Abstract
Biallelic mutations of the DNA annealing helicase SMARCAL1 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a-like 1) cause Schimke immuno-osseous dysplasia (SIOD, MIM 242900), an incompletely penetrant autosomal recessive disorder. Using human, Drosophila and mouse models, we show that the proteins encoded by SMARCAL1 orthologs localize to transcriptionally active chromatin and modulate gene expression. We also show that, as found in SIOD patients, deficiency of the SMARCAL1 orthologs alone is insufficient to cause disease in fruit flies and mice, although such deficiency causes modest diffuse alterations in gene expression. Rather, disease manifests when SMARCAL1 deficiency interacts with genetic and environmental factors that further alter gene expression. We conclude that the SMARCAL1 annealing helicase buffers fluctuations in gene expression and that alterations in gene expression contribute to the penetrance of SIOD.
INTRODUCTION
With the discovery of incomplete penetrance in 1925 (1–3), it became apparent that not all individuals with identical mutant alleles necessarily manifest a trait. Incomplete penetrance has been attributed to genetic and environmental factors as well as to developmental stochastics (4–7). In its strictest sense, incomplete penetrance applies to the entire lifetime of the individual (8), although some traits initially considered incompletely penetrant are probably better described as having age-dependent penetrance (8,9).
Incomplete penetrance for autosomal recessive disorders is less commonly described than for autosomal dominant disorders, but it does occur (4,10–13). Examples of genetic and environmental modulators of incomplete penetrance in autosomal recessive disorders are illustrated by the following. The penetrance of the deafness phenotype in individuals with biallelic mutations of DFNB26 can be suppressed by a dominant allele of the DFNM1 locus (10). The penetrance of Bardet–Biedl syndrome can require the triallelic inheritance of mutations compromising BBSome complex assembly and vesicle trafficking to the ciliary membrane (11,14,15). In cystic fibrosis, the T7 polypyrimidine track variant in intron 8 of CFTR leads to more efficient splicing of exon 9 and reduces penetrance of the p.R117H mutation and cystic fibrosis to 0.03% for individuals with the compound heterozygous mutations p.[R117H; T7] + [ΔF508] (12,16,17). Suppression of phenylketonuria in individuals with biallelic mutations of PAH is possible through restriction of dietary phenylalanine (18). Lastly, the penetrance of hemochromatosis in individuals with biallelic mutations of HFE depends on a combination of genetic factors such as polymorphisms in iron metabolism genes and environmental factors affecting the iron load such as alcohol, dietary iron intake, age and gender (13,19). Despite this progress, however, the basis of incomplete penetrance remains undefined for most autosomal recessive disorders, including Schimke immuno-osseous dysplasia (SIOD).
Recently, two families have been reported in which siblings of affected individuals have incomplete penetrance of SIOD (20,21). SIOD is a multisystem disorder of renal failure, immunodeficiency, skeletal dysplasia, arteriosclerosis, migraine-like headaches, cerebral ischemia, bone marrow failure and hypothyroidism (22,23). It is associated with biallelic mutations in SMARCAL1 that cause loss of SMARCAL1 enzymatic function (24–26). The SMARCAL1 enzyme recognizes single-stranded (ss)-to-double-stranded (ds) transitions in DNA (27) and functions as an annealing helicase (26). It also serves as a DNA stress response protein that participates in the maintenance of genomic integrity at stalled replication forks (28–32). Since components of DNA repair pathways frequently participate in transcription (33–40) and since variations in transcription can modulate trait penetrance (7), we hypothesized that SMARCAL1 also modulates transcription and that penetrance of SMARCAL1 deficiency results from further alteration of gene expression.
To test these hypotheses, we explored the effect of deficiency of the human, Drosophila and mouse SMARCAL1 orthologs on gene expression and the interaction of such deficiency with environmental and genetic modifiers of transcription. We find that in humans, flies and mice, deficiency of the respective SMARCAL1 ortholog alters expression of many genes and that in flies and mice deficiency of the respective ortholog is insufficient to cause disease in the absence of additional environmental or genetic insults.
RESULTS
Deficiency of Marcal1 is insufficient for disease manifestations in Drosophila
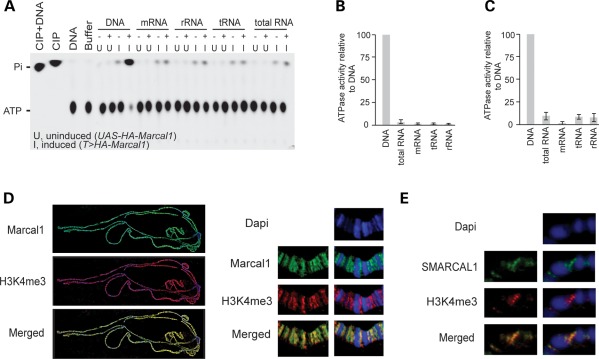
To investigate whether deficiency of SMARCAL1 orthologs is sufficient to cause disease, we developed a Drosophila melanogaster model. Similar to human SMARCAL1, the only fly ortholog Marcal1 also encodes a DNA-specific ATPase (NP_608883.1, Marcal1) (Fig. 1A and B); it shares 32% amino acid identity and 44% amino acid similarity with SMARCAL1 (24). Like human SMARCAL1 and mouse Smarcal1 (25,41–43), Marcal1 is a nuclear protein and is highly expressed in the early embryo and developing tissues and gonads (Supplementary Material, Fig. S1).
Figure 1.
DNA specificity and chromatin binding of Marcal1, SMARCAL1 and Smarcal1. (A) Radiograph of a thin layer chromatography plate showing that Marcal1 has DNA-dependent ATPase activity. Purified hemagglutinin (HA)-tagged Marcal1 was immunoprecipitated from transgenic flies expressing HA-tagged Marcal1 under induction (I) of the tubulin-GAL4 driver. Control immunoprecipitates were prepared in parallel from transgenic flies carrying the UAS-HA-Marcal1 transgene but not expressing it (U, uninduced). Both sets of immunoprecipitates were assayed for their ability to hydrolyze ATP to AMP and pyrophosphate (Pi) in the presence (+) or absence (−) of DNA, mRNA, rRNA, tRNA or total RNA. Calf intestinal phosphatase (CIP) was used as positive control, and the samples containing only DNA or buffer without immunoprecipitated enzyme were used as negative controls. (B and C) Plots showing the DNA-dependent ATPase activity of SMARCAL1 (B) and Smarcal1 (C) as measured by Kinase-Glo Luminescent Kinase Assay. For these assays, His-tagged SMARCAL1 and Smarcal1 were purified from HEK293 cells, using a nickel column, following the induction of expression with tetracycline. Nickel column elution fractions from uninduced cells were used as negative controls. Error bars represent 1 standard deviation. (D) Photographs showing immunofluorescence localization of HA-tagged Marcal1 on polytene chromosomes from tub-GAL4, UAS-HA-Marcal1 flies. Note that Marcal1 (green) binds the interband regions and co-localizes with trimethyl-K4-histone H3 (H3K4me3; red). (E) Photographs showing immunofluorescence localization of human SMARCAL1 on polytene chromosomes from MS1096-GAL4 and UAS-SMARCAL1 flies. Note that SMARCAL1 (green) also binds the interband regions and co-localizes with H3K4me3 (red).
By mobilizing P-element KG9850 and screening for deletions of the Marcal1 gene, we generated a loss-of-function Marcal1 mutant that lacked 679 bp extending from the middle of the first exon into the second intron (NM_135039.1:c. 673_1258delinsATGATGAAATAACATCATTATATCGATTAACACAG, p.G225MfsX3, Marcal1del, Supplementary Material, Fig. S2A). Marcal1del/del flies did not express Marcal1 mRNA and protein (Supplementary Material, Fig. S2B and C), and surprisingly, like the unaffected individuals with biallelic SMARCAL1 mutations (20,21), the mutant flies exhibited no morphologic differences when compared with wild-type flies and had a normal lifespan at 20°C.
Deficiency of murine Smarcal1 is insufficient to cause disease
To address whether the absence of disease in Marcal1-deficient flies was attributable to the absence of tissues such as bones, mammalian kidneys and T cells, we generated mice deficient for Smarcal1. Smarcal1 is the only SMARCAL1 ortholog in mouse and encodes a DNA-specific ATPase (NP_061287.2, Smarcal1) (Fig. 1C) that has 72% amino acid identity and 78% amino acid similarity to the human SMARCAL1 (24). Using standard homologous recombination knockout technology, we deleted the first two coding exons of Smarcal1 (NM_018817.2:c.172_989del, Smarcal1del/del). These exons include the replication protein A binding site, nuclear localization signal and the first HARP domain (Supplementary Material, Figs S3–S5); deletion of the HARP domains has been associated with defective annealing helicase activity and disease in humans (24,44). When maintained on a mixed 75% 129SvEv and 25% C57BL/6 background, the Smarcal1del/del mice did not show developmental, growth or physical abnormalities or other signs of disease through 24 months (Supplementary Material, Fig. S6A–L). Analysis of lymphatic tissues showed that Smarcal1del/del mice had a reduction in their B-cell count but no T-cell deficiency (Supplementary Material, Fig. S6M–P). Similarly, when the Smarcal1del allele was backcrossed onto a C57BL/6 background, the Smarcal1del/del mice did not show developmental, growth or physical abnormalities and had the same immunologic phenotype as the mice on the mixed background (data not shown). Therefore, like the incomplete penetrance reported for the two human families (20,21), deficiency of the respective SMARCAL1 ortholog in fruit flies and mice was insufficient to cause overt disease.
