Abstract
Inherited hearing loss in mice has contributed substantially to our understanding of inner-ear function. We identified a new allele at the Myo7a locus, Myo7ash1–8J; genomic characterization indicated that Myo7ash1–8J arose from complex deletion encompassing exons 38–40 and 42–46. Homozygous mutant mice had no detectable auditory brainstem response, displayed highly disorganized hair-cell stereocilia and had no detectable MYO7A protein. We generated mice that were digenic heterozygotes for Myo7ash1–8J and one of each Cdh23v-2J, Ush1gjs or Pcdh15av-3J alleles, or an Ush1c null allele. Significant levels of age-related hearing loss were detected in +/Myo7ash1–8J +/Ush1gjs, +/Myo7ash1–8J +/Cdh23v-2J and +/Myo7ash1–8J +/Pcdh15av-3J double heterozygous mice compared with age-matched single heterozygous animals, suggesting epistasis between Myo7a and each of the three loci. +/Pcdh15av-3J +/Ush1gjs double heterozygous mice also showed elevated hearing loss, suggesting Pcdh15–Ush1g epistasis. While we readily detected MYO7A, USH1C, CDH23 and PCDH15 using mass spectrometry of purified chick utricle hair bundles, we did not detect USH1G. Consistent with that observation, Ush1g microarray signals were much lower in chick cochlea than those of Myo7a, Ush1c, Cdh23 and Pcdh15 and were not detected in the chick utricle. These experiments confirm the importance of MYO7A for the development and maintenance of bundle function and support the suggestion that MYO7A, USH1G (Sans) and CDH23 form the upper tip-link complex in adult mice, likely in combination with USH1C (harmonin). MYO7A, USH1G and PCDH15 may form another complex in stereocilia. USH1G may be a limiting factor in both complexes.
INTRODUCTION
Mouse deafness mutants are exceptionally powerful in modeling human disease (1). In an outstanding example, mouse loci homologous to human Usher syndrome type I loci have been essential in determining the molecular basis for hearing loss in these patients (2).
All of the Usher I genes are expressed in hair cells, the sensory cells of the inner ear. Located at the apical surface of every hair cell, the hair bundle detects mechanical stimuli such as those produced by sound or head movement (2). Hair bundles are composed of ∼100 actin-filled stereocilia, which are coupled together with various links, including the tip links. Tip links are special; they sustain tension when a bundle is deflected in the excitatory direction and, in turn, gate mechanotransduction channels (2).
In mice, mutations in the Usher genes lead to severe disruption of hair-bundle morphology and profound deafness; these genes include those encoding the tip-link cadherins, protocadherin-15 (Pcdh15) and cadherin-23 (Cdh23); the scaffolding proteins, Sans (Ush1g) and harmonin (Ush1c); and the molecular motor myosin-VIIA (Myo7a) (2).
The molecular role for MYO7A protein has been controversial. In strong Myo7a alleles, which have very low levels of MYO7A (3), hair-bundle morphology is profoundly disrupted at early post-natal ages (4). Tip links are nonetheless present, and while mechanotransduction can be elicited at normal levels, much larger displacements than usual are required to initiate channel opening (5). Consistent with this model, in wild-type (WT) hair cells, MYO7A localizes to the upper tip-link insertion point (6). Although together these results suggest that MYO7A controls resting tension, control also has been attributed to myosin-1c (MYO1C) (7,8). An alternative or additional role for MYO7A may be in transporting those molecules that are essential for resting-tension control, rather than directly mediating climbing or slipping adaptation, consistent with requirement of MYO7A for localization of several Usher proteins (9).
In vitro experiments show that MYO7A forms a stable complex with USH1G (10), which binds tightly to USH1C (11); in turn, USH1C interacts with CDH23 (12). It is not clear whether any of these interactions are overlapping, i.e. whether a stable MYO7A–USH1G–USH1C–CDH23 complex forms in stereocilia, nor whether any of these interactions are relevant in mature stereocilia.
In addition, biochemical and genetic experiments have suggested that MYO7A and PCDH15 interact (13). Thus, an additional MYO7A-dependent complex could be present that includes not only PCDH15, but also USH1C and USH1G; indeed, a recent report suggested that USH1G was present at stereocilia tips, near the location of PCDH15 (14).
Because of the severe developmental defects seen with the Usher gene mutants, these mutant mice have been less useful for determining roles for the corresponding proteins in mature hair cells. However, the presence of a progressive hearing-loss phenotype in recessive-allele double heterozygotes would suggest functional interactions between the gene products. For example, a digenic interaction was demonstrated between the two tip-link cadherin genes, as Cdh23v–2J:Pcdh15av–3J double heterozygotes show age-related hearing loss that begins at high frequencies (15); these results are consistent with a CDH23–PCDH15 interaction not just during development, but in adult animals (16). Interactions between other Usher genes are less well characterized. In one example, double heterozygotes for several alleles each of Myo7a and Cdh23 did not have accelerated progressive hearing loss over single heterozygotes (17).
Because strong alleles of Myo7a are useful for experiments that directly test the post-natal role of MYO7A, we decided to test the suitability of a Myo7a allele identified at and available from The Jackson Laboratory. The Myo7ash1–8J allele has a complex genomic deletion; any protein it produces would lack two key C-terminal domains and was not detectable by protein immunoblot. Before ∼6 months of age, +/8J heterozygotes have no discernable inner-ear phenotype, while homozygotes have severely disrupted hair bundles and are deaf. While +/8J heterozygotes have modestly accelerated progressive hearing loss, double heterozygotes of Myo7ash1–8J and alleles of Ush1g and Cdh23, but not Pcdh15 and Ush1c, have substantially greater accelerated age-related hearing loss, indicating genetic interaction. Together the data suggest that interactions between MYO7A, USH1G, USH1C and CDH23 are required for auditory function in mature animals.
RESULTS
Mutant mouse phenotype and mapping
The Myo7a mutant mice described here were originally identified by their rapid circling movements, head tossing and hyperactivity. Unlike their normal littermates, mutant mice were unable to swim and did not respond to tapping or clicking sounds, which suggested hearing loss. The mutant allele was shown to be recessive, as an outcross to C57BL/6 (B6) mice produced only normal offspring. The new allele was shown to be deaf by auditory brainstem response (ABR) analysis at 4 weeks of age (see below).
F2 progeny (n = 100) were generated by intercrossing with CAST/Ei mice. The homozygous mutant F2 progeny (n = 21) were used for genetic mapping with a pooled DNA strategy (18). Once this approach efficiently localized the mutation to Chr 7, individual DNAs from the 21 affected F2 progeny were typed to refine the map position. Gene order, determined by minimizing the number of obligate cross-over events, and recombination frequency was calculated. The mutation was mapped distal to D7Mit31 (44 cM) with 2.5% recombination, in between D7Mit274 (at 26 cM with 27.7% recombination) and D7Mit43 (at 26 cM with 16% recombination).
