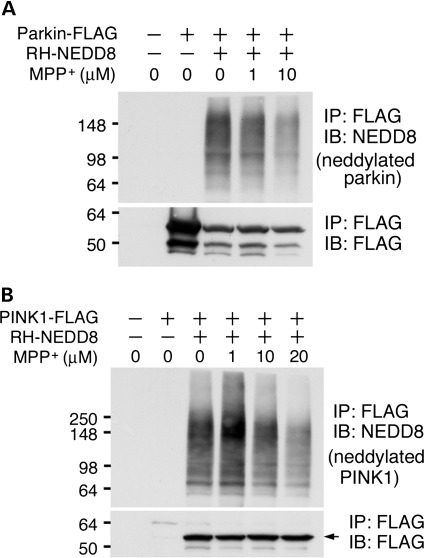
Regulation of parkin and PINK1 neddylation by MPP+ treatment
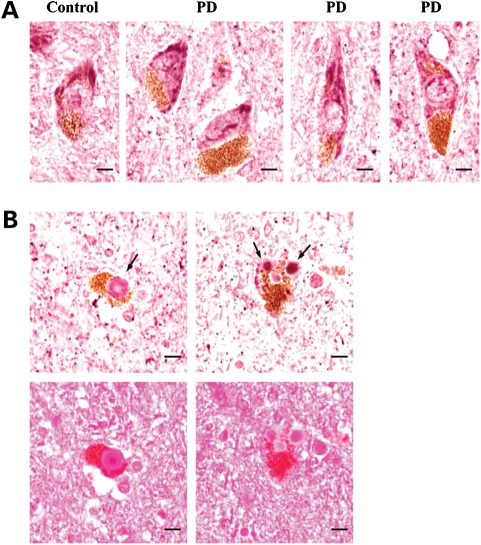
We also determined whether neddylation of parkin or PINK1 is regulated by PD-related neurotoxin. HEK293 cells were co-transfected NEDD8 with either parkin or PINK1 followed by 1-methyl-4-phenylpyridinium (MPP+) treatment, a PD-relevant toxic cation, at sub-lethal concentrations. The levels of both neddylated parkin and PINK1 were obviously reduced starting at 10 µm with dose dependence (Fig. 5). The results suggest that treatment of PD-related MPP+ alters the neddylation of parkin and PINK1. We further examined the distribution of NEDD8 in midbrain dopaminergic neurons of PD patients and their sex- and age-matched normal individuals using an anti-NEDD8 antibody. The NEDD8 immunoreactivity was visualized in cytoplasmic compartment in both PD and controls (Fig. 6A). NEDD8-immunopositive Lewy bodies in the dopaminergic neurons from the PD patient substantia nigra were also identified (Fig. 6B). Thus, NEDD8 or NEDD8-modified proteins are associated with PD pathology.
Figure 5.
MPP+ suppresses neddylation of parkin and PINK1. Cells transfected with the combination of parkin (A) or PINK1 (B) and NEDD8 (shown on top of the figures) were incubated with MPP+ at various concentrations (shown on top of the figures) for 18 h. Arrow indicates the 55 kDa PINK1 fragments. Note that neddylation of both parkin and PINK1 is suppressed by MPP+ at 10 µm.
Figure 6.
Detection of NEDD8-immunopositive Lewy bodies. (A) NEDD8 immunoreactivity was detected in the pigmented dopaminergic neurons in control and PD substantia nigra. The pictures are representative NEDD8 staining images from a control (male, 76years) and a PD patient (male, 78years). In total, four pairs of control and PD cases were examined. (B) Lewy bodies in pigmented dopaminergic neurons were stained with an anti-NEDD8 antibody (top panel) and eosin (bottom panel). Lewy bodies are indicated by arrows. Scale bar, 10 µm.
DISCUSSION
Neddylation is an important posttranslational modification affecting protein structure and function (46,47). In this study, we demonstrate that two PD-related proteins, parkin and PINK1, are neddylated. Neddylation modification results in increased ubiquitin E3 ligase activity of parkin and stabilization of a 55 kDa PINK1 proteolytic fragment. In vivo, enhanced neddylation by dAPP-BP1 overexpression rescues abnormal phenotypes induced by dPINK1 reduction in two independent dPINK1-RNAi Drosophila lines. NEDD8 or its conjugates are associated with the PD pathological hallmark Lewy bodies. PD-related neurotoxin MPP+ treatment inhibits neddylation of both parkin and PINK1. These findings identify a new posttranslational modification of parkin and PINK1 and suggest that neddylation abnormalities likely contribute to PD pathogenesis.