SMARCAL1 and Marcal1 bind open chromatin
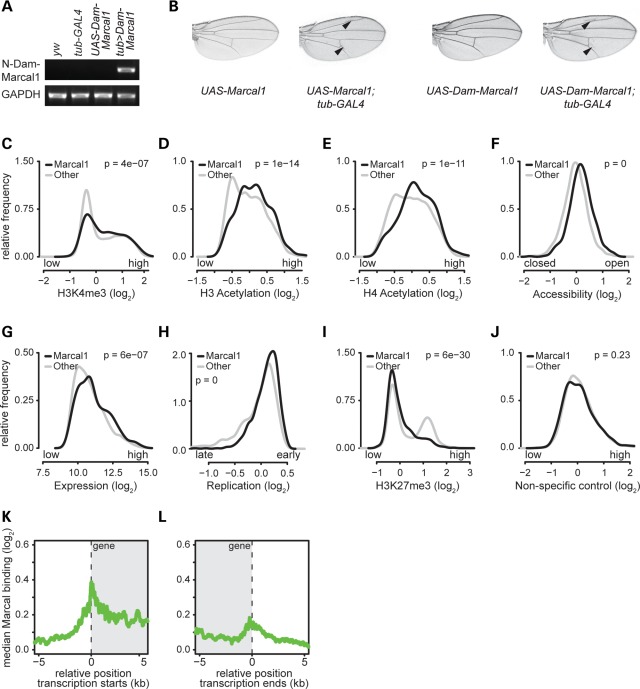
To better understand the function of proteins encoded by SMARCAL1 orthologs in the nucleus, we analyzed the association of SMARCAL1 and Marcal1 with chromatin. We used immunofluorescence to localize HA-tagged SMARCAL1 and Marcal1 on polytene chromosomes from third instar fruit fly larvae. Both SMARCAL1 and Marcal1 co-localize with trimethyl-K4 histone H3 and acetylated histone H4, which are markers of transcriptionally active chromatin (Fig. 1D and E and Supplementary Material, Fig. S7). To confirm and to define better this binding, we tagged Marcal1 on the N terminus with DNA adenine methyltransferase (Dam), which methylates adenine at GATC sequences (45); we judged the fusion protein to be functionally active since in vivo expression of Dam-Marcal1 induced extra wing veins similar to untagged Marcal1 or SMARCAL1 (25) (Fig. 2A and B and Supplementary Material, Fig. S8). Following transient expression of Dam-Marcal1 in Drosophila Kc167 cells, we found increased adenine methylation in regions enriched for trimethyl-K4 histone H3 and for acetylated histone H3 and H4 (Fig. 2C–E). The regions of increased adenine methylation also corresponded to accessible, transcriptionally active and early replicating chromatin (Fig. 2F–H and J). In contrast, adenine methylation was decreased in regions enriched for trimethyl-K27 histone H3 (Fig. 2I), a mark of inactive chromatin (46). The enrichment for adenine methylation spanned transcribed regions and was preferentially enriched in promoter regions compared with the rest of the gene (Fig. 2K and L).
Figure 2.
Expression of Marcal1 tagged with Dam on its N terminus methylates adenine in genomic regions of Kc167 cells with hallmarks of active transcription. (A) Photograph of an agarose gel showing that, by RT-PCR, tub-GAL4 specifically induces expression of Dam-Marcal1 in transgenic UAS-Dam-Marcal1;tub>GAL4 flies. (B) Expression of Dam-Marcal1 in UAS-Dam-Marcal1;tub>GAL4 flies induces extra wing veins similar to those observed in UAS-Marcal1;tub>GAL4 flies. (C–J) Density distribution plots showing relative frequencies (y-axes) of adenine methylation (Dam-Marcal1 target regions, black) and absence of adenine methylation (non-target regions, gray) for the indicated log2-transformed features: (C) trimethylation levels of lysine 4 of histone H3 (H3K4me3, a mark of actively transcribed chromatin), (D) acetylation levels of histone H3 (a mark of actively transcribed chromatin), (E) acetylation levels of histone H4 (a mark of actively transcribed chromatin), (F) chromatin accessibility (open and closed chromatin), (G) mRNA expression levels, (H) DNA replication timing, (I) trimethylation levels of lysine 27 of histone H3 (H3K27me3, a mark of non-transcribed genes) and (J) a non-specific antibody control (rabbit anti-IgG). The log2-transformed features were obtained from previously published genome-wide data (46,82–84). All P-values were calculated using the Mann–Whitney U test. (K and L) Alignment plots showing the log2-transformed median adenine methylation level (y-axes) relative to transcription start sites (K) and termination sites (L) of all target genes. The gray shading indicates the transcribed region. Transcription start and termination sites are at position 0 (dashed vertical lines). Note that Dam-Marcal1 preferentially targeted transcriptional start and termination sites.
To determine whether this preference for promoter regions might correlate with promoter proximal pausing or stalling of RNA polymerase II (RpII) (47), we compared our adenine methylation data with published RpII distribution in S2 cells (48) or Toll10b embryos (49). This showed that adenine methylation was enriched in genes bound by RpII regardless of whether RpII was stalled or active and was underrepresented in genes without RpII binding. In contrast, transcriptionally inactive polycomb targets (46) were underrepresented among actively transcribed RpII genes (Supplementary Material, Table S1). Thus, Marcal1 likely has a broader role in transcription than modulating promoter proximal pausing or stalling of RpII.
Marcal1 and SMARCAL1 genetically interact with transcriptional components
To obtain genetic evidence that Marcal1 and SMARCAL1 are involved in transcription, we conducted a screen in D. melanogaster for enhancers and suppressors of the wing vein phenotype induced by expressing Marcal1 or SMARCAL1 under the control of the UAS promoter and the tubulin-GAL4 or MS1096-GAL4 drivers, respectively (Supplementary Material, Table S2) (25). We observed epistatic interactions with mutations of transcriptional regulators, including chromatin proteins, mediator complex members, RpII complex components and transcription initiation, elongation and termination factors (Supplementary Material, Table S2 and Fig. S9). Generally, but not always, loss-of-function mutations of transcriptional enhancers suppressed the wing vein phenotype, whereas loss-of-function mutations of transcriptional repressors enhanced the wing vein phenotype (Supplementary Material, Table S2).
SMARCAL1 deficiency alters gene expression in SIOD skin fibroblasts
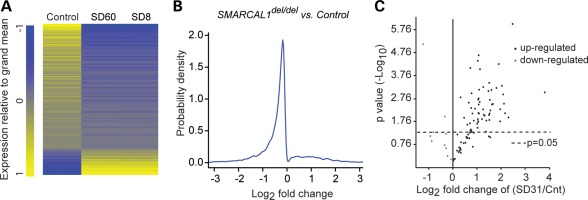
To clarify whether the SMARCAL1 chromatin binding and its genetic interactions with transcriptional components were indicative of function, we checked whether deficiency of SMARCAL1 altered gene expression. Using the Affymetrix Human Genome U133 Plus 2.0 Array, RNA derived from dermal fibroblasts of two SIOD patients (SD8 and SD60) showed significantly (q-value < 0.05) altered expression of 5644 genes (log2 median fold change of −0.255) (Fig. 3A and B; Supplementary Material, Fig. S10A and Table S3). Of these, 632 had >2-fold higher expression and 766 genes had >2-fold lower expression. The gene ontology (GO) biologic process annotations enriched among these differentially expressed genes included cellular and molecular metabolic processes, programmed cell death, cell cycle, signaling pathways and stress response (Supplementary Material, Tables S4 and S5).
Figure 3.
SMARCAL1 deficiency alters gene expression. (A) Heat map of the log2 fold differences in RNA levels (q-value <0.05) between control and SIOD patient (SD8 and SD60) skin fibroblasts. The RNA levels were measured using Affymetrix Human Genome U133 Plus 2.0 arrays and are the average of three biologic replicates. (B) Density plot showing the distribution of the log2 fold differences in RNA levels (q-value <0.05) between control and SIOD fibroblasts. (C) Volcano plot comparing the expression of stress genes in control and SIOD (SD31) skin fibroblast cell lines after 1 h incubation at 43°C followed by 1 h incubation at 37°C. The plot is derived from three biologic replicates.
Identification of the stress response was particularly intriguing since several patients with SMARCAL1 deficiency have developed severe migraine-like headaches, transient weakness and transient paraplegia during hot weather (C.F.B., unpublished data). To test whether SMARCAL1 modulated expression of heat shock genes, we heat-stressed skin fibroblasts from a control individual and three SIOD patients (SD31, SD120 and SD123) for 1 h at 43°C. Using quantitative reverse-transcriptase PCR (qRT-PCR), we found that many heat stress response genes were significantly over- or under-expressed in the SMARCAL1-deficient fibroblasts (Fig. 3C; Supplementary Material, Fig. S11 and Table S6).