Myo7ash1–8J is allelic with sh1
Before genetic mapping was completed, allelism tests were performed with several other similar circling and deaf mutants, including Myo15sh2/sh2, with negative results; no affected F1 offspring were born. Genetic mapping to a position distal to D7Mit31 suggested that this mutation might be allelic to sh1 (Myo7a). Accordingly, mice homozygous for the Myo7ash1–7J allele were mated to mice heterozygous for the new allele. Out of the 19 offspring, 6 displayed circling behavior by P20. All offspring were also tested for startle response as a preliminary screen for hearing impairment; offspring that displayed circling behavior also failed the startle response test. These data showed that the new allele did not complement another Myo7a allele; together with the linkage data, these complementation experiments confirmed the allelism with sh1 of the new allele, which we named Myo7ash1–8J (or 8J).
The 8J mutation is a complex genomic deletion
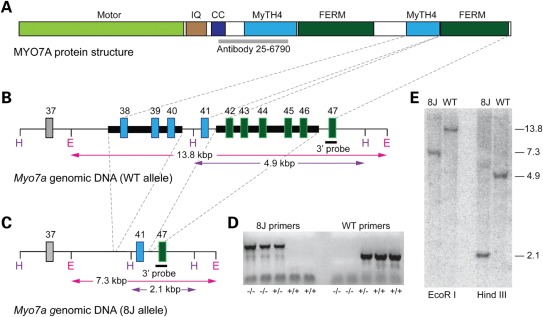
To characterize the genomic structure of the 8J allele, we took two parallel approaches (Fig. 1). In the first approach, we sequenced all exons of Myo7ash1–8J genomic DNA; in the second approach, we carried out Southern blot analysis of genomic DNA from homozygous Myo7ash1–8J mice and from the two parental mouse strains (C57BL/6J and SJL) on which the Myo7ash1–8J mutation arose spontaneously.
Figure 1.
Genomic structure of the 8J mutation. (A) MYO7A includes a motor head domain, five IQ domains (IQ region indicated) and a putative coiled-coil domain (CC); these are followed by two repeats of MyTH4 and FERM domains. The epitope recognized by the antibody used here (25–6790) is indicated in gray. (B) Structure of 3′ end of the WT Myo7a allele. Dashed lines between (A) and (B) indicate the relationship between Myo7a exons and protein structure. DNA missing in the 8J allele is highlighted with thick black bars. H and E indicate HindIII and EcoRI restriction sites; expected sizes resulting from HindIII or EcoRI restriction digests are indicated. (C) 8J allele. Dashed lines between (B) and (C) indicate deleted genomic DNA. (D) Genotyping for the 8J allele. DNA from five mice with indicated allele composition was separately subjected to PCR with 8J-specific and WT-specific primers. (E) Southern analysis; predicted band sizes (in kb) are indicated on right. The 3′ probe indicated in (A) and (B) was used.
To locate the 8J mutation, we amplified and sequenced individual exons from Myo7ash1–8J genomic DNA. Using the primers listed in Supplementary Material, Table S1, we successfully amplified exons 1–37 from Myo7ash1–8J genomic DNA; no mutations were identified in any of these exons. Using primers spanning individual exons over a region encompassing exons 38–46 (Supplementary Material, Table S2), we amplified polymerase chain reaction (PCR) products of the expected sizes from parental C57BL/6J and SJL genomic DNA but not when using Myo7ash1–8J genomic DNA as the template. This result suggested that a deletion spanning this region (exons 38–46) of Myo7a might be the causative element of the Myo7ash1–8J allele.
To test this possibility, we attempted to PCR amplify across the putative deletion and sequence the PCR products. Specific primers (Supplementary Material, Table S3), spaced apart by 500 bp, for the large intron upstream of exon 38 were used in conjunction with a primer specific for the 3′ untranslated region. One specific primer pair (47603 5′, EX47B 3′) amplified a PCR product of ∼1590 bp, which was cloned and sequenced.
Sequence analysis of this PCR product revealed a breakpoint occurring ∼1880 bp upstream of exon 38. The sequence skipped to the intron between exons 40 and 41 (∼55 bp downstream of exon 40) and ran uninterrupted to the next breakpoint located ∼20 bp downstream of exon 41 in the intron between exons 41 and 42. The sequence then skipped to the intron between exons 46 and 47 (Fig. 1).
A total of 6.4 kb appeared to be deleted from genomic DNA of 8J/8J homozygotes (Fig. 1A and B). The upstream deletion included exons 38 through 40 (3.6 kb), while the downstream deletion encompassed exons 42 through 46 (2.8 kb). To confirm these sequencing results, we performed Southern blots on genomic DNA from Myo7ash1–8J/sh1–8J homozygotes and from their two parental mouse strains, C57BL/6J and SJL. Mouse genomic DNA was cleaved with either EcoRI or HindIII and then separated by agarose gel electrophoresis. Using a 3′ probe specific for exon 47, we then probed blots of the digested genomic DNA from WT (C57BL/6J or SJL) or 8J/8J homozygous mice (Fig. 1E).
EcoRI digestion of WT and 8J/8J samples gave band sizes of ∼13.75 and ∼7.3 kb, respectively; HindIII digestion produced ∼4.9 kb WT band and ∼2.1 kb 8J/8J band, the expected sizes. The Southern analysis thus confirmed that a 6.412 kb sequence was deleted in the 8J allele (Fig. 1C). Mouse genotypes were readily distinguished by PCR (Fig. 1D).
Myo7ash1–8J phenotype
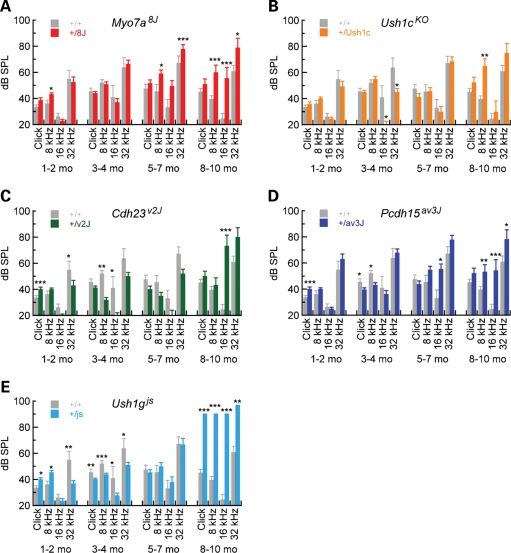
Circling mutants were tested for hearing sensitivity using ABR (Fig. 2). Even at the highest intensities, at all frequencies tested, homozygous 8J/8J mice showed no ABR even at the highest stimulus intensity [100 dB sound pressure level (SPL)]; unaffected littermate controls showed normal waveforms and thresholds at all frequencies tested. These data demonstrated that the 8J mice were profoundly deaf.