Several lines of in vitro and in vivo evidence indicate NEDD8 modification of parkin and PINK1. Parkin and PINK1 are specifically NEDD8 conjugated in transfected cells and in an in vitro neddylation assay using recombinant proteins. The neddylation of parkin and PINK1 is sensitive to a deneddylation enzyme but not a deubiquitination enzyme. In vivo, endogenous parkin purified from human brain is found modified at K76. Unlike ubiquitination, neddylation traditionally refers to conjugating a single NEDD8 molecule to a target protein. Nevertheless, recent studies identify NEDD8-immunopositive high-molecular-weight smear bands similar to hyperubiquitination with non-cullin targets, such as EGFR (35). Our results suggest that parkin and PINK1 are likely hyperneddylated based on the high-molecular-weight smears. Deletion experiments suggest that both parkin and PINK1 are neddylated at multiple sites (data not shown). Nonetheless, the exact sites and types (polyneddylation versus multiple mononeddylation) of NEDD8 modification on parkin and PINK1 remain to be determined.
The best-studied posttranslational NEDD8 modification is the neddylation of cullins. Cullins are a component of the SCF ubiquitin E3 ligase complex. Neddylation of cullins plays an essential role in regulating the E3 ligase activity of the SCF complex (48). Our results suggest that the neddylation of parkin potentiates its E3 ligase activity based on the observation of the increased ubiquitination of two parkin substrates, parkin and synphilin-1, in cells transfected with NEDD8. One of the biological consequences of neddylation is to alter the stability of target proteins. The ribosomal protein L11 becomes stabilized after NEDD8 modification, whereas Cul1 or Cul5 and EGFR were destabilized with NEDD8 modification (35,38,49). Consistent with the previous reports, PINK1 neddylation results in selected stabilization of a 55 kDa PINK1 fragment, leading to its accumulation in the cell. Interestingly, the 55 kDa PINK1 fragment is mainly detected in the cytosolic fraction and forms a complex with parkin (16). In contrast, the full-length PINK1 is mainly detected and likely functions in the mitochondrial fraction. Therefore, the sum of both parkin and PINK1 neddylation is to regulate the E3 ligase activity of the parkin-PINK1-DJ-1 complex. Regulation of PINK1 function by neddylation is further supported by an observation that neddylation E1 dAPP-BP1 subunit rescues the ommatidial degeneration, abnormal wing posture and male sterility induced by dPINK1 reduction. Moreover, knockdown of dAPP-BP1 results in the droopy wings resembling those found in flies with either parkin or PINK1 knockdown. These results are further supported by the findings from a recent genome-wide study identifying the lethal interaction of heterozygous deletion of a cytological region Df(3L)vin5 with both parkin knockdown and dPINK1 knockdown (45). The cytological region of Df(3L)vin5 contains dAPP-BP1. These findings suggest that neddylation regulates parkin and PINK1 in vivo.
Mutations of parkin and PINK1 are associated with the recessive familial form of PD. The pathogenic mutants of parkin and PINK1 have been shown to reduce the ubiquitin E3 ligase activity of parkin. The parkin and PINK1 complex promotes degradation of unfolded proteins via ubiquitin proteasomal pathway (16). Neddylation of parkin and PINK1 increases the ubiquitin E3 ligase activity of parkin. Impairment of neddylation likely results in reduced E3 ligase activity of the complex and leads to accumulation of mis/unfolded proteins in the cells. Consistent with this hypothesis, PD neurotoxin MPP+ treatment at sub-lethal concentrations results in reduced neddylation of both parkin and PINK1. Moreover, Lewy bodies are NEDD8 positive. These findings suggest that NEDD8 modification of parkin and PINK1 are involved in PD pathogenesis. The study also opens a new avenue to design potential treatment of PD via modulating neddylation of parkin and PINK1. Parkin and PINK1 play important roles in regulating the structure and function of mitochondria, including maintaining integrity, dynamics, quality control and transport (17,18,21,22,24). Further studies to address the effects NEDD8 modification of parkin and PINK1 on mitochondria are clearly warranted.