Marcal1 contributes to heat tolerance and modulates heat stress gene expression in Drosophila
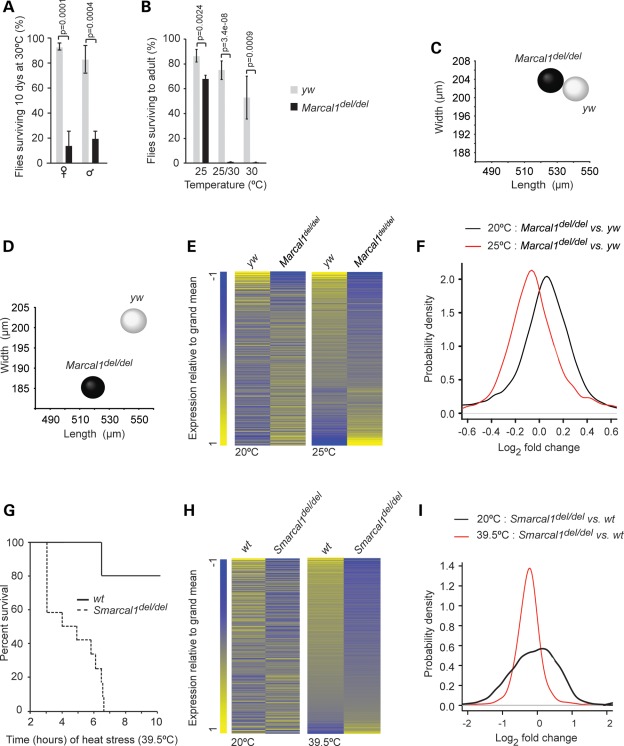
Given the abnormal response to heat stress by SIOD patients and SMARCAL1-deficient skin fibroblasts, we hypothesized that heat stress modifies the penetrance of Marcal1 deficiency. To test this, we reared Marcal1del/del and yellow white (yw) control flies at 20, 25 and 30°C. Although no differences were noted at 20°C, Marcal1del/del embryos and flies were significantly less viable at 25°C (<75% viability) and at 30°C (<20% viability) than yw control flies (Fig. 4A and B). Also, compared with yw flies, Marcal1del/del flies had abnormal expression of heat shock genes and proteins both at baseline and after 15 min at 37°C (Supplementary Material, Fig. S12).
Figure 4.
Marcal1 and Smarcal1 deficiencies increase susceptibility to heat stress. (A) Graph of the percent of yw control and Marcal1del/del flies surviving after 10 days at 30°C. Error bars represent 1 standard deviation. (B) Graph of the percent of yw control and Marcal1del/del embryos surviving through eclosion when raised at 25°C, when raised at 25°C for the first 5 days and then switched to 30°C or when raised at 30°C. Error bars represent 1 standard deviation. (C and D) Distribution plots of the dimensions of yw and Marcal1del/del eggs laid at 20°C (C) or at 25°C (D). (E) Heat maps of the log2 fold differences in all expressed mRNAs between yw and Marcal1del/del ovaries at 20°C and at 25°C. The RNA levels were measured using Affymetrix Drosophila Genome 2.0 Array and are the average of three biologic replicates. (F) Density plots showing the distribution of the log2 fold differences in gene expression between yw and Marcal1del/del ovaries at 20 and 25°C. (G) Survival curve for Smarcal1+/+ (n = 5) and Smarcal1del/del (n = 12) mice maintained for 10 h at 39.5°C. (H) Heat map of the log2 fold differences for all expressed RNAs between Smarcal1+/+ and Smarcal1del/del livers at 20 and 39.5°C. The RNA levels were derived from transcriptome sequencing and are the average of three biologic replicates. (I) Density plots showing the distribution of the log2 fold differences in RNA levels between Smarcal1+/+ and Smarcal1del/del livers at 20 and 39.5°C.
The Marcal1del/del flies also laid smaller eggs than yw controls at 25°C but not at 20°C (Fig. 4C and D). At 20°C, the mean (±SD) egg volume was 0.0116 ± 0.0018 mm3 for yw versus 0.0115 ± 0.0015 mm3 for Marcal1del/del (n = 100, P = 0.56). In contrast, at 25°C the mean (±SD) egg volume was 0.0117 ± 0.0013 mm3 for yw versus 0.0094 ± 0.0015 mm3 for Marcal1del/del (n = 100, P = 7.45e − 21).
To see whether this change in egg size correlated with altered gene expression, we compared RNA extracted from ovaries of yw and Marcal1del/del flies housed at 20°C with those housed at 25°C, using Affymetrix Drosophila Genome 2.0 Array. We found significant differences (q-value < 0.05) in the expression of 123 genes at 20°C and of 148 genes at 25°C; 81 genes were common in both groups. Furthermore, comparison of gene expression differences between yw and Marcal1del/del ovaries at 20 and 25°C showed a shift in the log2 median fold change from 0.01 at 20°C to −0.03 at 25°C (Fig. 4E and F). For both 20 and 25°C, the GO biologic process annotations enriched among genes differentially expressed between yw and Marcal1del/del ovaries included metabolic processes and response to stress (Supplementary Material, Tables S7–S9 and Fig. S10B).
Smarcal1 contributes to heat tolerance and modulates heat stress gene expression in mice
To test whether heat stress modified penetrance in the Smarcal1del/del mice, we housed the mice at 39.5°C. All Smarcal1del/del mice died within 6.5 h, whereas only 20% of wild-type (Smarcal1+/+) mice died in 10 h (Fig. 4G). We used Illumina Next Generation Sequencing to characterize the transcriptomes of livers from Smarcal1del/del and Smarcal1+/+ mice at an ambient temperature of 20°C or following 1 h at 39.5°C (Fig. 4H and I). At 20°C, 365 genes had >2-fold higher and 940 genes had >2-fold lower gene expression in Smarcal1del/del livers compared with Smarcal1+/+ livers, and the affected genes mapped to multiple GO biologic process annotations, including cellular and molecular metabolic processes and gene expression (Supplementary Material, Tables S10–S12). Following 1 h at 39.5°C, 107 genes had >2-fold higher and 309 genes had >2-fold lower gene expression in Smarcal1del/del livers compared with Smarcal1+/+ livers, and the median of gene expression differences between Smarcal1del/del liver and Smarcal1+/+ liver shifted from a log2 median fold change of −0.06 at 20°C to −0.24 at 39.5°C (Fig. 4H and I). The genes with altered expression mapped to multiple GO biologic process annotations, including cellular and molecular metabolic processes, gene expression, stress response and immune response (Supplementary Material, Tables S13 and S14, and Fig. S10C).
To confirm the altered stress response identified in the liver transcriptome data, we used qRT-PCR to analyze heat shock gene expression in RNA isolated from the livers and brains of the 3 h heat-stressed Smarcal1del/del and Smarcal1+/+ mice. This confirmed dysregulated expression of heat shock genes in both tissues (Supplementary Material, Table S15).
RpII inhibition decreases proliferation in SMARCAL1-deficient human fibroblasts
Having found that SMARCAL1, Marcal1 and Smarcal1 modulate gene expression during the heat stress response and are needed for heat tolerance, we hypothesized that other environmental and genetic factors altering transcription also induce penetrance when SMARCAL1 orthologs are deficient. To test this, we incubated skin fibroblasts from a control individual and three SIOD patients (SD31, SD120 and SD123) for 48 h with 1 μg/ml of α-amanitin, a toxin that preferentially inhibits RpII (50,51). At this dose, SIOD fibroblasts proliferated significantly slower than control fibroblasts as measured by MTT assay and EdU incorporation by the Click-iT EdU assay (Fig. 5A); there was no difference in apoptosis or necrosis as judged by TUNEL assay and trypan blue staining (data not shown). Also, knockdown of the largest subunits of RpII, using shRNAs against POLR2A or POLR2B in SIOD fibroblasts, resulted in significantly decreased proliferation compared with a non-targeting shRNA or with knockdown of RpII components in control fibroblasts (Supplementary Material, Fig. S13A and B). This interaction between RpII and SMARCAL1 did not result from protein–protein interactions since SMARCAL1 did not co-precipitate with RpII (data not shown).
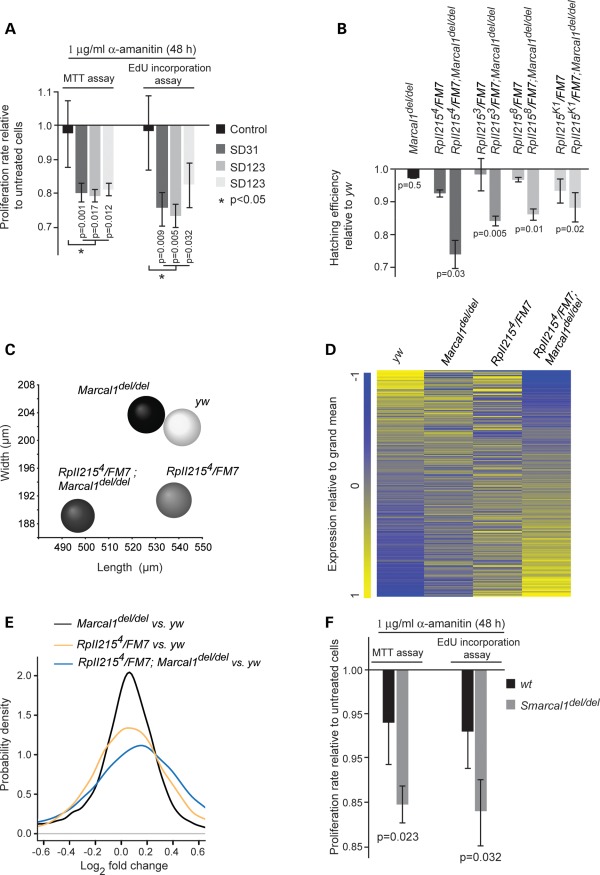
Figure 5.