Figure 2.
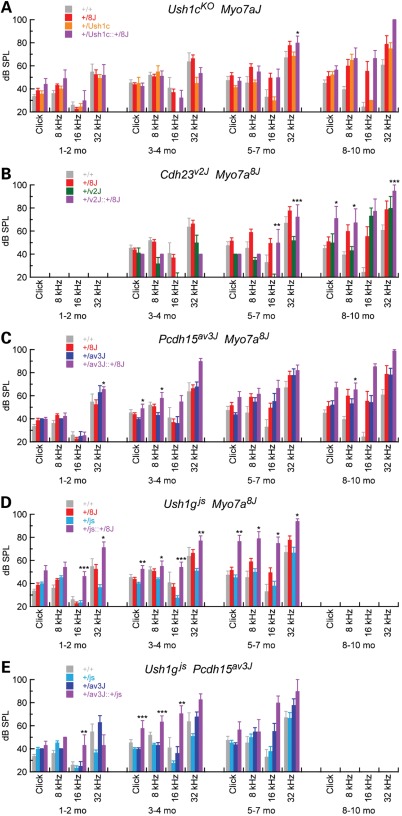
ABR for Usher I heterozygotes. ABR thresholds are plotted for 1–2, 3–4, 5–7 and 8–10 months for WT (n = 4–15) and the following heterozygous mice: (A) +/Myo7ash1–8J (n = 9–34), (B) +/Ush1cKO (n = 2–17), (C) +/Cdh23v-2J (n = 3–8), (D) +/Pcdh15av-3J (n = 5–20) and (E) +/Ush1gjs (n = 3–22). Significance for pairwise comparisons using two-tailed t-tests between WT and heterozygotes at individual ages and stimuli are indicated (*P < 0.05; **P < 0.01; ***P < 0.001). Mean ± SEM are plotted.
C57BL/6 mice are well known to have progressive hearing loss, which is severe by 12 months (19). Hearing thresholds in +/+ and +/8J mice increased over time, with +/8J threshold elevations being slightly higher; by two-tailed t-test, however, differences between +/8J and +/+ were only statistically significant at older ages. These results with +/8J heterozygotes were similar to those from the Myo7a4626SB allele, also severe; +/4626SB heterozygotes also showed no hearing loss compared with WT at 3 months (17).
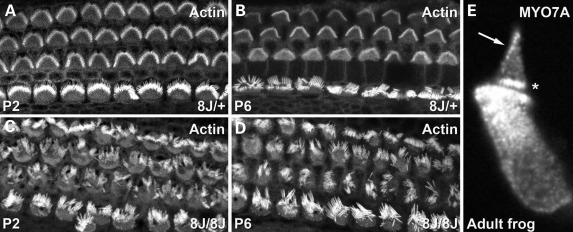
We examined hair-bundle morphology in Myo7ash1–8J hair cells from early post-natal mouse cochleas. Using fluorescein isothiocyanate (FITC)-phalloidin, we noted that bundles were morphologically normal in P2 and P6 +/8J heterozygotes (Fig. 3A and B). In contrast, hair bundles in 8J/8J homozygote cochleas were consistently disorganized (Fig. 3C and D), as were bundles from the utricle and the saccule (data not shown). These morphological results were consistent with the circling behavior and complete lack of auditory function seen in homozygotes.
Figure 3.
Disrupted hair bundles in Myo7ash1–8J homozygotes and adult MYO7A localization. (A–D) Actin detected with phalloidin. (A and B) Cochlear hair bundles from P2 and P6 +/8J heterozygote mice have normal morphology. (C and D) Cochlear hair bundles from P2 and P6 8J/8J mice are profoundly disrupted. (E) Isolated adult bullfrog hair cell stained for MYO7A. Note labeling at ankle region (asterisk) and at tips (arrow), site of mechanotransduction.
Since previous localization of MYO7A in adult bullfrog hair cells suggested that it was only located in hair bundles at the region of the ankle links (20), we re-examined its location there in light of the results of Grati and Kachar (6). Indeed, MYO7A could often be detected at tips of bullfrog stereocilia, consistent with a role in transduction (Fig. 3E).
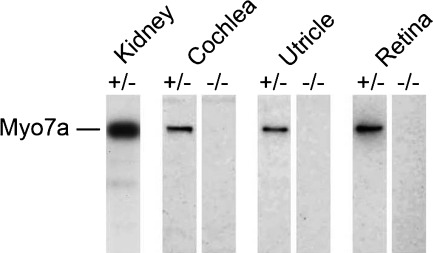
We also used protein immunoblotting of mouse tissue extracts to compare MYO7A expression between WT and 8J mice. The MYO7A antibody recognized a protein of ∼220 kDa, the size predicted for full-length MYO7A, in the kidney, cochlea, utricle and retina (Fig. 4). In contrast, MYO7A was not detectable in 8J/8J homozygotes (Fig. 4). The MYO7A antibody recognizes an epitope that would remain in a truncated protein expressed from the Myo7ash1–8J allele (Fig. 1), suggesting that any MYO7A protein expressed in 8J/8J homozygotes was degraded.
Figure 4.
No MYO7A protein in Myo7ash1–8J homozygotes. Equal protein concentrations of extracts from Myo7ash1–8J heterozygotes (±) or homozygotes (−/−) were separated by SDS–PAGE; MYO7A was detected by immunoblotting with the 25-6790 antibody. No MYO7A protein was detected in 8J/8J homozygous mutants.
Progressive hearing loss in double heterozygotes of Myo7ash1–8J and Usher genes
While hearing loss in +/8J mice is minimal compared with control, we sought to determine whether double heterozygotes of Myo7a and other Usher I genes might show synergistic hearing loss, indicating genetic interaction. Although homozygous mutants are profoundly deaf at early ages (data not shown), before 8–10 months, +/Ush1cKO, +/Cdh23v-2J, +/Pcdh15av-3J and +/Ush1gjs heterozygotes had hearing thresholds that are not substantially elevated over +/+ control mice (Fig. 1B–E). In contrast, thresholds were elevated over control for some frequencies at 8–10 months for +/Ush1cKO, +/Cdh23v-2J and +/Pcdh15av-3J heterozygotes. The results with Cdh23v-2J contrasted with those reported with a different Cdh23 allele (17); there, +/Cdh23v heterozygotes had elevated compound action-potential thresholds at 1–2 months. Overall, heterozygous mice at Usher I loci had near-normal hearing at least through 5–7 months.