MATERIALS AND METHODS
Materials
SH-SY5Y and HEK293 cells were purchased from ATCC and maintained as suggested. Human tissues were obtained from the NICHD Brain and Tissue Band for Developmental Disorders at the University of Maryland, Baltimore, MD, USA. Plasmids encoding FLAG-tagged, VSVG-tagged, myc-tagged parkin or PINK1 and HA-tagged ubiquitin were described previously (16). cDNAs encoding RH-NEDD8, FLAG-tagged synphilin-1 and NEDP1 were generated by polymerase chain reaction and subcloned into pcDNA3.1(-) (Invitrogen). Mutant NEDP1 was generated by site-directed mutagenesis (Stratagene) using mutagenic oligonucleotides as described previously (50). All plasmids are confirmed by sequencing. Rabbit monoclonal anti-NEDD8 antibody, rabbit polyclonal anti-His tag antibody, rabbit monoclonal anti-myc antibody and rabbit monoclonal anti-Bip antibody were from Cell Signaling Technology. Rabbit polyclonal anti-VSVG antibody was from QED Bioscience, Inc. Sheep polyclonal anti-NEDP1 antibody and goat polyclonal anti-APP-BP1 antibody were from Biomol International and Santa Cruz Biotechnology, Inc., respectively. All other primary antibodies were from Sigma-Aldrich. The secondary antibodies were from Jackson ImmunoResearch Laboratories. 1-methyl-4-phenyl-pyridinium iodide is from Sigma-Aldrich.
Protein neddylation or ubiquitination in transfected cells
Neddylation or ubiquitination was analyzed as previously described (16). Briefly, HEK293 cells were transfected with various plasmid combinations using calcium phosphate. Cells were lysed in 2% SDS buffer (2% SDS, 150 mm NaCl, 10 mm Tris–HCl, pH 8.0, 2 mm sodium orthovanadate, 5 mm sodium fluoride, 1× protease inhibitors) and boiled for 10 min followed by sonication. Lysates were diluted 1:10 in dilution buffer (10 mm Tris–HCl, pH 8.0, 150 mm NaCl, 2 mm EDTA, 1% Triton X-100), incubated at 4°C for 1 h with rotation and centrifuged at 20 800g for 30 min. The protein concentration of the resulting supernatants was determined by a modified Lowry assay (Dc Protein Assay, BioRad) and 500 μg of protein was used for immunoprecipitation unless otherwise stated. Immunoprecipitated proteins were washed with washing buffer (10 mm Tris–HCl, pH 8.0, 1 m NaCl, 1 mm EDTA, 1% NP-40), boiled in SDS sample buffer and separated on sodium dodecyl sulfate–polyacrylamide gel (SDS–PAGE). NEDD8, ubiquitin and precipitated proteins were immunodetected with respective antibodies.
In vitro deneddylation and neddylation assay
For the deneddylation assay, parkin-VSVG or PINK1-FLAG was coexpressed with NEDD8 in HEK293 cells. Immunoprecipitated parkin or PINK1 and recombinant NEDP1 (1 μg, Biomol) were added to 15 μl of deneddylation buffer (50 mm Tris–HCl, pH 7.5, 50 mm NaCl, 5 mm β-mercaptoethanol). Reactions proceeded overnight at 37°C and were stopped by addition of SDS–PAGE sample buffer. Proteins were separated on a 4–20% SDS–PAGE gel and immunoblotted with an anti-NEDD8 antibody. For the neddylation assay, recombinant parkin-FLAG-myc and PINK1-FLAG-VSVG were generated as described previously using a Bac-to-Bac baculovirus expressison system (Invitrogen) (16). Affinity-purified parkin or PINK1 was mixed with recombinant APP-BP1/Uba3 (0.165 μg, Biomol), His6-Ubc12 (0.5 μg, Biomol) and His6-NEDD8 (1 μg, Biomol) in 25 μl of neddylation buffer (20 mm Tris–HCl, pH 7.5, 2 mm ATP, 1 mm DTT, 5 mm MgCl2) in various combinations. E3 ligase is not necessarily required for NEDD8 conjugation in vitro (36,39). Neddylation reactions proceeded overnight at 37°C and were stopped by addition of SDS–PAGE sample buffer. Proteins were separated on a 4–20% SDS–PAGE gel and immunoblotted with respective antibodies.
Detection of neddylated parkin in human brain
Anti-parkin antibody and control mouse IgG were immobilized according to the manufacturer's protocol (Pierce, prod # 26147). Two micrograms lysates were prepared using 2% SDS buffer followed by diluting SDS as described above. Endogenous parkin was immunoprecipitated using the immobilized anti-parkin antibody. Samples were divided into two parts and resolved on 4–20% SDS–PAGE gels, separately. One part was used to detect NEDD8 and parkin, and the other part was used for silver staining and mass spectrometric analysis using a high-resolution hybrid LTQ-Orbitrap Velos (Thermo Fisher Scientific). Identification of neddylation sites was carried out with differential modification of lysine residues (GG, +114.1 kDa) in the database search.