Inhibition of RpII function causes penetrance of SMARCAL1, Marcal1 and Smarcal1 deficiency. (A) Graph showing the proliferation of α-amanitin-treated control and SMARCAL1del/del skin fibroblasts relative to untreated cells. The fibroblast cultures were treated with α-amanitin (1 μg/ml) for 48 h and proliferation was measured by the MTT and Click-iT EdU assays. (B) Graph showing the hatching rate at 20°C for Marcal1del/del, RpII2153 or 4 or 8 or K1/FM7 and RpII2153 or 4 or 8 or K1/FM7;Marcal1del/del embryos relative to the hatching rate of yw embryos. FM7 is an X chromosome balancer. (C) Distribution plot of egg dimensions showing that RpII2154/FM7;Marcal1del/del flies lay smaller eggs than yw, Marcal1del/del and RpII2154/FM7 flies. (D) Heat map comparing the log2 fold differences in all expressed mRNAs among yw, Marcal1del/del, RpII2154/FM7 and RpII2154/FM7;Marcal1del/del ovaries at 20°C. The RNA levels were measured using Affymetrix Drosophila Genome 2.0 Array and are the average of three biologic replicates. (E) Density plots showing the distribution of the log2 fold differences for transcripts depicted in (D). (F) Graph showing the proliferation of α-amanitin-treated Smarcal1+/+ and Smarcal1del/del MEFs relative to untreated MEFs. The MEFs were treated with α-amanitin (1 μg/ml) for 48 h and proliferation was measured by the MTT and Click-iT EdU assays. Error bars in (A), (B) and (F) represent 1 standard deviation.
RpII mutations decrease the viability and increase gene expression changes in Marcal1del/del flies
Given the sensitivity of SMARCAL1-deficient cells to RpII inhibition and knockdown, we tested for analogous epistatic interactions in Drosophila. We introduced a single mutant allele of the largest subunit of RpII (RpII2153, RpII2154, RpII2158 or RpII215K1) into the Marcal1del/del background (RpII2153/FM7;Marcal1del/del, RpII2154/FM7;Marcal1del/del, RpII2158/FM7;Marcal1del/del or RpII215K1/FM7;Marcal1del/del). Comparing the hatching of these embryos with Marcal1del/del and RpII heterozygous mutant embryos showed that these had a significantly reduced hatching rate at 20°C (Fig. 5B). This reduced hatching was not associated with increased apoptosis as measured by acridine orange staining or from altered protein–protein interactions between RpII215 and Marcal1 since Marcal1 did not co-precipitate with RpII215 (data not shown).
Besides reduced hatching, the introduction of a single mutant allele of RpII215 into the Marcal1del/del background reduced the size of eggs relative to those of Marcal1del/del and RpII215 heterozygous mutants (Fig. 5C). The mean (±SD) egg volume was 0.0116 ± 0.0018 mm3 for yw versus 0.0115 ± 0.0015 mm3 for Marcal1del/del (n = 100, P = 0.56), 0.0104 ± 0.0015 mm3 for RpII2154/FM7 (n = 100, P = 9.28e − 7) and 0.0094 ± 0.0016 mm3 for RpII2154/FM7;Marcal1del/del flies (n = 100, P = 1.03e − 13).
To test whether this reduced egg size was associated with changes in gene expression, we used the Affymetrix Drosophila Genome 2.0 Array to compare gene expression in the ovaries of Marcal1del/del, heterozygous RpII2154 (RpII2154/FM7), RpII2154/FM7;Marcal1del/del and yw flies. The introduction of the RpII2154 allele increased both the number of differentially expressed genes and the magnitude of the overall expression differences. A total of 744 genes were differentially (q-value < 0.05) expressed between RpII2154/FM7;Marcal1del/del and yw ovaries compared with 123 genes between Marcal1del/del and yw ovaries. The log2 median fold change of expression differences for Marcal1del/del versus yw ovaries was 0.01, whereas for RpII2154/FM7;Marcal1del/del versus yw ovaries, it was 0.07. The 744 differentially expressed genes mapped to multiple GO biologic process annotations, including cellular and metabolic processes, cell cycle and response to stress (Fig. 5D and E and Supplementary Material, Tables S7 and S16).
Reciprocally, the introduction of the biallelic deletion of Marcal1 into the background of the RpII2154 mutation also increased the number of differentially expressed genes and the magnitude of the overall expression differences. Comparison of gene expression in RpII2154/FM7 and yw ovaries identified 378 differentially (q-value < 0.05) expressed genes, and this increased to 744 for the comparison of RpII2154/FM7;Marcal1del/del and yw ovaries. Also, the log2 median fold change of expression differences was 0.01 for RpII2154/FM7 versus yw and 0.07 for RpII2154/FM7;Marcal1del/del versus yw ovaries. The 378 differentially expressed genes mapped to many GO biologic process annotations, including cellular and metabolic processes and gene expression (Fig. 5D and E and Supplementary Material, Tables S7 and S17).
RpII inhibition decreases proliferation in Smarcal1-deficient mouse embryonic fibroblasts
Having observed the interactions of SMARCAL1 and Marcal1 with RpII, we asked whether the inhibition of RpII function had a similar effect in Smarcal1del/del mouse embryonic fibroblasts (MEFs). When treated with 1 μg/ml α-amanitin, Smarcal1del/del MEFs proliferated significantly more slowly than Smarcal1+/+ fibroblasts as measured by the MTT assay and EdU incorporation by the Click-iT EdU assay (Fig. 5F). Also, they exhibited no difference in apoptosis or necrosis as judged by the TUNEL assay and trypan blue staining (data not shown). Furthermore, knockdown of the largest subunit of RpII, using siRNAs against Polr2a in Smarcal1del/del MEFs, resulted in a significantly decreased proliferation rate compared with knockdown of Polr2a in Smarcal1+/+ MEFs or with the treatment of Smarcal1del/del MEFs with a non-targeting siRNA (Supplementary Material, Fig. S13C and D).
RpII inhibition modifies the penetrance of Smarcal1 deficiency and partially recapitulates SIOD in Smarcal1-deficient mice
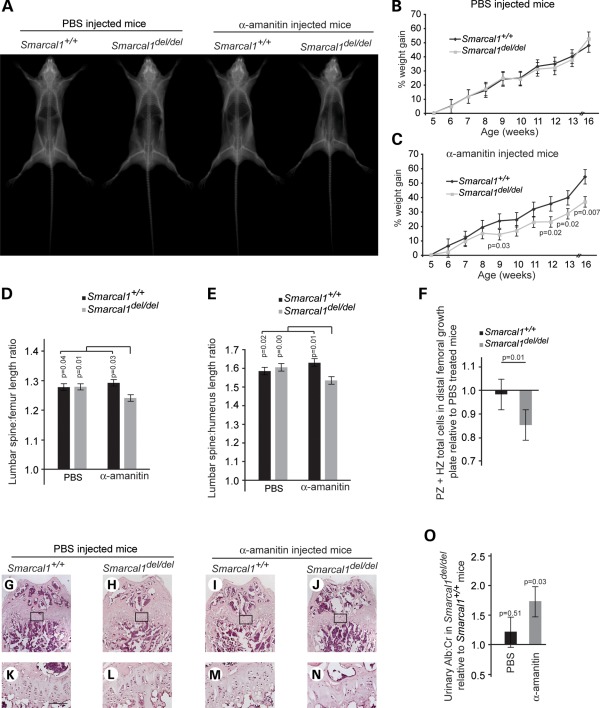
Based on the preceding findings, we hypothesized that variations in gene expression are critical for expression of SIOD disease features. To test this in vivo, we injected Smarcal1del/del and Smarcal1+/+ mice with either carrier (PBS, phosphate buffered saline) or α-amanitin (0.1 mg/kg/day) for 12 weeks. Although they did not develop T-cell deficiency (Supplementary Material, Fig. S14), the Smarcal1del/del mice injected with α-amanitin developed features of SIOD that were not observed in α-amanitin-treated Smarcal1+/+ mice or in PBS-treated Smarcal1del/del and Smarcal1+/+ mice (Fig. 6). First, they had length and weight growth restriction (Fig. 6A–C), and consistent with the growth restriction in SIOD, the mice had a disproportionately short spine (Fig. 6D and E). Second, as reported for SIOD growth plates (23,52), the distal femur growth plates were hypocellular, and chondrocytes in the proliferation and hypertrophic zones formed less organized columns (Fig. 6F–N). Finally, reminiscent of the early renal disease in SIOD, the treated Smarcal1del/del mice developed albuminuria (Fig. 6O).
Figure 6.