We generated doubly heterozygous mice for two Usher I alleles to examine digenic hearing loss. Synergistic threshold elevation was substantial for Myo7ash1–8J heterozygotes combined with +/Ush1gjs, small but significant for Myo7ash1–8J combined with either +/Cdh23v-2J or Pcdh15, but not significant for Myo7ash1–8J combined with Ush1cKO (Fig. 5). To assess statistical significance, we used analysis of covariance (ANCOVA) to determine whether the increase in threshold of double heterozygotes was significantly greater than the sum of the threshold elevations from each of the two individual heterozygotes while accounting for potential confounding effects of age, weight and gender. Complete tables of coefficients and P-values are reported in Supplementary Material, Tables S4–S8.
Figure 5.
Digenic inheritance of age-related deafness from Myo7a and other Usher genes. ABR for Usher I heterozygotes. ABR thresholds are plotted for 1–2, 3–4, 5–7 and 8–10 months for WT and the following heterozygous mice: (A) +/Ush1cKO +/Myo7ash1–8J (n = 3–7), (B) +/Cdh23v-2J +/Myo7ash1–8J (n = 4), (C) +/Pcdh15av-3J +/Myo7ash1–8J (n = 6–18), (D) +/Ush1gjs +/Myo7ash1–8J (n = 7–22) and (E) +/Ush1gjs +/Pcdh15av-3J (n = 3–7). Significance was assessed for four genotypes at each time point and stimulus using ANOVA (*P < 0.05; **P < 0.01; ***P < 0.001). Significance is only indicated for double heterozygous mice; values for significance for other mice are listed in Supplementary Material Tables S4–S8. WT and heterozygous data from Figure 1 are replotted here, as the model tested incorporates all four genotypes at each stimulus and age. Mean ± SEM are plotted.
Hearing loss in +/Myo7ash1–8J +/Ush1gjs mice was particularly striking; double heterozygotes exhibited synergistic hearing loss at 1–2 and 5–7 months, with excess ABR threshold elevation over the individual heterozygotes of >20 dB SPL in many cases (Fig. 5D). Ush1g also interacted genetically with Pcdh15; hearing loss in +/Pcdh15av-3J +/Ush1gjs double heterozygotes exhibited highly significant elevated ABR thresholds, particularly at 3–4 months (Fig. 5E). These results show genetic interaction of Myo7a and Ush1g, as well as Ush1g and Pcdh15.
+/Myo7ash1–8J +/Cdh23v-2J double heterozygotes had modest but significant ABR threshold elevation at 5–7 and 8–10 months, albeit only at several frequencies (Fig. 5C). These results diverged from those seen previously, albeit with a different Cdh23 mutation and a different genetic background (17). Likewise, +/Myo7ash1–8J8J +/Pcdh15av-3J double heterozygotes showed synergistic threshold elevation at some ages and frequencies (Fig. 5C). These results show a weak genetic interaction of Myo7a and Cdh23 or Pcdh15.
In contrast, +/Myo7ash1–8J8J +/Ush1cKO double heterozygotes showed no hearing loss that was greater than the sum of the threshold elevation of the individual alleles, except at 32 kHz at 5–7 months (Fig. 5B). We therefore have no evidence for the genetic interaction of Myo7a and Ush1c.
Abundance of Usher proteins
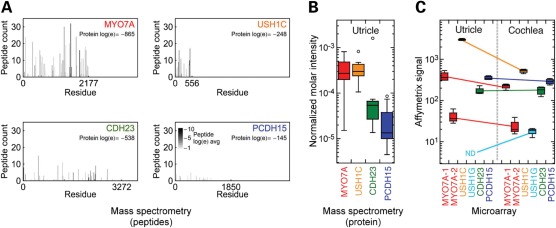
Using hair bundles purified from E20 chicken utricles (21), we determined the abundance of the Usher proteins by mass spectrometry to estimate their stoichiometric relationships (Fig. 6A). CDH23, PCDH15 and USH1C were readily detected with extremely low log(e) statistical scores and moderate-to-high spectral counts. At ∼200 molecules per stereocilium, MYO7A was similar in concentration to USH1C (∼200 molecules) and in excess over CDH23 (∼40) and PCDH15 (∼10).
Figure 6.
Expression of Usher I genes in the chick utricle and cochlea. (A) Mass spectrometry sequence coverage of Usher I proteins MYO7A, USH1C, CDH23 and PCDH15 in purified hair bundles from the chick utricle. Each mark corresponds to a detected peptide, with its width equal to the corresponding sequence length and height indicating the number of different times that peptide was counted. The shade of the mark indicates the average log(e) score of the detected peptides, indicating confidence in the identification. Data are summed from 13 independent experiments. (B) Quantitation of Usher proteins. Data from 13 experiments were plotted with box plots; outliers are indicated by open circles. The median is indicated as a horizontal line within the box, while the box bottom and top correspond to the 25th and 75th percentiles; whiskers correspond to the data range exclusive of outliers. (C) Affymetrix microarray analysis of Usher I transcripts. Results from four utricle and four cochlea hybridizations are presented. Two probesets for Myo7a were present on the chicken genome chip; all other genes were represented by single probesets. Ush1g was not detected in the chick utricle, consistent with the inability to detect it by protein mass spectrometry.
Although we did not detect USH1G in chick vestibular hair bundles, analysis of Affymetrix microarray data from the chick utricle and cochlea indicated that Ush1g transcripts were not detectable in the utricle, although they were in the cochlea (Fig. 6B). Two Affymetrix probesets detected Myo7a; one of them (MYO7A-2 in Fig. 6B; GgaAffx.469.5.S1_s_at) was directed against a sequence in the most 5′-coding exon, however, and may be less suitable for mRNA quantitation. Myo7a transcripts detected with the MYO7A-1 probeset (GgaAffx.469.1.S1_s_at) were present at similar concentrations in the utricle and cochlea and were similar in concentration to Cdh23 and Pcdh15 transcripts. Ush1c transcripts were notably higher in the utricle than in the cochlea, but cochlea Ush1c transcripts were similar in abundance to Myo7a, Cdh23 and Pcdh15 in the cochlea (Fig. 6B).