Cycloheximide chase experiments
To determine the half-life of the PINK1 protein, HEK293 cells were cotransfected with PINK1-FLAG and NEDD8 followed by cycloheximide treatment (50 µg/ml) for 0, 2, 4 and 6 h. PINK1 protein levels were detected by immunoprecipitation. Cells cotransfected with PINK1 and an empty vector were included as a control.
Drosophila experiments
Flies expressing dPINK1 RNAi and GMR-GAL4 were described previously (43). Flies expressing UAS-dAPP-BP1 were a gift from J. Yim (51). Flies expressing how24B-GAL4 were obtained from the Bloomington Drosophila Stock Center. Flies expressing UAS-dcr-2, UAS-dAPP-BP1 RNAi and tubulin-GAL4 were from the Vienna Drosophila RNAi Center. External eye phenotypes were examined and photographed as described (43). An additional fly line expressing Mhc-GAL4 and dPINK1 RNAi was a gift from Bingwei Lu (44). For the analysis of wing phenotypes, the crosses were performed at room temperature and the F1 adults were kept at 29°C immediately after eclosion. After 7 days, flies displaying drooped wing postures were counted. To examine the fertility of males, tubulin-GAL4/dPINK1-RNAi flies were crossed with either UAS-dAPP-BP1 flies, w1118 flies or UAS-lacZ flies. The progenies were incubated at 29°C to optimize RNAi-mediated knockdown. Each individual F1 male was crossed to three w1118 virgin females. After 10 days, the number of vials with larvae or pupae was counted.
Immunohistochemistry
We examined NEDD8 distributions in dopaminergic neurons using midbrain tissues from four PD patients and four sex- and age-matched normal individuals. Ten percent formalin-fixed midbrain tissues were paraffin embedded. Five-micrometer-thin sections were prepared and deparaffinized/rehydrated prior to being microwaved for antigen retrieval for 10 min, three times in a 10 mm citrate buffer (pH 6.0). Sections were pretreated with avidin/biotin blocking solution (Vector laboratories, Inc.) to mask endogenous biotin prior to being treated with hydrogen peroxide and blocked with 5% goat serum. Further, sections were stained with anti-NEDD8 antibody according to the antibody manufacturer's instruction. The NEDD8 immunoreactivity was visualized using the avidin–biotin–peroxidase complex method (ABC Elite, Vector laboratories, Inc.) with VIP (Vector laboratories, Inc.) as the substrate. To verify NEDD8-specific immunoreactivity, non-immunized rabbit IgG was used. Sections were scanned at a magnification of ×20 using the Aperio ScanScope XT system (Aperio Technologies). After saving the scanned images, the sections were further stained with eosin to verify Lewy bodies.
Statistics
The unpaired t-test and the Chi-square test in Figure 3 were performed using GraphPad Prism 5 (GraphPad Software, Inc.).
FUNDING
This work was supported by National Institutes of Health (RO1NS057289 and PO1ES016738 to Z.Z.); CIRM (RS1-00331-1 and RL1-00682-1 to Z.Z.); grants from National Natural Science Foundation of China (to Z.Z. and D.W. ); a 973 project from Ministry of Science and Technology of China (to Z.Z.); and fellowships from the Parkinson's Disease Foundation (to G.V.); and from American Parkinson Disease Association (to Y.S.C.).