Treatment of Smarcal1del/del mice with α-amanitin partially recapitulates SIOD. (A) Radiographs of representative male mice after 12 weeks of daily intra-peritoneal (IP) injections with PBS or 0.1 mg/kg α-amanitin. (B) Growth curve showing that Smarcal1+/+ (n = 7) and Smarcal1del/del (n = 9) mice gain weight equally when given daily IP injections of PBS. (C) Growth curve showing that Smarcal1del/del (n = 9) mice gain less weight than Smarcal1+/+ (n = 7) mice when given daily ip-injections of 0.1 mg/kg α-amanitin. (D and E) Graphs showing the ratio of lumbar spine (L1–L6) length to femur length (D) or humerus length (E) for α-amanitin- and PBS-treated mice. Note that the α-amanitin treatment disproportionately shortened the lumbar spine of the Smarcal1del/del mice. (F) Plot of fold change in chondrocyte number in the proliferative (PZ) and hypertrophic (HZ) zones in the distal femoral growth plate of α-amanitin-treated mice, Smarcal1del/del (n = 7) and Smarcal1+/+ (n = 7), relative to PBS-treated mice, Smarcal1del/del (n = 7) and Smarcal1+/+ (n = 6). (G–N) Photographs of representative H&E staining of the distal femoral growth plate of Smarcal1+/+ and Smarcal1del/del male mice treated with PBS or α-amanitin for 12 weeks. (K)–(N) Higher magnifications of the boxed areas on (G)–(J), respectively. Note the hypocellular growth plate and poorly organized columns of chondrocytes in the growth plate of the α-amanitin-treated Smarcal1del/del mouse. Bar = 100 μm. (O) Graph showing urine albumin excretion by Smarcal1del/del mice relative to Smarcal1+/+ mice, following PBS or α-amanitin treatment. Bars in (B)–(F) and (O) represent standard errors.
DISCUSSION
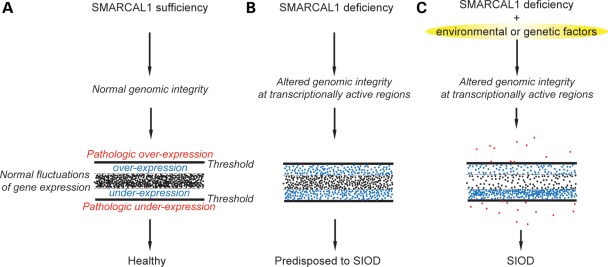
We have shown that the proteins encoded by SMARCAL1 orthologs localize to transcriptionally active chromatin, modulate gene expression and have epistatic interactions with transcription factors. We also found that, similar to the lack of penetrance for biallelic SMARCAL1 mutations in humans, deficiency of the orthologs in fruit flies and mice is insufficient to cause disease in these organisms and that penetrance is associated with environmental or genetic insults that further modify gene expression. From these observations, we hypothesize that the annealing helicase function of SMARCAL1, Marcal1 or Smarcal1 maintains DNA topology to buffer variability in gene expression and thereby mitigates penetrance of pathologic traits arising from environmental and genetic insults (Fig. 7).
Figure 7.
Model depicting the contribution of thresholded variations in gene expression to the penetrance of SIOD. (A) SMARCAL1 orthologs buffer random fluctuations in gene expression by modulating DNA helicity within the promoter and across transcribed regions. (B) Deficiency of the SMARCAL1 orthologs impairs maintenance of DNA structure within the transcriptionally active regions, and thereby alters gene expression. These alterations in gene expression are within a threshold of tolerance and compensated for such that few or no phenotypic features are apparent in humans and model organisms. (C) However, when transcription is further compromised by environmental or genetic factors that cause gene expression to pass a threshold, the organism is unable to compensate and manifests a phenotype.
As an annealing helicase, SMARCAL1 resolves ss-to-ds DNA transitions (26). Such transitions occur during DNA replication, repair, recombination and transcription, and recent studies have shown that SMARCAL1 participates in the DNA stress response both at stalled replication forks and at double-strand DNA breaks repaired by recombination or end joining (28–32). However, since defects of DNA repair, replication and recombination have not been detected clinically in SIOD patients (53), we reasoned that, like many DNA repair enzymes (33–35,37,40,54), SMARCAL1 contributes to transcription and that its deficiency results in gene expression changes contributing to the pathophysiology of SIOD.
There are at least three non-exclusive models by which SMARCAL1 deficiency can alter gene expression. First, unrepaired DNA damage impedes RpII progression and impairs transcription (55). Second, like ERCC6, SMARCAL1 could be part of the transcriptional complex and thus its deficiency directly affects RpII transcription (34). Third, the DNA structure maintained by SMARCAL1 modulates gene expression. Evidence against the first is the findings that (i) SIOD patients do not have ultraviolet light hypersensitivity (53), (ii) SMARCAL1-deficient fibroblasts do not have delayed recovery of mRNA synthesis following exposure to ultraviolet light or illudin S (C.F.B., unpublished data), and (iii) the proliferation rate of SMARCAL1-deficient cells is not detectably perturbed relative to control cells even though the transition from G2 to M phase is slightly delayed (28,31). Evidence against the second proposed mechanism includes (i) failure of SMARCAL1 homologues to co-purify or co-immunoprecipitate with RpII and transcription complexes studied (56–58) and (ii) failure of RpII to co-purify or co-immunoprecipitate with SMARCAL1 or Marcal1. We hypothesize, therefore, that the SMARCAL1 orthologs influence gene expression through the maintenance of DNA structure.
SMARCAL1 could modulate RpII transcription through the maintenance of the topology and helicity of duplex DNA. In prokaryotes, changes in DNA helicity cause gene expression to be enhanced, repressed or unchanged (59), and the equilibrium between duplex DNA and strand opening modulates transcription factor binding and production of full-length RNA (60,61). Similarly, in eukaryotes, in vitro and in vivo studies of MYC expression show that transcription-induced supercoiling melts MYC far upstream element (FUSE) to enable binding by structure-sensitive regulatory proteins such as FUSE-binding protein (FBP) and FBP-interacting repressor (FIR) (62,63). Binding of FBP and FIR to FUSE modifies the rate of MYC promoter firing (62,63). Since negative supercoiling and positive supercoiling are generated upstream and downstream of the transcription bubbles (64,65), respectively, SMARCAL1, Marcal1 or Smarcal1 might contribute to the maintenance of DNA topology within and adjacent to transcribed regions. Deficiency of these orthologs would then alter gene expression as a consequence of the changes in DNA helicity or topology.
This model could also explain why, in contrast to transcription factors that bind conserved promoter elements, SMARCAL1, Marcal1 and Smarcal1 deficiencies do not consistently affect expression of homologous genes. Homologous genes in human, flies and mice frequently reside in different genomic neighborhoods or chromatin regions (66); therefore, the need for the SMARCAL1, Marcal1 and Smarcal1 to maintain duplex DNA around homologous genes might vary among species and thereby have differing effects on expression of homologous genes.
In summary, SMARCAL1 deficiency is insufficient to cause SIOD, but the addition of environmental and genetic insults affecting transcription does cause penetrance and partial recapitulation of SIOD in model organisms. At both the molecular and genetic levels, SMARCAL1 plays a role in modulating gene expression. One model for the penetrance of biallelic mutations of SMARCAL1 orthologs is shown in Figure 7. In this model, SMARCAL1 orthologs modulate random fluctuations in gene expression and thereby potentiate or capacitate phenotypic changes induced by genetic or non-genetic insults, whereas the absence of the SMARCAL1 orthologs creates a chromatin environment permissive for such insults, pushing gene expression beyond a threshold of disease (Fig. 7).
MATERIALS AND METHODS
Drosophila genetic studies
The y1;P{SUPorP}Marcal1KG09850/CyO flies, which have a P element insertion in the Drosophila homologue of SMARCAL1 (Marcal1) at 25B4, were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN, USA). The loss-of-function mutant was obtained by imprecise P element excision (67); this deleted 679 bp nucleotides extending from the middle of the first exon into the second intron. The Marcal1del flies were crossed to w1118 flies for eight generations, and the specificity of the small egg phenotype to the homozygous deletion of Marcal1 in Marcal1del/del flies was confirmed by failure of Df(2L)Excel7793, which has a breakpoint of 25B1;25B8, and Df(2L)Excel7795, which has a breakpoint of 25B3;25B9, to complement the Marcal1del allele (68).
For expression of tagged and untagged Marcal1 cDNAs, we generated five transgenic lines in w1118 flies, using the pUAST and pUASP vectors: (i) UAST-Marcal1, (ii) UAST-HA-GFP-Marcal1, (iii) UAST-SMARCAL1, (iv) UASP-HA-GFP-Marcal1 and (v) UAST-N-Dam-Marcal1. All the other UAS lines, insertions and GAL4 lines used in this study were obtained from the Bloomington Drosophila Stock Center.
Using the SMARCAL1 and Marcal1 overexpression lines MS1096-GAL4/MS1096-GAL4; +/+; pUAST-SMARCAL1/pUAST-SMARCAL1 and pUAST-Marcal1/CyO; tubulin-GAL4/TM3, Sb1 the F1 genetic screen was carried out at 28°C; all other crosses were performed at 20°C unless indicated otherwise in the text. For the wing phenotype analysis, images from 10 wings for each genotype were acquired using a Zeiss Axiovert 200 microscope and scored by two independent readers. The reference for the enhancement or the suppression of the wing veins was the average of SMARCAL1 or Marcal1 overexpression flies crossed to w1118 mutants of three different genetic backgrounds (Bloomington stocks 3605, 5905, 6326).
For egg hatching efficiency, three groups of 100 yw, Marcal1del/del, RpII2153 or 4 or 8 or K1/FM7 and RpII2153 or 4 or 8 or K1/FM7;Marcal1del/del embryos were collected at 20°C. Hatching rates were calculated 45–48 h after egg deposition.
ATPase activity assay
The ATPase assay was performed with the purified Marcal1, SMARCAL1 and Smarcal1 proteins as previously described (25,69). See Supplementary Material for more details.