DISCUSSION
Complementation testing revealed that the 8J mutation is an allele of Myo7a produced by a 6.4 kb deletion within Myo7a, which included part of the region encoding the MyTH4 and FERM domains of repeat 2. MYO7A protein was not detected in 8J/8J homozygous mice by immunoblotting or immunocytochemistry, while hair bundles in the cochlea and the vestibular system were profoundly disrupted. Homozygous 8J/8J mice were deaf, as indicated by ABR analysis, and displayed the shaker-1 vestibular phenotype, which included hyperactivity, head bobbing and circling. As the Myo7ash1–8J allele is essentially a Myo7a null, 8J/8J homozygous mice provide an ideal background on which to breed Myo7a transgenics to test for rescue of the structural and behavioral phenotypes. The Myo7ash1–8J mice, available from Jackson Laboratories, will thus be useful for further dissecting MYO7A function.
The 8J allele of Myo7a is severe
Myo7a mutations in both mouse and zebrafish highlight the need for expression of sufficient levels of MYO7A. For example, the 26SB and 3336SB shaker-1 alleles express MYO7A at <20% of WT levels, leading to severely disrupted hair bundles (3). Similarly, MYO7A cannot be detected in 8J/8J homozygotes. The second MyTH4 and FERM domains of MYO7A are disrupted in the 8J allele. The 26SB mutation lies within the second MyTH4 domain, changing a conserved Phe residue to Ile, while 3336SB is missing the last 35 amino acids, partially deleting the second FERM domain. The most similar allele of zebrafish Myo7a, C2144X (marinertn4503), results in the profound splaying of larval inner-ear hair bundles (22). Interestingly, marinertr202b, a second mariner mutant allele with a G2073D substitution in the C-terminus of the second FERM domain, had mostly intact hair bundles (22); while the second FERM domain may not be necessary for bundle formation, mechanically elicted hair-cell extracellular potentials were dramatically reduced in marinertr202b, suggesting that mechanotransduction requires the second FERM domain.
Taken together, these observations suggest that the loss of the C-terminus of MYO7A may trigger protein degradation. Alternatively, as with the Myo7apolka allele (23), Myo7a mRNA may be destabilized by the 8J mutation. In either case, as a consequence of insufficient MYO7A levels, hair bundles form improperly and cannot support normal hair-cell function.
Functional interactions between Usher I genes
In vitro experiments suggest that MYO7A, USH1G and USH1C are capable of forming a stable complex (6,10,11). Moreover, this complex interacts with CDH23 through CDH23–MYO7A and CDH23–USH1C interactions (12,24). In hair cells, MYO7A is located in several locations in the hair bundle, but has been recently shown to be present at the upper tip-link density, interacting with CDH23, USH1C and USH1G (6). PCDH15 is also part of the tip-link complex (16), albeit physically distant from the upper tip-link density. Moreover, MYO7A has also been suggested to interact with PCDH15, and the two proteins co-localize at some developmental times at stereocilia tips (9) and ankles (13). Localization evidence in Usher I homozygotes also has provided evidence for interaction of USH1C with CDH23, PCDH15 and USH1G (9,25), as well as USH1G–CDH23 and USH1G–PCDH15 interactions (14), although the severity of the homozygous phenotype means that all of these interactions can only be assessed in developing hair cells.
Within experimental error, MYO7A and USH1C are at approximately the same concentration in E20–21 chicks, a late developmental age (26). Both proteins are in excess over CDH23, consistent with immunocytochemical quantitation of these three proteins at tip links (6), and PCDH15. These results support a role for USH1C in increasing the number of motor molecules coupled to CDH23 and PCDH15, which would increase processivity and force production of the motor complex.
At a molecular level, digenic inheritance classically arises from mutations in two genes that contribute to the same protein complex (27–31). Moreover, in contrast with mutations that affect development of the hair bundle, alleles expressed in hair cells that cause progressive hearing loss by definition interrogate mature hair cells. Taken together, digenic inheritance of progressive hearing loss provides prima facie evidence that the proteins encoded by the two genes interact physically in hair cells.
While heterozygotes with single functional copies of Myo7a, Ush1g or Ush1c have normal hearing with minimal age-related threshold elevation, double heterozygotes of Myo7a and Ush1g, in particular, or to a lesser extent Cdh23 or Pcdh15 have elevated hearing thresholds that progressively worsen over several months. These genetic results provide evidence that MYO7A–USH1G, MYO7A–CDH23 and MYO7A–PCDH15 interactions are important for mature hair cells; they also suggest that USH1G and PCDH15 are present in the same complex. While genetic interactions do not necessarily establish physical connections, such as the interaction between Cdh23 alleles and the Ca2+-ATPase Atp2b2 (32), the strong in vitro and developmental evidence for the MYO7A–USH1G–USH1C–CDH23 complex together with our genetics results suggests that this complex persists into adult hair cells. Moreover, our results support the assignment of USH1G and PCDH15 to a different complex, perhaps also with MYO7A. Digenic progressive deafness caused by the combination of recessive mutations in Myo7a and other Usher genes thus reinforces the suggestion that MYO7A is part of tip-link complexes in mature hair bundles.
MATERIALS AND METHODS
Mice
All of the mutant strains were backcrossed for more than 10 generations to a C57BL/6J (B6) background; all mice were thus on the same B6 genetic background, which includes a synonymous single-nucleotide polymorphism in exon 7 of the Cdh23 (753A) allele, causing in-frame skipping of exon 7. The new Myo7a allele described in this report arose as a spontaneous mutation on the B6;SJL-Tg (c177-lacZ)226Bri/J background at The Jackson Laboratory (TJL) and was transferred to the B6 background. The colony was genotyped for TgN (c177lacZ) 226Bri in March 2000 and verified free of the transgene. It was designated as B6.Cg-Myo7ash1–8J/J and is available from TJL (stock #003184). The following strains from TJL were used for the remaining mice: B6.A-Ush1gjs/J (stock #000783), B6 (V)-Cdh23v–2J/J (stock #002552), C57BL/6J-Pcdh15av–3J/J (stock #002072) and B6.129-Ush1ctm1Xzl/Kjn (referred to here as Ush1cKO; stock #006853). B6.Cg-Myo7ash1–7J/J (stock #002919), which was identified as a Myo7a allele at TJL by complementation with the original sh1 allele, was used for complementation tests. Animal Care and Use Committees at TJL, CWRU and OHSU approved the mouse husbandry and experiments at the respective universities.
Double heterozygous mice were generated in two ways. In the first, we crossed mice heterozygous in one gene (e.g. +/Myo7ash1–8J) with mice homozygous in a second gene (e.g. Ush1gjs/js). In this example, the cross generated equal numbers of +/Myo7ash1–8J +/Ush1gjs double heterozygotes and +/Ush1gjs heterozygotes. Most double heterozygote data were obtained from similar crosses. In the second, we crossed heterozygotes in one gene with heterozygotes in another gene; this cross yielded WT, individual heterozygotes and double heterozygotes in 1:1:1:1 ratio. For comparisons between genotypes, data from mice of appropriate genotype were pooled together regardless of the cross that generated them.