ACKNOWLEDGEMENTS
We thank Drs Sung-Il Yoon, Toshiya Tsuji and Rena Baek for technical advice and Christopher Wu for manuscript proofreading.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bekris L.M., Mata I.F., Zabetian C.P. The genetics of Parkinson disease. J. Geriatr. Psychiatry Neurol. 2010;23:228–242. doi: 10.1177/0891988710383572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 3.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 4.Leroy E., Boyer R., Auburger G., Leube B., Ulm G., Mezey E., Harta G., Brownstein M.J., Jonnalagada S., Chernova T., et al. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 5.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 6.Bonifati V., Rizzu P., van Baren M.J., Schaap O., Breedveld G.J., Krieger E., Dekker M.C., Squitieri F., Ibanez P., Joosse M., et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 7.Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B., et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez A., Heimbach A., Grundemann J., Stiller B., Hampshire D., Cid L.P., Goebel I., Mubaidin A.F., Wriekat A.L., Roeper J., et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 10.Lautier C., Goldwurm S., Durr A., Giovannone B., Tsiaras W.G., Pezzoli G., Brice A., Smith R.J. Mutations in the GIGYF2 (TNRC15) gene at the PARK11 locus in familial Parkinson disease. Am. J. Hum. Genet. 2008;82:822–833. doi: 10.1016/j.ajhg.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strauss K.M., Martins L.M., Plun-Favreau H., Marx F.P., Kautzmann S., Berg D., Gasser T., Wszolek Z., Muller T., Bornemann A., et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum. Mol. Genet. 2005;14:2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 12.Paisan-Ruiz C., Bhatia K.P., Li A., Hernandez D., Davis M., Wood N.W., Hardy J., Houlden H., Singleton A., Schneider S.A. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann. Neurol. 2009;65:19–23. doi: 10.1002/ana.21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Fonzo A., Dekker M.C., Montagna P., Baruzzi A., Yonova E.H., Correia Guedes L., Szczerbinska A., Zhao T., Dubbel-Hulsman L.O., Wouters C.H., et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–245. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 14.Nalls M.A., Plagnol V., Hernandez D.G., Sharma M., Sheerin U.M., Saad M., Simon-Sanchez J., Schulte C., Lesage S., Sveinbjornsdottir S., et al. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wenzel D.M., Lissounov A., Brzovic P.S., Klevit R.E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong H., Wang D., Chen L., Choo Y.S., Ma H., Tang C., Xia K., Jiang W., Ronai Z., Zhuang X., et al. Parkin, PINK1, and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J. Clin. Invest. 2009;119:650–660. doi: 10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark I.E., Dodson M.W., Jiang C., Cao J.H., Huh J.R., Seol J.H., Yoo S.J., Hay B.A., Guo M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 18.Deng H., Dodson M.W., Huang H., Guo M. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl Acad. Sci. USA. 2008;105:14503–14508. doi: 10.1073/pnas.0803998105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feany M.B., Pallanck L.J. Parkin: a multipurpose neuroprotective agent. Neuron. 2003;38:13–16. doi: 10.1016/s0896-6273(03)00201-0. [DOI] [PubMed] [Google Scholar]
- 20.Moore D.J. Parkin: a multifaceted ubiquitin ligase. Biochem. Soc. Trans. 2006;34:749–753. doi: 10.1042/BST0340749. [DOI] [PubMed] [Google Scholar]
- 21.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J., Lee S.B., Lee S., Kim Y., Song S., Kim S., Bae E., Kim J., Shong M., Kim J.M., et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 23.Ulusoy A., Kirik D. Can overexpression of parkin provide a novel strategy for neuroprotection in Parkinson's disease? Exp. Neurol. 2008;212:258–260. doi: 10.1016/j.expneurol.2008.04.026. [DOI] [PubMed] [Google Scholar]
- 24.Wang X., Winter D., Ashrafi G., Schlehe J., Wong Y.L., Selkoe D., Rice S., Steen J., Lavoie M.J., Schwarz T.L. PINK1 and parkin target miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011;147:893–906. doi: 10.1016/j.cell.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cesari R., Martin E.S., Calin G.A., Pentimalli F., Bichi R., McAdams H., Trapasso F., Drusco A., Shimizu M., Masciullo V., et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc. Natl Acad. Sci. USA. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veeriah S., Taylor B.S., Meng S., Fang F., Yilmaz E., Vivanco I., Janakiraman M., Schultz N., Hanrahan A.J., Pao W., et al. Somatic mutations of the Parkinson's disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat. Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murata H., Sakaguchi M., Jin Y., Sakaguchi Y., Futami J.I., Yamada H., Kataoka K., Huh N.H. A new cytosolic pathway from a Parkinson's disease-associated kinase, BRPK/PINK1: activation of AKT via MTORC2. J. Biol. Chem. 2010;286:7182–7189. doi: 10.1074/jbc.M110.179390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plun-Favreau H., Klupsch K., Moisoi N., Gandhi S., Kjaer S., Frith D., Harvey K., Deas E., Harvey R.J., McDonald N., et al. The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat. Cell Biol. 2007;9:1243–1252. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 29.Pridgeon J.W., Olzmann J.A., Chin L.S., Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang D.T., Ayrault O., Hunt H.W., Taherbhoy A.M., Duda D.M., Scott D.C., Borg L.A., Neale G., Murray P.J., Roussel M.F., et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol. Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurz T., Chou Y.C., Willems A.R., Meyer-Schaller N., Hecht M.L., Tyers M., Peter M., Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Meyer-Schaller N., Chou Y.C., Sumara I., Martin D.D., Kurz T., Katheder N., Hofmann K., Berthiaume L.G., Sicheri F., Peter M. The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc. Natl Acad. Sci. USA. 2009;106:12365–12370. doi: 10.1073/pnas.0812528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broemer M., Tenev T., Rigbolt K.T., Hempel S., Blagoev B., Silke J., Ditzel M., Meier P. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol. Cell. 2010;40:810–822. doi: 10.1016/j.molcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 35.Oved S., Mosesson Y., Zwang Y., Santonico E., Shtiegman K., Marmor M.D., Kochupurakkal B.S., Katz M., Lavi S., Cesareni G., et al. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J. Biol. Chem. 2006;281:21640–21651. doi: 10.1074/jbc.M513034200. [DOI] [PubMed] [Google Scholar]
- 36.Xirodimas D.P., Saville M.K., Bourdon J.C., Hay R.T., Lane D.P. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 37.Gao F., Cheng J., Shi T., Yeh E.T. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFkappaB-dependent transcription. Nat. Cell Biol. 2006;8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 38.Xirodimas D.P., Sundqvist A., Nakamura A., Shen L., Botting C., Hay R.T. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9:280–286. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dil Kuazi A., Kito K., Abe Y., Shin R.W., Kamitani T., Ueda N. NEDD8 protein is involved in ubiquitinated inclusion bodies. J. Pathol. 2003;199:259–266. doi: 10.1002/path.1283. [DOI] [PubMed] [Google Scholar]
- 40.Mori F., Nishie M., Piao Y.S., Kito K., Kamitani T., Takahashi H., Wakabayashi K. Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol. Appl. Neurobiol. 2005;31:53–61. doi: 10.1111/j.1365-2990.2004.00603.x. [DOI] [PubMed] [Google Scholar]
- 41.Baker R.T., Catanzariti A.M., Karunasekara Y., Soboleva T.A., Sharwood R., Whitney S., Board P.G. Using deubiquitylating enzymes as research tools. Methods Enzymol. 2005;398:540–554. doi: 10.1016/S0076-6879(05)98044-0. [DOI] [PubMed] [Google Scholar]
- 42.Ryu K.Y., Baker R.T., Kopito R.R. Ubiquitin-specific protease 2 as a tool for quantification of total ubiquitin levels in biological specimens. Anal. Biochem. 2006;353:153–155. doi: 10.1016/j.ab.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 43.Wang D., Qian L., Xiong H., Liu J., Neckameyer W.S., Oldham S., Xia K., Wang J., Bodmer R., Zhang Z. Antioxidants protect PINK1-dependent dopaminergic neurons in Drosophila. Proc. Natl Acad. Sci. USA. 2006;103:13520–13525. doi: 10.1073/pnas.0604661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J.W., Yang L., Beal M.F., Vogel H., Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl Acad. Sci. USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandes C., Rao Y. Genome-wide screen for modifiers of Parkinson's disease genes in Drosophila. Mol. Brain. 2011;4:17. doi: 10.1186/1756-6606-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duda D.M., Borg L.A., Scott D.C., Hunt H.W., Hammel M., Schulman B.A. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xirodimas D.P. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem. Soc. Trans. 2008;36:802–806. doi: 10.1042/BST0360802. [DOI] [PubMed] [Google Scholar]
- 48.Pan Z.Q., Kentsis A., Dias D.C., Yamoah K., Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23:1985–1997. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 49.Wu J.T., Lin H.C., Hu Y.C., Chien C.T. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat. Cell Biol. 2005;7:1014–1020. doi: 10.1038/ncb1301. [DOI] [PubMed] [Google Scholar]
- 50.Mendoza H.M., Shen L.N., Botting C., Lewis A., Chen J., Ink B., Hay R.T. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem. 2003;278:25637–25643. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 51.Kim H.J., Kim S.H., Shim S.O., Park E., Kim C., Kim K., Tanouye M.A., Yim J. Drosophila homolog of APP-BP1 (dAPP-BP1) interacts antagonistically with APPL during Drosophila development. Cell Death Differ. 2007;14:103–115. doi: 10.1038/sj.cdd.4401935. [DOI] [PubMed] [Google Scholar]