Immunofluorescence
Immunostaining of the Drosophila tissues and detection of human SMARCAL1 and mouse Smarcal1 were performed according to standard procedures as previously described (25,41,70,71). See Supplementary Material for more details.
High-dimensional (11-color) flow cytometry
Lymphoid tissues (spleen and thymus) from Smarcal1del/del and Smarcal1+/+ mice were harvested in RPMI-1640 medium containing 10% FCS. The tissues were pressed through a 70 μm nylon cell strainer, suspended in 10 ml of PBS and incubated with red cell lysis buffer (150 mm NH4CI) for 5 min. After washing with 10 volumes of PBS, single-cell suspensions were stained with fluorochrome-conjugated anti-mouse B220, TCRaβ, CD4, CD8, CD44 and CD25 antibodies (BD Biosciences) (Supplementary Material, Table S18). ‘Fluorescence-minus-one’ controls were included to determine the level of non-specific staining and autofluorescence associated with subsets of cells in each fluorescence channel. Propidium iodide was added to all samples before data collection to identify dead cells. High-dimensional flow cytometry data were collected on an LSRII FACS instrument (BD Biosciences). The FLOWJO (TreeStar, San Carlos, CA, USA) software was used for fluorescence compensation and analysis.
Generation of Smarcal1 knock-out mice
Using standard homologous recombination knockout technology, we deleted the first two coding exons of Smarcal1 (NM_018817.2:c.172_989del) and generated Smarcal1del/del mice (see Supplementary Material for more details). Mice used in this study were housed, bred and euthanized according to accepted ethical guidelines approved by the Institutional Review Board of Baylor College of Medicine (Houston, TX, USA, IRB protocol: AN-2983) or the University of British Columbia (Vancouver, British Columbia, Canada, Animal Care Certificate: A10-0296).
Cell cultures
Human dermal fibroblasts from SIOD patients and unaffected individuals and fibroblasts from Smarcal1+/+ and Smarcal1del/del embryos were cultured in DMEM (Gibco/Invitrogen) containing 15% fetal bovine serum (Gibco/Invitrogen) and 1% antibiotic/antimycotic (Gibco/Invitrogen) at 37°C and 5% CO2. The SIOD patient fibroblasts had the following mutations in the SMARCAL1 protein or gene: SD8: p.[L397fsX40] + [?]; SD31: homozygous deletion of the first 5 exons of SMARCAL1 (NT_005403.17:g.[67482574_67497178del] + [67482574_67497178del]); SD60: p.[E848X] + [E848X]; SD120: p.[R764Q] + [E848X]; and SD123: p.[R17X] + [R17X].
Heat stress
Drosophila: For assessing survival at 30°C, 200 flies of each genotype and sex (yw and Marcal1del/del) were maintained in a temperature- and humidity-controlled incubator for 10 days. Surviving flies were tallied on day 10. To assess reproductive capacity, three groups of 100 yw and Marcal1del/del embryos (300 in total for each genotype) were reared at different temperatures: group 1 was reared at 25°C; group 2 was reared at 25°C for the first 5 days and then shifted to 30°C (25/30°C); and group 3 was reared at 30°C. Flies surviving to adulthood were counted. For assessing gene or protein expression, ten 1–3-day-old female flies from each genotype (yw and Marcal1del/del) were heat-stressed at 37°C in a water bath for 15, 30, 45 and 60 min with or without recovery for 1 h at room temperature. Mouse: Three groups of 4–5 Smarcal1+/+ and 4–12 Smarcal1del/del female mice at 3–4 months of age were heat-stressed at 39.5°C for 1, 3 and 10 h as previously described (72). Human dermal fibroblasts: SIOD (SD31, SD120 and SD123) and control fibroblasts were heat-stressed for 1 h at 43°C followed by 1 h of recovery at 37°C as previously described (73).
α-Amanitin treatment
Mice: Beginning at the age of 30 days, Smarcal1del/del and Smarcal1+/+ mice (seven to nine per group) were injected intra-peritoneally daily with either PBS (n = 9 Smarcal1del/del and n = 7 Smarcal1+/+) or 0.1 mg/kg α-amanitin (Sigma) (n = 9 Smarcal1del/del and n = 7 Smarcal1+/+) diluted in PBS for 12 weeks. At the end of treatment, radiography was performed using the Faxitron X-ray cabinet. Human dermal fibroblasts: SIOD (SD31, SD120 and SD123) and control dermal fibroblasts were cultured in 96-well plates (3 × 103 cells/well). After 24 h, the medium was supplemented with 0 or 1 µg/ml α-amanitin and the cells were analyzed 48 h later for cell proliferation and viability, using the MTT assay as previously described (74). To measure proliferation with Click-iT EdU assay (Invitrogen), fibroblasts were cultured in the Lab-Tek 8 chambered cover glass system (7.5 × 103 cells/well). After 24 h, the medium was supplemented with 0 or 1 µg/ml α-amanitin, and after another 24 h, 10 μm EdU was added to each well. EdU detection and analysis was performed after 24 h, using Alexa Fluor 555 and Zeiss Axiovert 200 microscope, according to the manufacturer's protocol. Murine embryonic fibroblasts: Sensitivity of Smarcal1del/del and Smarcal1+/+ fibroblasts to α-amanitin (0 and 1 µg/ml) was performed as described above for human fibroblasts.
RNA extraction and RT-PCR
RNA was extracted from the indicated tissue, using the RNeasy Mini Kit (Qiagen). cDNA synthesis was performed using SuperScript III First Strand Kit (Invitrogen) or qScript cDNA SuperMix (Quanta). Quantitative real-time RT-PCR was performed using the ABI 7500 Fast system, using the primers listed in Supplementary Material, Table S19 (see Supplementary Material for more details).
Urinary protein and creatinine measurement
Mouse urine samples were analyzed for total protein (Lowry Protein Assay Kit, Bio-Rad), creatinine (Creatinine Assay Kit, Cayman) or albumin (bromocresol green method). See Supplementary Material for more details.
Histopathology
Smarcal1del/del and Smarcal1+/+ mice tissues were fixed in 4% PFA in PBS, paraffin-embedded and cut into 5 μm sections according to standard protocols (75). After H&E staining, tissues were analyzed using a Zeiss Axiovert 200 microscope. For distal femoral growth plate analysis, we counted all chondrocytes within the proliferative and hypertrophic zones and within 250 μm of the vertical midline.
Apoptotic analysis
Acridine orange staining for the detection of apoptotic cells in 20–23 h Drosophila embryos was performed as previously described (76). For human fibroblasts and MEFs, apoptotic cells were detected by TUNEL assay, using ApopTag Peroxidase In Situ Apoptosis Detection Kit (S7100, Chemicon).
RpII knock-down
Knock-down of the RpII genes encoding for the largest subunits of RpII in human fibroblasts and MEFs was carried out according to the Amaxa Biosystems optimized protocols (U-023 or A-023), using GFP-tagged shRNAs targeting POLR2A or POLR2B (SABiosciences) or Alexa Fluor 488-tagged siRNAs targeting Polr2a (Qiagen). See Supplementary Material for more details.
RNA sequencing and data analysis
For RNA sequencing, three samples of liver RNA were extracted from each group of 3–4-month-old Smarcal1del/del and Smarcal1+/+ female mice at 20°C and after 1 h at 39.5°C. The RNA samples for each group were pooled, and RNA sequencing libraries were constructed and sequenced using the whole transcriptome shotgun sequencing procedure, as previously described (77–79). See Supplementary Material for details.
Microarray gene expression analyses
For gene expression array analysis of human cells, two samples of RNA were extracted from each of two SIODs (SD8 and SD60) and three of control skin fibroblast cell lines, labeled and hybridized to Affymetrix Human Genome U133 Plus 2.0 Arrays. For expression array analysis of Drosophila ovaries, three samples of RNA were extracted from Drosophila ovaries for each genotype at 20 and 25°C, labeled and hybridized to Affymetrix Drosophila Genome 2.0 Array. See Supplementary Material for details.
GO analysis
GO analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (80,81) (See Supplementary Material for more details).
Dam analysis of Marcal1 binding
We performed DamID as previously described (46) (see Supplementary Material for more details).
Statistics
Data are presented as mean ± SD unless otherwise stated. Comparisons were made using the two-tailed Student's t-test and Mann–Whitney U test. Differences were considered significant at a P-value of <0.05 or a q-value of <0.05.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by a Ruth L. Kirschstein National Research Service Award (L.I.E.) and grants from the March of Dimes (6-FY02–136 to C.F.B.); the Gillson Longenbaugh Foundation (C.F.B.); the Dana Foundation (C.F.B. and D.B.L.); the New Development Award, Microscopy, and Administrative Cores of the Mental Retardation and Developmental Disabilities Research Center at Baylor College of Medicine (C.F.B.); the Burroughs Wellcome Foundation (1003400 to C.F.B.); the National Institute of Diabetes, Digestive, and Kidney Diseases, National Institute of Health (R03 DK062174 and R21DK065725 to C.F.B.); the Association Autour D'Emeric et D'Anthony (C.F.B.); the Michael Smith Foundation for Health Research (CI-SCH-O1899(07-1) to C.F.B.); Little Giants Foundation (C.F.B.); and the Child & Family Research Institute of British Columbia Children's Hospital (C.F.B.). C.F.B. and M.A.M. are scholars of the Michael Smith Foundation for Health Research.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Jan M. Friedman, David Cortez, Eberhard Passarge, Louis Lefebvre, Millan Patel, Shirin Kalyan and Matt Larouche for critical review of this manuscript. In addition, we would like to thank Hans Teunissen, Joke van Bemmel, Bas van Steensel, the NKI Central Microarray Facility and Lixin Xu in Child & Family Research Institute Flow Core Facility for technical support and reagents. The GEO reference number for the series of all microarray and transcriptome data is GSE35554. The GEO accession numbers for the human cell line microarray data subseries, the Drosophila melanogaster microarray data subseries and the mouse transcriptome data subseries are GSE35551, GSE35552 and GSE35553, respectively.