Auditory-evoked brain stem response
ABRs were conducted as described previously (33). Briefly, mice were anesthetized and their body temperature was maintained at 37–38°C by placing them on a heating pad in a soundproof chamber during testing. An Intelligent Hearing System (Miami, FL, USA) was used to generate acoustic stimuli and ABR measurements. Platinum subdermal needle electrodes were inserted at the vertex (active), ventrolaterally to the right ear (reference) and the left ear (ground). Alternating click stimuli of 50 µs duration and tone bursts with 3 ms duration (1.5 ms rise–fall time with no plateau) of 8, 16 and 32 kHz were presented to both ears of the animals. The ABR threshold was obtained for each animal by reducing the stimulus intensity from 100 dB SPL in 10 dB steps and finally in 5 dB steps to the lowest intensity that can evoke a reproducible ABR pattern on the computer screen.
Statistical analysis of ABR data
Two sample t-tests were used to test differences in ABR threshold between WT and each type of heterozygous mice for a given sound stimulus and age group separately. ANCOVA models were employed to test for interaction effects of double heterozygotes in ABR threshold. For each combination of sound frequency and age group of double heterozygotes, an ANCOVA model was fitted separately. Demographic variables (age in days, weight and gender) were included in each model to account for potential confounding effects. The model tested was:
| (1) |
where G1 and G2 refer to genes 1 and 2 and can take values of 0 or 1 depending on whether the WT or mutant allele is present. The coefficient b corresponds to increments of ABR threshold, where the threshold is b0 for +/+, b0+b1 for +/G1, b0+b2 for +/G2, and b0+b1+b2+b3 for +/G1 +/G2. The coefficient b3 therefore indicates the synergistic effect of the double heterozygous combination of G1 and G2. Three other demographic variables (age, weight and gender) were also included in the model but are omitted in Eq. (1) for clarity. Statistical results are reported in Supplementary Material, Tables S4–S8.
8J genotyping
Genomic DNA was purified from mouse tails and used for PCR amplification. A ∼1590 bp region of Myo7a flanking the 8J mutation was amplified using 8J primers 47603 5′ (forward: 5′-ATTTGGGACAGATGTGATTGGTGGG-3′) and EX 47 3′ (reverse: 5′-CAAAAGAGGTTGCATTGATGGAGCTG-3′) (Supplementary Material, Tables S2 and S3). The predicted PCR product was amplified only in samples containing the 8J allele. To avoid amplification of a large PCR product (7.97 kb) from WT samples, a separate WT reaction amplified a PCR product of ∼850 bp only in samples containing exon 44 of Myo7a; primers used were EX 44 5′ (forward: 5′-AGGGGTTGAGACAGCAAGTGCTGG-3′) and EX 44 3′ (reverse: 5′-CCACAGCCTCAAGGGTCCCTATAAG-3′). For quick initial screening, we observed that we could tell apart a +/8J heterozygote from an 8J/8J homozygote at P5 by flipping the pups over on their backs; +/8J heterozygotes righted themselves in 1–2 s, whereas 8J/8J homozygotes took 5–10 s or longer.
Genotyping of double heterozygote mice
Genomic DNA was amplified by PCR and sequenced. PCR primers for the Cdh23v-2J mutation were V2-F (5′-GCCAAAGCCCTCTTCAAGAT-3′) and V2-R (5′-GGGAAAGCGTTTAGCTTGTT-3′), which generated a fragment of 215 bp using a standard PCR protocol (15). For Pcdh15av-3J, the single-nucleotide addition in this allele (34) introduced a new MboII restriction site. PCR primers used for screening for this allele were AV3-F (5′-GACGGCAAACTGCTCGATA-3′) and AV3-R (5′-GGGATGCAACAGAGGATGAT-3′). Amplification of WT and av-3J each produced a band of 190 bp; digestion with MboII cleaved the sequence harboring the av-3J mutation into 120 and 70 bp fragments and left the WT sequence undigested. With this approach, +/av-3J mice show three bands (190, 120 and 70 bp) and WT mice show a single band (190 bp). For genotyping Ush1gjs, we used primers Js-L2 (5′-TCCAGAAGAAGCTGGAGAGG-3′) and Js-R127 (5′-GCGCACAAACATCACATCAC-3′), which produced a product of 126 bp. This product was digested with the restriction enzyme MwoI (New England Biolabs), then run on 3% agarose gel; the WT band was 108 bp, while Ush1gjs gave a 94 bp band. For the Ush1c knockout (−/−) mouse genotyping, methods were as detailed previously (35). The forward and reverse primers were, respectively, 5′-CGCAAGCATTACAAACATCG-3′ and 5′-TCCATCCCAGACTCTTGCTT-3′; bands of 431 bp (WT) and 276 bp (mutant) were amplified.
Immunoblotting
Mouse tissues (cochlea, utricle, retina and kidney) were harvested and snap-frozen. Equal protein amounts from tissue extracts were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to a polyvinylidine fluoride membrane. To detect mouse MYO7A, membranes were incubated with 1 µg/ml of an affinity-purified rabbit anti-human MYO7A polyclonal antibody (Proteus BioSciences, #25-6790) (36), which is specific for amino acids 880–1077. Blots were developed with donkey anti-rabbit HRP (Jackson ImmunoResearch) and SuperSignal West Pico (Pierce, Rockford, IL, USA) using a Fuji chemiluminescence imager. MYO7A band intensities were quantified using ImageJ.
Immunocytochemistry and phalloidin labeling of cochlear hair cells
Cochleas were dissected out of temporal bones from P0 to P6 mice in Dulbecco's modified Eagle's medium with 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (25 mm), pH 7.5. The stria vascularis and tectorial membrane were removed and then cochleas were fixed for 20 min in 4% formaldehyde. Tissues were washed twice in phosphate-buffered saline (PBS), 10min each. To permeabilize, tissues were transferred to a solution containing 0.2% saponin/blocking cocktail (10 mg/ml bovine serum albumin, 3% normal donkey serum, in PBS) for 10 min. Subsequently, tissues were incubated overnight at 4°C with 5 µg/ml anti-MYO7A antibody in blocking solution. Tissues were washed three times in PBS, 10min each. Subsequently, tissues were incubated at room temperature for 2 h in blocking solution containing donkey anti-rabbit Cy5 (Molecular Probes, Eugene, OR, USA), diluted 1/10 000 and phalloidin-FITC (0.25 µm). Tissues were then washed three times in PBS for 10min each. Tissues were mounted on glass slides in Vectashield (Vector Laboratories Inc., Burlingame, CA, USA) with glass coverslips and examined by confocal microscopy. Isolated bullfrog hair cells were prepared and labeled with an anti-MYO7A antibody as described (20).