Conflict of Interest statement. None declared.
References
- 1.Romaschoff D.D. Über die Variabilität in der Manifestierung eines erblichen Merkmales (Abdomen abnormalis) bei Drosophila funebris F. J. Psychol. Neurol. 1925;31:323–325. [Google Scholar]
- 2.Timoféeff-Ressovsky N.W. Über den Einfluss des Genotypus auf das phänotypen Auftreten eines einzelnes Gens. J. Psychol. Neurol. 1925;31:305–310. [Google Scholar]
- 3.Vogt O. Psychiatrisch wichtige Tatsachen der zoologisch-botanischen Systematik. Zeitschrift für die gesamte. Neurol. Psychiatr. (Bucur) 1926;101:805–832. [Google Scholar]
- 4.Zlotogora J. Penetrance and expressivity in the molecular age. Genet. Med. 2003;5:347–352. doi: 10.1097/01.gim.0000086478.87623.69. [DOI] [PubMed] [Google Scholar]
- 5.Kurnit D.M., Layton W.M., Matthysse S. Genetics, chance, and morphogenesis. Am. J. Hum. Genet. 1987;41:979–995. [PMC free article] [PubMed] [Google Scholar]
- 6.Strobeck M.W., Reisman D.N., Gunawardena R.W., Betz B.L., Angus S.P., Knudsen K.E., Kowalik T.F., Weissman B.E., Knudsen E.S. Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J. Biol. Chem. 2002;277:4782–4789. doi: 10.1074/jbc.M109532200. [DOI] [PubMed] [Google Scholar]
- 7.Raj A., Rifkin S.A., Andersen E., van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khoury M.J., Beaty T.H., Cohen B.H. Fundamentals of Genetic Epidemiology. New York: Oxford University Press; 1993. [Google Scholar]
- 9.Strachan T., Read A.P. Human Molecular Genetics. 2nd edn. New York: Wiley-Liss; 1999. [Google Scholar]
- 10.Riazuddin S., Castelein C.M., Ahmed Z.M., Lalwani A.K., Mastroianni M.A., Naz S., Smith T.N., Liburd N.A., Friedman T.B., Griffith A.J., et al. Dominant modifier DFNM1 suppresses recessive deafness DFNB26. Nat. Genet. 2000;26:431–434. doi: 10.1038/82558. [DOI] [PubMed] [Google Scholar]
- 11.Katsanis N., Ansley S.J., Badano J.L., Eichers E.R., Lewis R.A., Hoskins B.E., Scambler P.J., Davidson W.S., Beales P.L., Lupski J.R. Triallelic inheritance in Bardet–Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293:2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- 12.Kiesewetter S., Macek M., Jr., Davis C., Curristin S.M., Chu C.S., Graham C., Shrimpton A.E., Cashman S.M., Tsui L.C., Mickle J., et al. A mutation in CFTR produces different phenotypes depending on chromosomal background. Nat. Genet. 1993;5:274–278. doi: 10.1038/ng1193-274. [DOI] [PubMed] [Google Scholar]
- 13.Sachot S., Moirand R., Jouanolle A.M., Mosser J., Fergelot P., Deugnier Y., Brissot P., le Gall J.Y., David V. Low penetrant hemochromatosis phenotype in eight families: no evidence of modifiers in the MHC region. Blood Cells Mol. Dis. 2001;27:518–529. doi: 10.1006/bcmd.2001.0394. [DOI] [PubMed] [Google Scholar]
- 14.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 15.Seo S., Baye L.M., Schulz N.P., Beck J.S., Zhang Q., Slusarski D.C., Sheffield V.C. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc. Natl Acad. Sci. USA. 2010;107:1488–1493. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu C.S., Trapnell B.C., Curristin S., Cutting G.R., Crystal R.G. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nat. Genet. 1993;3:151–156. doi: 10.1038/ng0293-151. [DOI] [PubMed] [Google Scholar]
- 17.Thauvin-Robinet C., Munck A., Huet F., Genin E., Bellis G., Gautier E., Audrezet M.P., Ferec C., Lalau G., Georges M.D., et al. The very low penetrance of cystic fibrosis for the R117H mutation: a reappraisal for genetic counselling and newborn screening. J. Med. Genet. 2009;46:752–758. doi: 10.1136/jmg.2009.067215. [DOI] [PubMed] [Google Scholar]
- 18.Scriver C.R., Sly W.S., Childs B., Beaudet A.L., Valle D., Kinzler K.W., Vogelstein B. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. [Google Scholar]
- 19.Weiss G. Genetic mechanisms and modifying factors in hereditary hemochromatosis. Nat. Rev. Gastroenterol. Hepatol. 2010;7:50–58. doi: 10.1038/nrgastro.2009.201. [DOI] [PubMed] [Google Scholar]
- 20.Bokenkamp A., deJong M., van Wijk J.A., Block D., van Hagen J.M., Ludwig M. R561C missense mutation in the SMARCAL1 gene associated with mild Schimke immuno-osseous dysplasia. Pediatr. Nephrol. 2005;20:1724–1728. doi: 10.1007/s00467-005-2047-x. [DOI] [PubMed] [Google Scholar]
- 21.Dekel B., Metsuyanim S., Goldstein N., Pode-Shakked N., Kovalski Y., Cohen Y., Davidovits M., Anikster Y. Schimke immuno-osseous dysplasia: expression of SMARCAL1 in blood and kidney provides novel insight into disease phenotype. Pediatr. Res. 2008;63:398–403. doi: 10.1203/PDR.0b013e31816721cc. [DOI] [PubMed] [Google Scholar]
- 22.Schimke R.N., Horton W.A., King C.R. Chondroitin-6-sulphaturia, defective cellular immunity, and nephrotic syndrome. Lancet. 1971;2:1088–1089. doi: 10.1016/s0140-6736(71)90400-4. [DOI] [PubMed] [Google Scholar]
- 23.Spranger J., Hinkel G.K., Stoss H., Thoenes W., Wargowski D., Zepp F. Schimke immuno-osseous dysplasia: a newly recognized multisystem disease. J. Pediatr. 1991;119:64–72. doi: 10.1016/s0022-3476(05)81040-6. [DOI] [PubMed] [Google Scholar]
- 24.Boerkoel C.F., Takashima H., John J., Yan J., Stankiewicz P., Rosenbarker L., Andre J.L., Bogdanovic R., Burguet A., Cockfield S., et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat. Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- 25.Elizondo L.I., Cho K.S., Zhang W., Yan J., Huang C., Huang Y., Choi K., Sloan E.A., Deguchi K., Lou S., et al. Schimke immuno-osseous dysplasia: SMARCAL1 loss-of-function and phenotypic correlation. J. Med. Genet. 2009;46:49–59. doi: 10.1136/jmg.2008.060095. [DOI] [PubMed] [Google Scholar]
- 26.Yusufzai T., Kadonaga J.T. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muthuswami R., Truman P.A., Mesner L.D., Hockensmith J.W. A eukaryotic SWI2/SNF2 domain, an exquisite detector of double-stranded to single-stranded DNA transition elements. J. Biol. Chem. 2000;275:7648–7655. doi: 10.1074/jbc.275.11.7648. [DOI] [PubMed] [Google Scholar]
- 28.Bansbach C.E., Betous R., Lovejoy C.A., Glick G.G., Cortez D. The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 2009;23:2405–2414. doi: 10.1101/gad.1839909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J., Ghosal G., Chen J. The annealing helicase HARP protects stalled replication forks. Genes Dev. 2009;23:2394–2399. doi: 10.1101/gad.1836409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yusufzai T., Kong X., Yokomori K., Kadonaga J.T. The annealing helicase HARP is recruited to DNA repair sites via an interaction with RPA. Genes Dev. 2009;23:2400–2404. doi: 10.1101/gad.1831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciccia A., Bredemeyer A.L., Sowa M.E., Terret M.E., Jallepalli P.V., Harper J.W., Elledge S.J. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postow L., Woo E.M., Chait B.T., Funabiki H. Identification of SMARCAL1 as a component of the DNA damage response. J. Biol. Chem. 2009;284:35951–35961. doi: 10.1074/jbc.M109.048330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann A.R. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 2001;15:15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 34.Proietti-De-Santis L., Drane P., Egly J.M. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 2006;25:1915–1923. doi: 10.1038/sj.emboj.7601071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elmayan T., Proux F., Vaucheret H. Arabidopsis RPA2: a genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr. Biol. 2005;15:1919–1925. doi: 10.1016/j.cub.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 36.Faucher D., Wellinger R.J. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010;6:e1001082. doi: 10.1371/journal.pgen.1001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guzder S.N., Sung P., Bailly V., Prakash L., Prakash S. RAD25 is a DNA helicase required for DNA repair and RNA polymerase II transcription. Nature. 1994;369:578–581. doi: 10.1038/369578a0. [DOI] [PubMed] [Google Scholar]
- 38.Coin F., Proietti De Santis L., Nardo T., Zlobinskaya O., Stefanini M., Egly J.M. p8/TTD-A as a repair-specific TFIIH subunit. Mol. Cell. 2006;21:215–226. doi: 10.1016/j.molcel.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 39.Liu H., Herrmann C.H., Chiang K., Sung T.L., Moon S.H., Donehower L.A., Rice A.P. 55K isoform of CDK9 associates with Ku70 and is involved in DNA repair. Biochem. Biophys. Res. Commun. 2010;397:245–250. doi: 10.1016/j.bbrc.2010.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu D.S., Cortez D. A role for cdk9-cyclin k in maintaining genome integrity. Cell Cycle. 2011;10:28–32. doi: 10.4161/cc.10.1.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deguchi K., Clewing J.M., Elizondo L.I., Hirano R., Huang C., Choi K., Sloan E.A., Lucke T., Marwedel K.M., Powell R.D., Jr., et al. Neurologic phenotype of Schimke immuno-osseous dysplasia and neurodevelopmental expression of SMARCAL1. J. Neuropathol. Exp. Neurol. 2008;67:565–577. doi: 10.1097/NEN.0b013e3181772777. [DOI] [PubMed] [Google Scholar]