Mass spectrometry
Protein digests were analyzed by liquid chromatography tandem mass spectrometry using an Agilent 1100 series capillary LC system and an LTQ linear ion trap mass spectrometer (Thermo Scientific, West Palm Beach, FL, USA). Electrospray ionization was performed with an Ion Max source fitted with a 34 G metal needle and 2.7 kV source voltage. Samples were applied at 20 ml/min to a Michrom trap cartridge, then switched onto a 0.5 × 250 mm Zorbax SB-C18 column with 5 mm particles (Agilent) using a mobile phase containing 0.1% formic acid, 7–30% acetonitrile gradient over 195 min and 10 µl/min flow rate. Data-dependent collection of MS/MS spectra used dynamic exclusion (repeat count of 1, exclusion list of 50, exclusion duration of 30 s and exclusion mass width of −1 to +4) to obtain MS/MS spectra of the three most abundant parent ions following each survey scan from m/z 400 to 2000. Raw data from the LTQ mass spectrometer were converted to DTA files using DTA Extract in Bioworks (version 3.3; Thermo Fisher). DTA files were converted to .mgf format for X! Tandem analysis using merge.pl (Matrix Science). The Ensembl Chicken v59 (22 194 entries) database was used, with 179 common contaminant entries added and all forward entries (proteins plus contaminants) appended with sequence-reversed entries to facilitate error estimates. X! Tandem parameters were: parent ion tolerance of +2.5 and −1 Da, fragment ion tolerance of 0.4 Da, average parent ion masses, monoisotopic fragment ion masses, static cysteine modification of +57 Da, potential peptide N-terminal modification of +42 Da, potential methionine modifications of +16 and +32 Da, potential tryptophan modifications of +16 and +32 Da, potential asparagine modification of +1 Da, potential glutamine modification of +1 Da, maximum of two missed cleavages and trypsin cleavage specificity. Proteins were quantified by intensity factor (21) and converted to molecules per stereocilium using a normalized molar intensity for actin of 0.56 and by assuming 400 000 actin monomers per stereocilium (Shin et al., submitted for publication).
Microarray
Affymetrix microarray experiments and data analysis have been described previously (37); raw and processed data can be downloaded from the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo under accession no. GSE32272.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health (R01 DC009246 to Q.Y.Z., R01 DC007392 to Q.Y.Z., R01 DC002368 to P.G.G. and P30 DC005983 to P.G.G.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Ken Johnson for comments on the manuscript; he and Dr Xue Z. Liu participated in helpful initial discussions of the digenetic crosses. David Corey provided the EGFP-mMyo7a-C1 plasmid.
Conflict of Interest statement. None declared.
REFERENCES
- 1.McGary K.L., Park T.J., Woods J.O., Cha H.J., Wallingford J.B., Marcotte E.M. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl Acad. Sci. USA. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson G.P., de Monvel J.B., Petit C. How the genetics of deafness illuminates auditory physiology. Annu. Rev. Physiol. 2011;73:311–334. doi: 10.1146/annurev-physiol-012110-142228. [DOI] [PubMed] [Google Scholar]
- 3.Hasson T., Walsh J., Cable J., Mooseker M.S., Brown S.D., Steel K.P. Effects of shaker-1 mutations on myosin-VIIa protein and mRNA expression. Cell Motil. Cytoskeleton. 1997;37:127–138. doi: 10.1002/(SICI)1097-0169(1997)37:2<127::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Self T., Mahony M., Fleming J., Walsh J., Brown S.D., Steel K.P. Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development. 1998;125:557–566. doi: 10.1242/dev.125.4.557. [DOI] [PubMed] [Google Scholar]
- 5.Kros C.J., Marcotti W., van N.S.M., Self T.J., Libby R.T., Brown S.D., Richardson G.P., Steel K.P. Reduced climbing and increased slipping adaptation in cochlear hair cells of mice with Myo7a mutations. Nat. Neurosci. 2002;5:41–47. doi: 10.1038/nn784. [DOI] [PubMed] [Google Scholar]
- 6.Grati M., Kachar B. Myosin VIIa and sans localization at stereocilia upper tip-link density implicates these Usher syndrome proteins in mechanotransduction. Proc. Natl Acad. Sci. USA. 2011;108:11476–11481. doi: 10.1073/pnas.1104161108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt J.R., Gillespie S.K., Provance D.W., Shah K., Shokat K.M., Corey D.P., Mercer J.A., Gillespie P.G. A chemical-genetic strategy implicates myosin-1c in adaptation by hair cells. Cell. 2002;108:371–381. doi: 10.1016/s0092-8674(02)00629-3. [DOI] [PubMed] [Google Scholar]
- 8.Stauffer E.A., Scarborough J.D., Hirono M., Miller E.D., Shah K., Mercer J.A., Holt J.R., Gillespie P.G. Fast adaptation in vestibular hair cells requires myosin-1c activity. Neuron. 2005;47:541–553. doi: 10.1016/j.neuron.2005.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lefevre G., Michel V., Weil D., Lepelletier L., Bizard E., Wolfrum U., Hardelin J.P., Petit C. A core cochlear phenotype in USH1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development. 2008;135:1427–1437. doi: 10.1242/dev.012922. [DOI] [PubMed] [Google Scholar]
- 10.Wu L., Pan L., Wei Z., Zhang M. Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science. 2011;331:757–760. doi: 10.1126/science.1198848. [DOI] [PubMed] [Google Scholar]
- 11.Yan J., Pan L., Chen X., Wu L., Zhang M. The structure of the harmonin/sans complex reveals an unexpected interaction mode of the two Usher syndrome proteins. Proc. Natl Acad. Sci. USA. 2010;107:4040–4045. doi: 10.1073/pnas.0911385107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siemens J., Kazmierczak P., Reynolds A., Sticker M., Littlewood-Evans A., Müller U. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proc. Natl Acad. Sci. USA. 2002;99:14946–14951. doi: 10.1073/pnas.232579599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Senften M., Schwander M., Kazmierczak P., Lillo C., Shin J.B., Hasson T., Geleoc G.S., Gillespie P.G., Williams D., Holt J.R., Müller U. Physical and functional interaction between protocadherin 15 and myosin VIIa in mechanosensory hair cells. J. Neurosci. 