- 42.Kilic S.S., Donmez O., Sloan E.A., Elizondo L.I., Huang C., Andre J.L., Bogdanovic R., Cockfield S., Cordeiro I., Deschenes G., et al. Association of migraine-like headaches with Schimke immuno-osseous dysplasia. Am. J. Med. Genet. A. 2005;135:206–210. doi: 10.1002/ajmg.a.30692. [DOI] [PubMed] [Google Scholar]
- 43.Elizondo L.I., Huang C., Northrop J.L., Deguchi K., Clewing J.M., Armstrong D.L., Boerkoel C.F. Schimke immuno-osseous dysplasia: a cell autonomous disorder? Am. J. Med. Genet. A. 2006;140:340–348. doi: 10.1002/ajmg.a.31089. [DOI] [PubMed] [Google Scholar]
- 44.Ghosal G., Yuan J., Chen J. The HARP domain dictates the annealing helicase activity of HARP/SMARCAL1. EMBO Rep. 2011;12:574–580. doi: 10.1038/embor.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Steensel B., Delrow J., Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- 46.Tolhuis B., de Wit E., Muijrers I., Teunissen H., Talhout W., van Steensel B., van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- 47.Gilchrist D.A., Nechaev S., Lee C., Ghosh S.K., Collins J.B., Li L., Gilmour D.S., Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muse G.W., Gilchrist D.A., Nechaev S., Shah R., Parker J.S., Grissom S.F., Zeitlinger J., Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeitlinger J., Stark A., Kellis M., Hong J.W., Nechaev S., Adelman K., Levine M., Young R.A. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindell T.J., Weinberg F., Morris P.W., Roeder R.G., Rutter W.J. Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science. 1970;170:447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- 51.Ljungman M., Zhang F., Chen F., Rainbow A.J., McKay B.C. Inhibition of RNA polymerase II as a trigger for the p53 response. Oncogene. 1999;18:583–592. doi: 10.1038/sj.onc.1202356. [DOI] [PubMed] [Google Scholar]
- 52.Clewing J.M., Antalfy B.C., Lücke T., Najafian B., Marwedel K.M., Hori A., Powel R.M., Safo Do A.F., Najera L., SantaCruz K., et al. Schimke immuno-osseous dysplasia: a clinicopathological correlation. J. Med. Genet. 2007;44:122–130. doi: 10.1136/jmg.2006.044313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boerkoel C.F., O'Neill S., Andre J.L., Benke P.J., Bogdanovic R., Bulla M., Burguet A., Cockfield S., Cordeiro I., Ehrich J.H., et al. Manifestations and treatment of Schimke immuno-osseous dysplasia: 14 new cases and a review of the literature. Eur. J. Pediatr. 2000;159:1–7. doi: 10.1007/s004310050001. [DOI] [PubMed] [Google Scholar]
- 54.Merino C., Reynaud E., Vazquez M., Zurita M. DNA repair and transcriptional effects of mutations in TFIIH in Drosophila development. Mol. Biol. Cell. 2002;13:3246–3256. doi: 10.1091/mbc.E02-02-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tornaletti S. Transcription arrest at DNA damage sites. Mutat. Res. 2005;577:131–145. doi: 10.1016/j.mrfmmm.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 56.Aygun O., Svejstrup J., Liu Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8580–8584. doi: 10.1073/pnas.0804424105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeronimo C., Langelier M.F., Zeghouf M., Cojocaru M., Bergeron D., Baali D., Forget D., Mnaimneh S., Davierwala A.P., Pootoolal J., et al. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol. Cell. Biol. 2004;24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robert F., Blanchette M., Maes O., Chabot B., Coulombe B. A human RNA polymerase II-containing complex associated with factors necessary for spliceosome assembly. J. Biol. Chem. 2002;277:9302–9306. doi: 10.1074/jbc.M110516200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hatfield G.W., Benham C.J. DNA topology-mediated control of global gene expression in Escherichia coli. Annu. Rev. Genet. 2002;36:175–203. doi: 10.1146/annurev.genet.36.032902.111815. [DOI] [PubMed] [Google Scholar]
- 60.Lim H.M., Lewis D.E., Lee H.J., Liu M., Adhya S. Effect of varying the supercoiling of DNA on transcription and its regulation. Biochemistry. 2003;42:10718–10725. doi: 10.1021/bi030110t. [DOI] [PubMed] [Google Scholar]
- 61.Pruss G.J., Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989;56:521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- 62.Kouzine F., Sanford S., Elisha-Feil Z., Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 63.Liu J., Kouzine F., Nie Z., Chung H.J., Elisha-Feil Z., Weber A., Zhao K., Levens D. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006;25:2119–2130. doi: 10.1038/sj.emboj.7601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rahmouni A.R. Z-DNA as a probe for localized supercoiling in vivo. Mol. Microbiol. 1992;6:569–572. doi: 10.1111/j.1365-2958.1992.tb01502.x. [DOI] [PubMed] [Google Scholar]
- 65.Wang Z., Droge P. Differential control of transcription-induced and overall DNA supercoiling by eukaryotic topoisomerases in vitro. EMBO J. 1996;15:581–589. [PMC free article] [PubMed] [Google Scholar]
- 66.Brown J.M., Leach J., Reittie J.E., Atzberger A., Lee-Prudhoe J., Wood W.G., Higgs D.R., Iborra F.J., Buckle V.J. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robertson H.M., Preston C.R., Phillis R.W., Johnson-Schlitz D.M., Benz W.K., Engels W.R. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parks A.L., Cook K.R., Belvin M., Dompe N.A., Fawcett R., Huppert K., Tan L.R., Winter C.G., Bogart K.P., Deal J.E., et al. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 69.Zhao K., Wang W., Rando O.J., Xue Y., Swiderek K., Kuo A., Crabtree G.R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 70.Zink D., Paro R. Drosophila polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 1995;14:5660–5671. doi: 10.1002/j.1460-2075.1995.tb00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ashburner M. Drosophila: A Laboratory Handbook and Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 72.Leon L.R., DuBose D.A., Mason C.W. Heat stress induces a biphasic thermoregulatory response in mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R197–R204. doi: 10.1152/ajpregu.00046.2004. [DOI] [PubMed] [Google Scholar]
- 73.Miyakoda M., Suzuki K., Kodama S., Watanabe M. Activation of ATM and phosphorylation of p53 by heat shock. Oncogene. 2002;21:1090–1096. doi: 10.1038/sj.onc.1205196. [DOI] [PubMed] [Google Scholar]
- 74.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 75.Hirano R., Interthal H., Huang C., Nakamura T., Deguchi K., Choi K., Bhattacharjee M.B., Arimura K., Umehara F., Izumo S., et al. Spinocerebellar ataxia with axonal neuropathy: consequence of a Tdp1 recessive neomorphic mutation. EMBO J. 2007;26:4732–4743. doi: 10.1038/sj.emboj.7601885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arama E., Steller H. Detection of apoptosis by terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling and acridine orange in Drosophila embryos and adult male gonads. Nat. Protoc. 2006;1:1725–1731. doi: 10.1038/nprot.2006.235. [DOI] [PubMed] [Google Scholar]
- 77.Shah S.P., Morin R.D., Khattra J., Prentice L., Pugh T., Burleigh A., Delaney A., Gelmon K., Guliany R., Senz J., et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 78.Shah S.P., Kobel M., Senz J., Morin R.D., Clarke B.A., Wiegand K.C., Leung G., Zayed A., Mehl E., Kalloger S.E., et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N. Engl. J. Med. 2009;360:2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 79.Morin R.D., Johnson N.A., Severson T.M., Mungall A.J., An J., Goya R., Paul J.E., Boyle M., Woolcock B.W., Kuchenbauer F., et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 81.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 82.Greil F., van der Kraan I., Delrow J., Smothers J.F., de Wit E., Bussemaker H.J., van Driel R., Henikoff S., van Steensel B. Distinct HP1 and Su(var)3–9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes Dev. 2003;17:2825–2838. doi: 10.1101/gad.281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schubeler D., MacAlpine D.M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D.E., O'Neill L.P., Turner B.M., Delrow J., et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schubeler D., Scalzo D., Kooperberg C., van Steensel B., Delrow J., Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.