2006;26:2060–2071. doi: 10.1523/JNEUROSCI.4251-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caberlotto E., Michel V., Foucher I., Bahloul A., Goodyear R.J., Pepermans E., Michalski N., Perfettini I., Alegria-Prevot O., Chardenoux S., et al. Usher type 1G protein sans is a critical component of the tip-link complex, a structure controlling actin polymerization in stereocilia. Proc. Natl. Acad. Sci. USA. 2011;108:5825–5830. doi: 10.1073/pnas.1017114108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Q.Y., Yan D., Ouyang X.M., Du L.L., Yu H., Chang B., Johnson K.R., Liu X.Z. Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum. Mol. Genet. 2005;14:103–111. doi: 10.1093/hmg/ddi010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazmierczak P., Sakaguchi H., Tokita J., Wilson-Kubalek E.M., Milligan R.A., Müller U., Kachar B. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 17.Holme R.H., Steel K.P. Progressive hearing loss and increased susceptibility to noise-induced hearing loss in mice carrying a Cdh23 but not a Myo7a mutation. J. Assoc. Res. Otolaryngol. 2004;5:66–79. doi: 10.1007/s10162-003-4021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor B.A., Navin A., Phillips S.J. PCR-amplification of simple sequence repeat variants from pooled DNA samples for rapidly mapping new mutations of the mouse. Genomics. 1994;21:626–632. doi: 10.1006/geno.1994.1323. [DOI] [PubMed] [Google Scholar]
- 19.Johnson K.R., Erway L.C., Cook S.A., Willott J.F., Zheng Q.Y. A major gene affecting age-related hearing loss in C57BL/6J mice. Hear Res. 1997;114:83–92. doi: 10.1016/s0378-5955(97)00155-x. [DOI] [PubMed] [Google Scholar]
- 20.Hasson T., Gillespie P.G., Garcia J.A., MacDonald R.B., Zhao Y., Yee A.G., Mooseker M.S., Corey D.P. Unconventional myosins in inner-ear sensory epithelia. J. Cell Biol. 1997;137:1287–1307. doi: 10.1083/jcb.137.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin J.B., Streijger F., Beynon A., Peters T., Gadzala L., McMillen D., Bystrom C., Van der Zee C.E., Wallimann T., Gillespie P.G. Hair bundles are specialized for ATP delivery via creatine kinase. Neuron. 2007;53:371–386. doi: 10.1016/j.neuron.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ernest S., Rauch G.J., Haffter P., Geisler R., Petit C., Nicolson T. Mariner is defective in myosin VIIA: a zebrafish model for human hereditary deafness. Hum. Mol. Genet. 2000;9:2189–2196. doi: 10.1093/hmg/9.14.2189. [DOI] [PubMed] [Google Scholar]
- 23.Schwander M., Lopes V., Sczaniecka A., Gibbs D., Lillo C., Delano D., Tarantino L.M., Wiltshire T., Williams D.S., Müller U. A novel allele of myosin VIIa reveals a critical function for the C-terminal FERM domain for melanosome transport in retinal pigment epithelial cells. J. Neurosci. 2009;29:15810–15818. doi: 10.1523/JNEUROSCI.4876-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahloul A., Michel V., Hardelin J.P., Nouaille S., Hoos S., Houdusse A., England P., Petit C. Cadherin-23, myosin VIIa and harmonin, encoded by Usher syndrome type I genes, form a ternary complex and interact with membrane phospholipids. Hum. Mol. Genet. 2010;19:3557–3565. doi: 10.1093/hmg/ddq271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan D., Kamiya K., Ouyang X.M., Liu X.Z. Analysis of subcellular localization of Myo7a, Pcdh15 and Sans in Ush1c knockout mice. Int. J. Exp. Pathol. 2011;92:66–71. doi: 10.1111/j.1365-2613.2010.00751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodyear R.J., Gates R., Lukashkin A.N., Richardson G.P. Hair-cell numbers continue to increase in the utricular macula of the early posthatch chick. J. Neurocytol. 1999;28:851–861. doi: 10.1023/a:1007070121751. [DOI] [PubMed] [Google Scholar]
- 27.Kusch M., Edgar R.S. Genetic studies of unusual loci that affect body shape of the nematode Caenorhabditis elegans and may code for cuticle structural proteins. Genetics. 1986;113:621–639. doi: 10.1093/genetics/113.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stearns T., Botstein D. Unlinked noncomplementation: isolation of new conditional-lethal mutations in each of the tubulin genes of Saccharomyces cerevisiae. Genetics. 1988;119:249–260. doi: 10.1093/genetics/119.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hays T.S., Deuring R., Robertson B., Prout M., Fuller M.T. Interacting proteins identified by genetic interactions: a missense mutation in alpha-tubulin fails to complement alleles of the testis-specific beta-tubulin gene of Drosophila melanogaster. Mol. Cell. Biol. 1989;9:875–884. doi: 10.1128/mcb.9.3.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kajiwara K., Berson E.L., Dryja T.P. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264:1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- 31.Vockley J. Encyclopedia of Life Sciences. Chichester: John Wiley & Sons, Ltd; 2011. Digenic inheritance. doi:10.1002/9780470015902.a0005560.pub2. [Google Scholar]
- 32.Johnson K.R., Zheng Q.Y., Noben-Trauth K. Strain background effects and genetic modifiers of hearing in mice. Brain Res. 2006;1091:79–88. doi: 10.1016/j.brainres.2006.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng Q.Y., Johnson K.R., Erway L.C. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alagramam K.N., Murcia C.L., Kwon H.Y., Pawlowski K.S., Wright C.G., Woychik R.P. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat. Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- 35.Tian C., Liu X.Z., Han F., Yu H., Longo-Guess C., Yang B., Lu C., Yan D., Zheng Q.Y. Ush1c gene expression levels in the ear and eye suggest different roles for Ush1c in neurosensory organs in a new Ush1c knockout mouse. Brain Res. 2010;1328:57–70. doi: 10.1016/j.brainres.2010.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasson T., Heintzelman M.B., Santos-Sacchi J., Corey D.P., Mooseker M.S. Expression in cochlea and retina of myosin VIIa, the gene product defective in Usher syndrome type 1B. Proc. Natl Acad. Sci. USA. 1995;92:9815–9819. doi: 10.1073/pnas.92.21.9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spinelli K.J., Klimek J.E., Wilmarth P.A., Shin J.B., Choi D., David L.L., Gillespie P.G. Distinct energy metabolism of auditory and vestibular sensory epithelia revealed by quantitative mass spectrometry using MS2 intensity. Proc. Natl Acad. Sci. USA. 2012;109:E268–E277. doi: 10.1073/pnas.1115866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.